Background: The plasma membrane Na+/K+-ATPase undergoes internalization upon ouabain binding.
Results: The internalization time course, intracellular location of internalized pumps, and sensitivity to various inhibitors have been characterized.
Conclusion: The ouabain-induced internalized pumps are destined for lysosomal degradation.
Significance: Ouabain-induced internalization may serve to eliminate Na+/K+-ATPase molecules bound with cardiac glycosides.
Keywords: Endocytosis, Lysosomes, Na+/K+-ATPase, PI3-kinase (PI3K), Src, Cardiac Glycosides, Ouabain
Abstract
Internalization of the Na+/K+-ATPase (the Na+ pump) has been studied in the human lung carcinoma cell line H1299 that expresses YFP-tagged α1 from its normal genomic localization. Both real-time imaging and surface biotinylation have demonstrated internalization of α1 induced by ≥100 nm ouabain which occurs in a time scale of hours. Unlike previous studies in other systems, the ouabain-induced internalization was insensitive to Src or PI3K inhibitors. Accumulation of α1 in the cells could be augmented by inhibition of lysosomal degradation but not by proteosomal inhibitors. In agreement, the internalized α1 could be colocalized with the lysosomal marker LAMP1 but not with Golgi or nuclear markers. In principle, internalization could be triggered by a conformational change of the ouabain-bound Na+/K+-ATPase molecule or more generally by the disruption of cation homeostasis (Na+, K+, Ca2+) due to the partial inhibition of active Na+ and K+ transport. Overexpression of ouabain-insensitive rat α1 failed to inhibit internalization of human α1 expressed in the same cells. In addition, incubating cells in a K+-free medium did not induce internalization of the pump or affect the response to ouabain. Thus, internalization is not the result of changes in the cellular cation balance but is likely to be triggered by a conformational change of the protein itself. In physiological conditions, internalization may serve to eliminate pumps that have been blocked by endogenous ouabain or other cardiac glycosides. This mechanism may be required due to the very slow dissociation of the ouabain·Na+/K+-ATPase complex.
Introduction
The Na+/K+-ATPase is a ubiquitously expressed P-type ATPase. Its main role is to maintain Na+ and K+ gradients across the plasma membrane by ATP-driven active transport of 3Na+ ions in exchange for 2K+. The pump consists of a catalytic α subunit, a regulatory β subunit, and an auxiliary FXYD polypeptide. Four α (α1–4), three β (β1–3), and seven FXYD isoforms exist in mammalian organisms, resulting in considerable tissue-specific variability in pump kinetics (1, 2).
Cardiac glycosides (CGs)2 are a large family of clinically relevant specific inhibitors of the Na+/K+-ATPase, used classically to treat heart failure (3). Although most CGs are derived from plants, several CGs have been identified in mammalian tissues including ouabain, marinobufagenin, and digoxin (3, 4). It is clear today that CGs are synthesized in the adrenal glands and function as endogenous hormones, but their exact roles and cellular functions are poorly understood (5).
A considerable volume of research suggests that in addition to its classical role of maintaining ionic homeostasis, the Na+/K+-ATPase mediates a CG-triggered (and more specifically, ouabain-triggered) cascade of signal transduction (6). Nontoxic concentrations of ouabain have been shown to trigger activation of the tyrosine kinase Src and initiate multiple signal transduction pathways. These include: PLC/IP3/CICR, PI3K, reactive oxygen species, PLC/DG/PKC/Raf/MEK/ERK1/2, and Ras/Raf/MEK/ERK1/2 (7). Such effects could also be isoform-specific (8).
Independently, a number of studies have documented internalization of the Na+/K+-ATPase upon incubation of cells with nontoxic doses of ouabain and other CGs. This effect was suggested to be triggered by ouabain-induced activation of Src and to be involved in the signal transduction involved in translocation of the pump to the nucleus (9, 10). Alternatively, it was proposed that ouabain-induced internalization may be a means of regulating endosomal pH (11) or a natriuresis mechanism induced by endogenous CG (12).
Additional extracellular stimuli have been shown to induce internalization of the Na+/K+-ATPase. These include dopamine (13), parathyroid hormone (14), hypoxia (15, 16), hypercapnia (17), and sepsis (18). The underlying mechanisms appear to involve activation of several protein kinases, phosphorylation of the N terminus of α-Na+/K+-ATPase, and its subsequent ubiquitination. The time course of internalization processes induced by the different stimuli ranged from minutes to many hours.
The current study characterizes ouabain-induced internalization of the Na+/K+-ATPase in the human non-small cell lung carcinoma cell line H1299. We have taken advantage of the availability of a cell clone in which fluorescently tagged α1 is expressed from its normal genomic location, to study real-time internalization kinetics and intracellular location of the endogenous pump. In this system, α1 is internalized in a time scale of several hours and is directed to lysosomal degradation. Unlike previous reports (9) the effect of ouabain was not attenuated by removing external K+. Taken together with the finding that internalization of human α1 is not inhibited by transfection with ouabain-resistant rat α1, we conclude that internalization is not secondary to the effect of ouabain on the ionic balance or energetics of the treated cells. A possible mechanism and the physiological role of the ouabain-induced internalization are discussed.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
An H1299 cell clone expressing endogenous YFP-tagged α1-Na+/K+-ATPase that was generated by exon tagging was obtained from the library of annotated reporter cell clones. This library was created by first generating an H1299 clone with strong nuclear and weak cytoplasmic mCherry fluorescence and then random integration of enhanced yellow fluorescent protein (YFP) flanked by splicing sites into the H1299 genome (19, 20). In the cell clone identified and used in this study, the YFP construct was incorporated into the first α1 intron, generating an α1 protein in which YFP is incorporated between the fourth and fifth amino acids. Cells were cultured in RPMI 1640 medium (Biological Industries, Beit Haemek, Israel) supplemented with 10% FCS, 1 mm sodium pyruvate, penicillin, and streptomycin. In some experiments cells were maintained in a K+-free RPMI 1640 medium (Biological Industries). Rat α1-Na+/K+-ATPase-expressing cell clones were produced by transfecting the above YFP-α1 cells with rat α1 tagged in its N terminus by the hemagglutinin A (HA) epitope subcloned into pcDNA-3 (21). Transfected cells were cultivated in the presence of 10 μm ouabain, and stably transfected ouabain-resistant cell clones were isolated.
Cell Viability Assay
Effects of ouabain on cell viability were determined using a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT)-based cell proliferation assay kit (Biological Industries) according to the manufacturer's instructions.
Imaging and Quantification of Na+/K+-ATPase Internalization
Cells were plated on a noncoated 8-well μ-slide (ibidi, Martinsried, Germany). The imaging stage was prewarmed to 37 °C, and CO2 was supplied. The next day cells were treated with ouabain or digoxin and monitored for 18 h by time-lapse microscopy. Images were taken at 30-min intervals using an Olympus FV10i-LIV microscope with a ×60 water immersion lens. For each time point four adjacent fields of view (FOVs) having 20–50 cells total were recorded. Three channels were imaged: red (mCherry), green (YFP-α1), and phase contrast. Each stack of images was combined using maximal intensity projection, and a montage of the four adjacent FOVs was created using ImageJ 1.42q freeware. Background was removed from the mCherry channel using the rolling ball method, and images were further processed by a dedicated Matlab program as follows. First, nuclei were identified by high mCherry fluorescence. The cytoplasm and plasma membrane of each cell were segmented, and areas of high intracellular YFP were identified. Next, total cell and internal YFP intensities were measured for each cell. Intracellular YFP-α1 was then determined from the ratio intracellular/total YFP signal. For each time point all cells visualized in all FOVs were averaged.
For fluorescence recovery after photobleaching, cells were seeded on an 8-well μ-slide at low abundance to allow minimal cell-cell contact. Groups of 1–3 cells were maximally photobleached using the “tornado” function of the scanning confocal microscope (Olympus FV1000) through a ×60 oil immersion objective. Fluorescent recovery was monitored for ∼14 h. At least three bleached areas were monitored, and data were normalized to the prebleach fluorescence.
Colocalization of Internalized Na+/K+-ATPase with Subcellular Markers
Cells were seeded in μ-Slide VI0.4 (ibidi) and cultivated for 24 h to reach confluence of ∼80%. Na+/K+-ATPase internalization was induced by the application of 100 nm ouabain, and monolayers were fixed at −20 °C in methanol either 2 or 4.5 h later. The fixed monolayers were washed in PBS plus Ca2+ and Mg2+, blocked with 3% BSA, and incubated overnight at 4 °C in a humidified chamber with either anti GRASP65 or anti LAMP1 antibodies. Samples were washed and incubated for 2 h at room temperature with Cy5-labeled secondary antibody diluted in 1% BSA PBS solution. Slides were visualized using an Olympus FV1000 confocal scanning microscope with a ×60 oil immersion lens. For each treatment several different FOVs were photographed and quantified. Quantification of the amount of Na+/K+-ATPase in the LAMP1- and GRASP65-labeled areas has been determined as follows using ImageJ. For each FOV, a median filter was applied, followed by background subtraction. Then using automatic otsu threshholding, the Cy5 channel was turned into a “mask,” and the amount of YFP-Na+/K+-ATPase in this area was measured. Similar colocalization of mCherry and YFP fluorescence has been used to assay for nuclear expression of Na+/K+-ATPase.
Internalization Analysis Using Surface Biotinylation
Biotinylation of cell surface proteins was done as reported previously (22). In brief, cell monolayers at ∼90% confluence were incubated for 15 min at 4 °C on a rocker with 1 mg/ml EZ-Link sulfo-succinimido-biotin (sulfo-NHS-SS-biotin; Pierce). Cells were washed twice in PBS plus Ca2+ and Mg2+, and free unreacted sulfo-NHS-SS-biotin was quenched by a 20-min incubation with 0.1% BSA (w/v). Biotinylated cells were then incubated in cell culture medium ± ouabain and other reagents for 7 h. At the end of this period cells were washed four times in a reducing solution (100 nm Tris, pH 8.6, 100 mm NaCl, 2.5 mm EDTA, 50 mm Na+-MES) and further incubated for 10 min at 4 °C to allow complete cleavage of surface-exposed S-S bonds. Effective removal of surface-exposed biotin was verified by reducing one plate immediately after the biotinylation. After two rinses in PBS, cells were lysed in a buffer composed of 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 5 mm EDTA, and protease inhibitors (20 μg/ml leupeptin, 10 milli-trypsin inhibitory units/ml aprotinin, and 2 mm PMSF). Cell lysates were centrifuged for 10 min (14,000 × g, 4 °C), and the supernatants were incubated with streptavidin-agarose beads (Immunopure immobilized streptavidin; Pierce) diluted in 50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm EDTA plus protease inhibitors. Following an overnight incubation at 4 °C, the beads were washed twice with rinsing solution (20 mm Tris-HCl, pH 7.4, 500 mm NaCl) and once with 10 mm Tris-HCl, pH 7.4. Washed beads were pelleted and resuspended in Laemmli buffer plus 50 mm dithiothreitol (DTT), and biotinylated internalized proteins were eluted by incubation at 55 °C for 30 min. Samples were resolved on 7.5% SDS-PAGE and transferred to polyvinylidine difluoride membranes (Immobilon-P; Millipore). Membranes were blocked with PBS containing 0.1% Tween 20 (PBS-T) and 3% BSA for 1 h at room temperature. The blot was cut into several regions and incubated overnight at 4 °C with different primary antibodies in PBS-T containing 1% BSA. Membranes were washed three times in PBS-T and then incubated for 1 h at room temperature with an appropriate HRP-conjugated secondary antibody in PBS-T. Following three washes in PBS-T, protein expression was assayed by enhanced chemiluminescence and quantified using ImageQuant LAS 4000 mini (GE Healthcare).
Statistics
Data are expressed as means ± S.E., and significance was calculated by the two-tailed t test.
Antibodies
A monoclonal antibody to the N-terminal sequence of the α1 subunit of Na,K-ATPase (6H) was kindly provided by Dr. M. J. Caplan, Yale University School of Medicine. A polyclonal anti-phospho-Src (Tyr-418) was from MBL International Corporation (Nagoya, Japan), and a monoclonal anti-ubiquitin antibody was from Covance (Princeton, NJ). A monoclonal anti-LAMP1 was from the Hybridoma Bank of the University of Iowa. Monoclonal anti-HA and anti-GFP antibodies were purchased from Santa Cruz Biotechnology, monoclonal anti-β-tubulin was from Sigma-Aldrich, and rabbit polyclonal anti-GRASP65 was from Abcam (Cambridge, MA). Cy5-coupled secondary antibodies were from Jackson ImmunoResearch Laboratories.
RESULTS
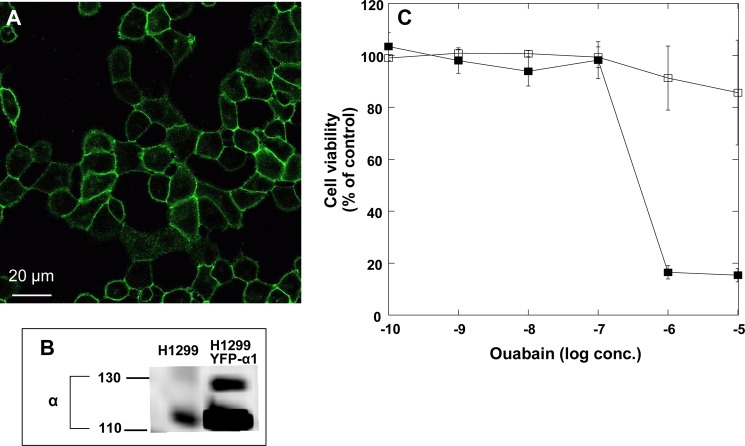
CG-induced internalization of the Na+/K+-ATPase was studied in an H1299 cell clone stably expressing YFP-tagged α1 from the normal α1 locus in the genome. Because the YFP-tagged protein is expressed from the normal chromosomal location of α1, an adequate level of expression and genomic regulation is assured. In addition, these cells express mCherry that gives strong nuclear and weak cytoplasmic fluorescence that assists with computerized segmentation of cells (see below). The YFP-tagged α1 is properly directed to the plasma membrane (Fig. 1A). It migrates as ∼140-kDa band which can be discriminated from the untagged 110-kDa α1 protein expressed from the other allele (Fig. 1B). In the parental H1299 line only the 110-kDa band is detected.
FIGURE 1.
H1299 cells expressing YFP-α1 Na+/K+-ATPase. A, image of a confluent monolayer of H1299 cell clone expressing YFP-α1Na+/K+-ATPase from its endogenous chromosomal location. B, Western blot of H1299 cells expressing YFP-α1-Na+/K+-ATPase and parental nontransfected H1299 cells with anti-α-Na+/K+-ATPase (6H). C, sensitivity of H1299 cells to ouabain. Confluent monolayers of cells were incubated with the indicated concentration of ouabain (filled squares) or diluent (empty squares) for 24 h. The percentage of viable cells was determined by the XTT assay. Means ± S.E. (error bars) of five independent experiments are depicted.
To determine the ouabain concentrations that can be used to study internalization with no toxic effect, confluent monolayers of H1299 cells were incubated for 24 h with increasing amounts of ouabain, and cell viability was determined by the XTT assay. As shown in Fig. 1C, a prolonged incubation with up to 100 nm ouabain had little effect on cell viability. Na+/K+-ATPase activity and ouabain binding of the human enzymes measured under similar conditions suggest that at 100 nm ouabain <50% of the pumps are blocked (e.g. Refs. 23–25).
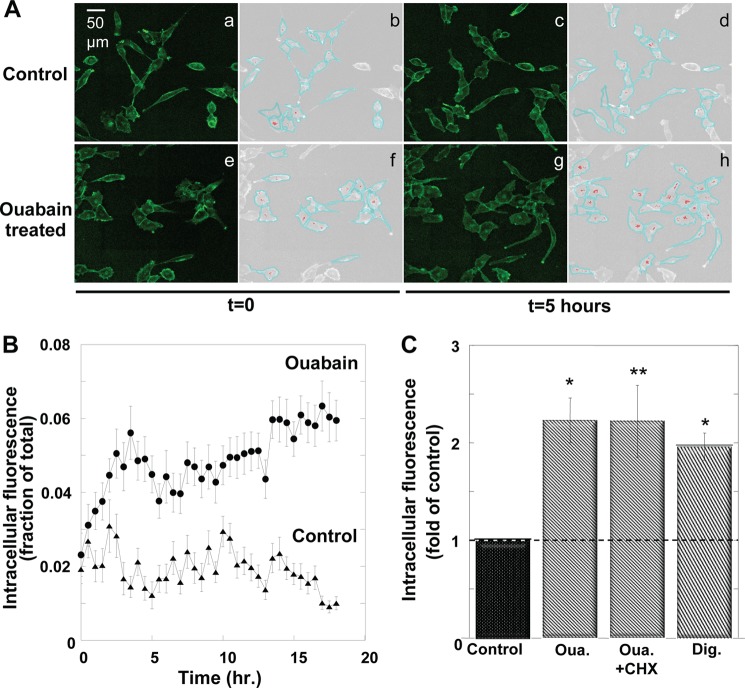
The fluorescence tag provides a convenient way to monitor ouabain-induced internalization of α1 but requires a means to quantify changes in intracellular versus plasma membrane fluorescence. Accordingly, ouabain-induced internalization was imaged over several hours by time-lapse microscopy, and time-dependent changes in intracellular YFP-α1 were analyzed and quantified as detailed under “Experimental Procedures.” The application of 100 nm ouabain induced substantial internalization of α1 that developed over several hours, and most of the internalization took place within 5 h (red patches in Fig. 2, A). On the average, 100 nm ouabain doubled the amount of intracellular α1, and its effect could be mimicked by digoxin, another CG with similar affinity for the pump (Fig. 2C) (25).
FIGURE 2.
Ouabain induced internalization of the Na+/K+-ATPase. A, representative confocal (a, c, e, and g) and computer-assisted segmented (b, d, f, and h) images of YFP-α1 H1299 cells. Cells received either 100 nm ouabain or diluent (t = 0). Images were taken at t = 0 and 5 h later and processed as described under “Experimental Procedures.” YFP fluorescence was segmented to membrane (cyan) and internal (red) regions. a and b, untreated cells at t = 0. c and d, untreated cells, t = 5 h. e and f, cells treated with 100 nm ouabain, t = 0. g and h, cells treated with 100 nm ouabain, t = 5 h. B, time course of YFP-α1 internalization. Intracellular fluorescence was quantified at 30-min intervals following the application of 100 nm ouabain (t = 0). Intracellular fluorescence is expressed as the fraction of the total cell fluorescence and averaged over all the cells in the imaged field. Means ± S.E. (error bars) of 20–40 cells are depicted. C, cells were treated with 100 nm ouabain or digoxin. One batch of cells received 100 μg/ml CHX prior to the addition of ouabain. Plateau values of intracellular α1-YFP were determined at t >4 h and normalized to values in control cells that receive diluent. Means ± S.E. of three different experiments are depicted. *, p < 0.005; **, p < 0.02.
In principle, the intracellular fluorescence may also include contribution from newly synthesized pumps “en route” to the plasma membrane. Two experiments were designed to exclude this possibility. In the first, the ouabain-induced intracellular accumulation of YFP-α1 was measured in the presence of the translation inhibitor cycloheximide (CHX). As shown in Fig. 2C, CHX did not lower accumulation of YFP-α1 within the cell. In the second, the rate of YFP-α1 synthesis was directly measured by monitoring the appearance of fluorescence in cells following a complete photobleaching of YFP-α1. Experiments such as that depicted in Fig. 3 demonstrate that ouabain is without effect on the rate of α1 synthesis measured as an increase in total fluorescence. The experiment also demonstrates that at the concentration used in the experiment of Fig. 2C, CHX almost completely inhibits α1 synthesis.
FIGURE 3.
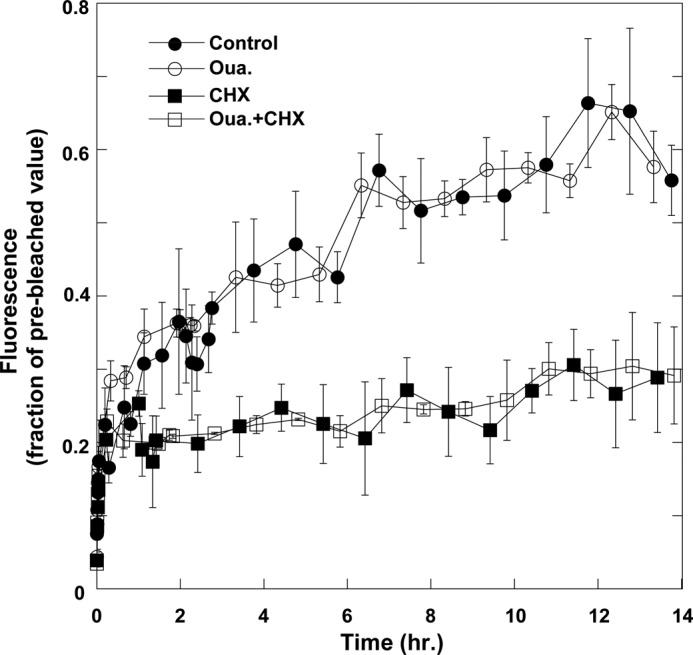
Na+/K+-ATPase synthesis in H1299 cells. Confluent cell monolayers were treated with diluent (control, filled circles), 100 nm ouabain (open circles), 100 μg/ml CHX (filled squares), and ouabain + CHX (open squares). De novo Na+/K+-ATPase synthesis was monitored by bleaching a field of ∼1–4 cells and monitoring the rate of fluorescence recovery. Data were normalized to the total fluorescence of the recorded field shortly before the bleaching. The values are averages of at least three fields for every treatment. Means ± S.E. (error bars) are depicted.
An alternative way to demonstrate and quantify ouabain-induced internalization is by surface biotinylation and detection of internalized biotin. In this protocol, the cell surface is biotinylated with sulfo-NHS-SS-biotin and the cells are then incubated with ouabain. Surface-exposed biotin is removed by incubation with an impermeable reducing agent, and the remaining biotinylated α1 represents the internalized biotinylated protein. Fig. 4A demonstrates such an experiment. It further confirms that the incubation with ouabain induces internalization of both the YFP-tagged α1 (140 kDa) and the untagged α1 (110 kDa). Efficiency of the cleavage of cell surface biotin is verified by reducing one plate prior to the incubation with ouabain. A dose response of ouabain-induced α1 internalization is depicted in Fig. 4B. Some effect was apparent already at 10 nm ouabain, although it is statistically insignificant, but a significant effect required at least 100 nm, consistent with the concentration required to occupy the ouabain site on human α1 in physiological conditions. The data also demonstrate that at 100 nm ouabain >15% of the surface-exposed (biotinylated) pumps are internalized, indicating a highly significant process. This number is in fact an underestimate because part of the internalized proteins will undergo lysosomal degradation (see below).
FIGURE 4.
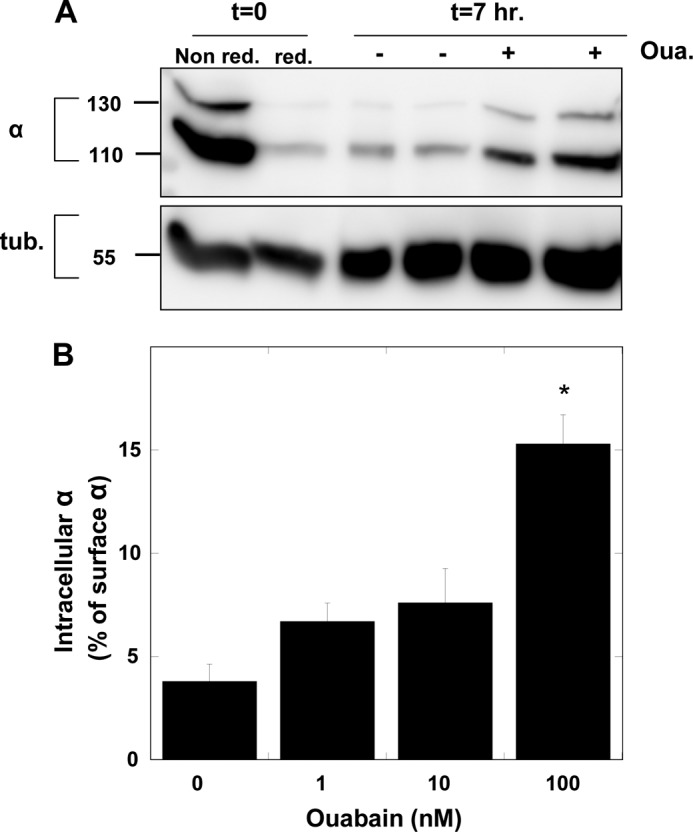
Na+/K+-ATPase internalization measured by surface biotinylation. A, surface proteins in confluent monolayers of H1299 cells were biotinylated by incubation with 1 mg/ml sulfo-NHS-SS-biotin for 15 min. Different plates received either ouabain (100 nm) or diluent, and the plates were incubated for an additional 7 h. At the end of the incubation period, bound surface-exposed biotin was cleaved by incubation with 50 mm Na+-MES. Two additional control plates were either not reduced (Non red.) or reduced shortly after the biotinylation (red.). Cells were lysed, and biotinylated proteins were purified and resolved on SDS-polyacrylamide gel as described under “Experimental Procedures.” Blots were probed with antibodies to α-Na+/K+-ATPase (6H) and tubulin (loading control). B, cells were treated as above with different concentrations of ouabain. Internalized α1 is expressed as percentage of the surface-expressed (i.e. biotinylated) α1 at t = 0. Means ± S.E. (error bars) of three independent experiment are depicted. *, p < 0.001.
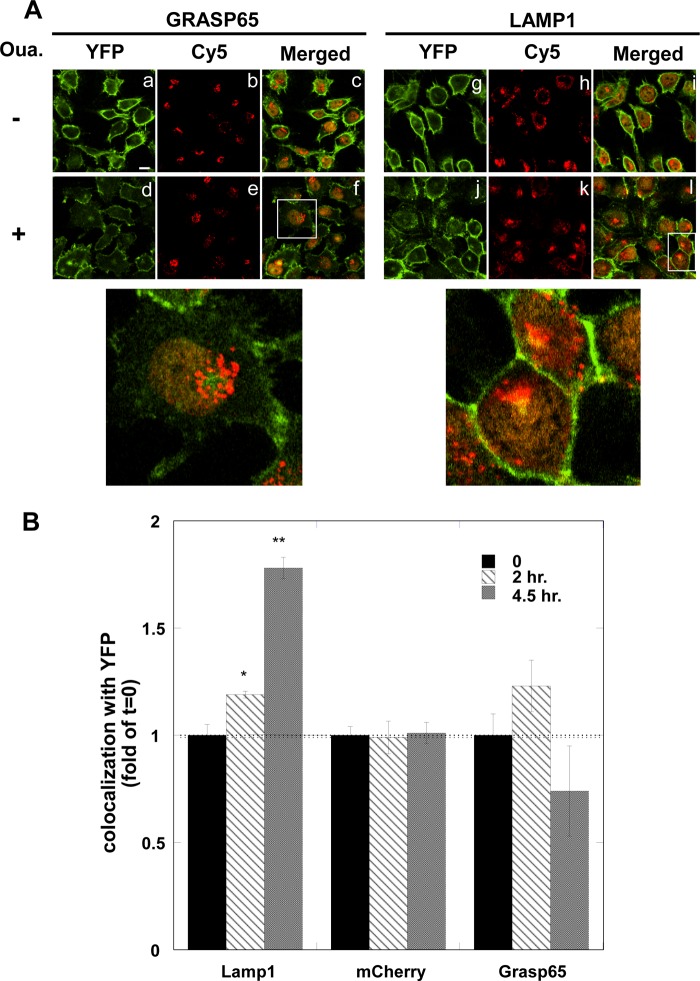
Next, we aimed to identify the cellular location of the internalized α1 using markers for specific organelles. Fig. 5A depicts images of YFP-α1 H1299 cells that were fixed and stained with the lysosomal and Golgi markers LAMP1 and GRASP65, respectively. The data indicate colocalization of internalized α1 with LAMP1 but not with GRASP65. Other experiments showed lack of colocalization with nuclear mCherry fluorescence. Quantification of the above colocalization was done as described under “Experimental Procedures” and is illustrated in Fig. 5B. The results clearly demonstrate the ouabain-induced increase in the colocalization of YFP-α1 and LAMP1 with no significant change in colocalization with GRASP65 or mCherry.
FIGURE 5.
Co-localization of YFP-α1 and lysosomal marker. A, top, cell monolayers were incubated for 4.5 h with (d–f, j–l) and without (a–c, g–i) 100 nm ouabain and stained for the lysosome and Golgi markers LAMP1 (g–l) and GRASP65 (a–f). Images were acquired using Olympus FV1000 confocal scanning microscope at ×60 magnification. Green, YFP-α1; red, LAMP1 or GRASP65. Bottom, enlargements show the areas marked by squares in images f and l. B, quantification of the colocalization of YFP-α1 and various intracellular markers is shown. The amount of α1 in the nucleus, lysosome and Golgi has been determined by colocalization of the YFP signal with that obtained from mCherry, LAMP1, and GRASP65, respectively. Means ± S.E. (error bars) of data from five to nine FOVs imaged 0, 2, and 4.5 h after the application of ouabain are depicted. *, p < 0.02; **, p < 0.005.
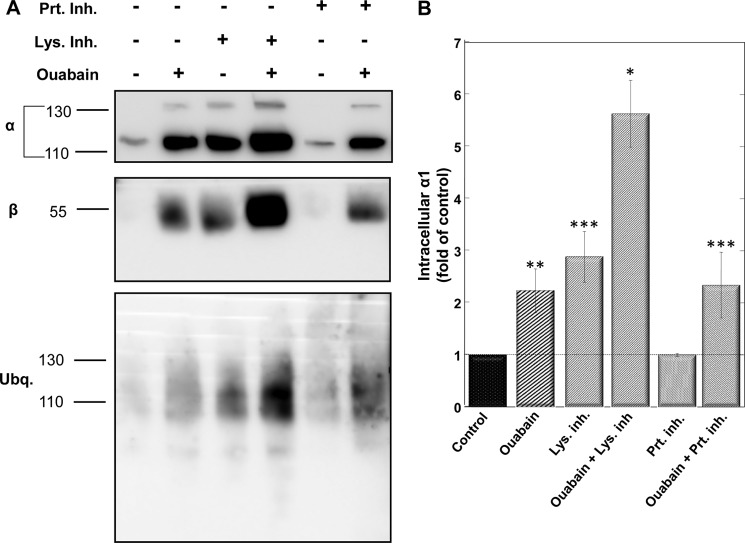
Lysosomal localization of the internalized YFP-α1 suggests that these pumps are destined for degradation. One can predict further that (i) inhibition of lysosomal degradation should increase the amount of α1 accumulated in lysosomes and (ii) the internalized pump is likely to be ubiquitinated. Fig. 6 summarizes effects of lysosomal inhibitors (chloroquine, NH4Cl, and leupeptin) and proteosomal inhibitor (bortezomib) on the ouabain-induced internalization of Na+/K+-ATPase. As predicted, inhibiting lysosomal degradation evoked by itself an increase in the intracellular pool of α1 and greatly augmented the response to ouabain. However, the proteosomal inhibitor had no effect on internalized α1 irrespective of ouabain treatment. An additional finding seen in Fig. 6A is that both α and β are internalized. We have also tested the possibility that the internalized pump is ubiquitinated. Blots of the streptavidin pulled-down proteins with an anti-ubiquitin antibody identified a single band with the same molecular mass of α1 that is augmented by ouabain and the lysosomal inhibitor but not the proteosomal inhibitor. Thus, it appears that ouabain treatment does indeed induce ubiquitination of α1.
FIGURE 6.
Effects of lysosomal and proteosomal inhibitors on the intracellular pool of α1. A, plates of confluent monolayers of H1299 were surface-biotinylated and then incubated for 7 h ± 100 nm ouabain and either lysosomal inhibitors (20 μg/ml leupeptin, 10 nm chloroquine, and 20 mm NH4Cl) or proteosomal inhibitor (2.5 μm bortezomib). Internalized proteins were isolated on streptavidin beads, resolved electrophoretically, and blotted with antibodies to α- and β-Na+/K+-ATPase and ubiquitin. B, quantification of biotinylated (internalized) α1 is shown. Means ± S.E. (error bars) of three to five experiments using the above procedure are averaged. *, p < 0.0005; **, p < 0.002; ***, p < 0.05.
Next, we addressed possible mechanisms by which ouabain induces internalization of the Na+/K+-ATPase. In principle, this process could be triggered by one of several events. The first possibility is the normal dynamics of membrane protein trafficking which undergo “quality control” in endosomes (26), so that proteins are either recycled back to the membrane or are directed for degradation. By this mechanism ouabain-bound pumps are sensed by endosomal chaperones as nonfunctional and are therefore sent for lysosomal degradation. The second option is that α1 internalization is secondary to a signaling cascade triggered by ouabain binding and involves the activation of protein kinases (27). Indeed, it was reported that in LLC-PK1 cells the ouabain-induced decrease in surface α is sensitive to PI3K and Src inhibitors (9). Finally, it is possible that α1 internalization is a more general response to inhibition of active Na+ and K+ transport by which partial inhibition of plasma membrane Na+/K+-ATPase results in an increase in cell Na+, a subsequent increase in cell Ca2+ (due to coupling with plasma membrane and mitochondrial Na+/Ca2+ exchange) and, possibly, an elevated ATP/ADP ratio. Any of these or other consequences of the disrupted ionic balance may be associated with signaling events leading to internalization of Na+/K+-ATPase.
In principle, it should be possible to determine whether internalization involves inhibition of active Na+ and K+ transport by looking at internalization of the human YFP-α1 pumps in cells transfected also with a ouabain-insensitive rat α1. The Ki for inhibition of rat kidney Na+/K+-ATPase (α1β1) is approximately 100 μm (28). With a sufficiently high level of expression of rat α1, incubation of the cells with 100 nm ouabain should have no significant effect on the overall cellular energetics and ionic balance, whereas effects due to ouabain binding to a particular human α1 molecule or intracellular signaling should not be altered. Moreover, if ouabain-induced signaling is involved, rat α may also be internalized, even though the amount of ouabain bound to the rat pump is very small.
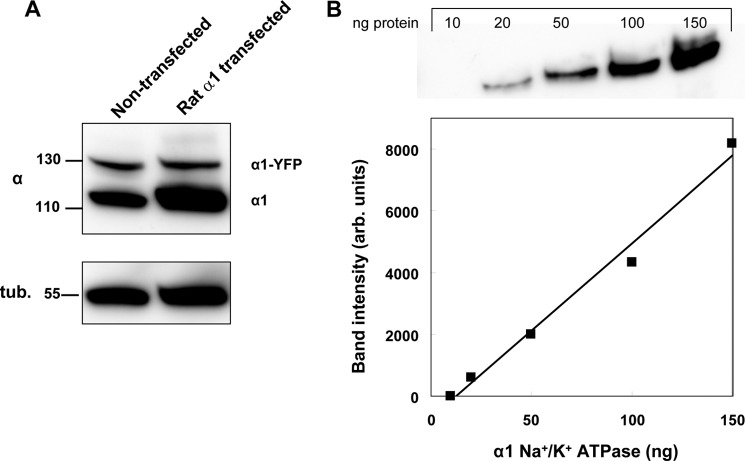
Accordingly, YFP-α1-expressing H1299 cells were transfected with HA-tagged rat α1, and cells stably expressing functional ouabain-resistant pumps were selected by cultivation in 10 μm ouabain. Overexpression of rat α1 was found to suppress expression of the endogenous human α1, which precludes determination of its ouabain-induced internalization. Therefore, we selected cell clones in which the amount of rat α1 expressed is sufficiently high to compensate for the inhibitory effect of 100 nm ouabain but is low enough to allow detection of internalized human α1. Such a cell clone is depicted in Fig. 7. The expression of rat α1 is indicated by the increased intensity of the ∼110-kDa band, whereas the existence of the ∼140-kDa band indicates substantial expression of human YFP-α1. Quantification of the relative amounts of human and rat α1 was done assuming that the ratio between the 140-kDa and 110-kDa bands of human α1 is independent of the expression of rat α1. Also, we quantified pump abundance by calibrating the immunostaining intensity of known amounts of α1 expressed and purified from Pichia pastoris (29) (Fig. 7B). The resulting amounts of human and rat α1 are given in Table 1 and indicate that the amount of rat α1 introduced is sufficient to compensate for suppression by ouabain of expression of 66% of the amount of human α1, and presumably also cation pumping activity, assuming the same Na,K-ATPase turnover rates of the rat and human proteins. In addition, these cells grow well in 10 μm ouabain, suggesting that the amount of rat α1 transfected is more than sufficient to sustain viability.
FIGURE 7.
Expression and quantification of rat α1 in H1299 cells. A, Western blot of nontransfected and rat α1-transfected H1299 cells. The image depicts the 140-kDa (human YFP-α1), 110-kDa (human α1 in nontransfected cells and human + rat α1 in transfected cells), and 55-kDa (tubulin loading control) bands. B, upper, different segment of the same blot in A in which 10–150 ng of purified human α1 was blotted with the anti-α1 antibody. Lower, correlation between the chemiluminesence signal and the amount of α1 protein.
TABLE 1.
Quantification of human and rat α1 in H1299 cells
Intensities of the 140- and 110-kDa α1 bands in Fig. 7A were first translated to absolute amounts of α1 protein (in ng/mg of protein of whole cell lysate) by means of the calibration curve of Fig. 7B. Contributions of rat and human α1proteins to the 110-kDa band of transfected cells were then calculated assuming that the ratio of YFP-α1 to untagged human α1 is the same for the nontransfected and rat α1-transfected cells.
| Na+/K+-ATPase isoform | α1 protein |
|
|---|---|---|
| Nontransfected cells | Rat α1-transfected cells | |
| ng/mg cell protein | ||
| Human YFP-α1 | 64.3 | 56.6 |
| YFPα1 + untagged human α1 | 243.1 | 198.9 |
| Rat α1 | 131.0 | |
| Rat/human α1 ratio | 0.66 | |
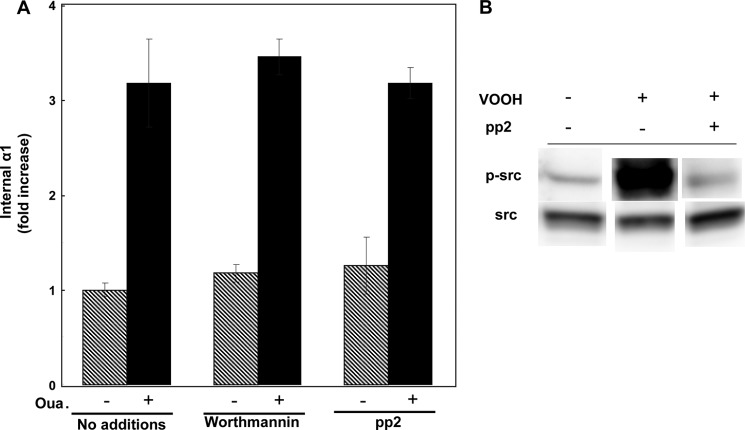
Fig. 8 quantifies three experiments measuring internalization of YFP-α1 in rat α1-expressing H1299 cells. The key observation is that overexpression of ouabain-insensitive α1 did not prevent internalization of human YFP-α1 by 100 nm ouabain, i.e. this process is not likely to be triggered by the effect of ouabain on the overall cation homeostasis. The data also argue against the possibility of an effect that is secondary to ouabain-induced signaling and mediated by the activated protein kinases. Such a mechanism is not expected to discriminate between human and rat α1 because at a sufficiently high dose of ouabain both proteins are internalized (data not shown). To further exclude a mediating role of the ouabain-induced signaling, we tested the effects of inhibitors of two key protein kinases participating in this cascade (Fig. 9). Neither the PI3K inhibitor wortmannin nor the Src blocker PP2 appears to affect internalization (Fig. 9A). The ability of PP2 to inhibit Src activation was confirmed by demonstrating its effect on Src activation by vanadate (Fig. 9B). Thus, the current process is different from the one reported in Ref. 9. Also ineffective was the clathrin inhibitor Pitstop 2 (data not shown).
FIGURE 8.
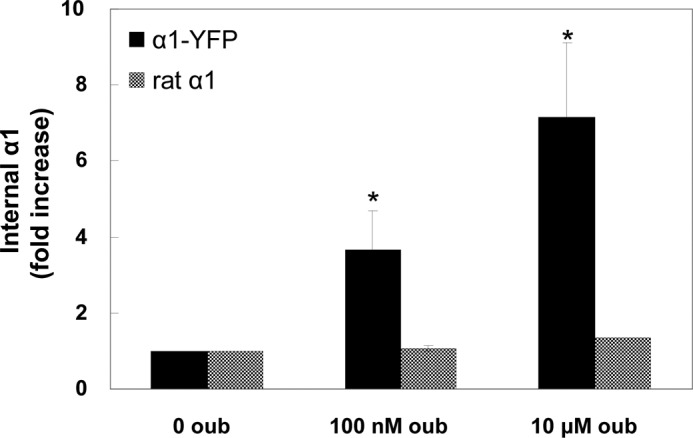
α1 internalization in rat α1-transfected cells. Internalization was measured by surface biotinylation as above in rat α1-expressing cells. The internalized human and rat α1 were discriminated using anti-YFP and anti-HA antibodies. Data are expressed as -fold increase in intracellular biotinylated α1. Means ± S.E. (error bars) of three experiments are depicted. *, p < 0.05.
FIGURE 9.
Effects of protein kinase inhibitors on α1 internalization. A, cells were treated for 7 h ± 100 nm ouabain and either 1 μm wortmannin or 10 μm PP2. Intracellular biotinylated α1 was quantified and expressed as -fold increase relative to the untreated sample. Means ± S.E. (error bars) of two to four independent experiments is depicted. It all cases the effect of ouabain was highly significant (p < 0.005). B, effect of PP2 on Src phosphorylation. Near-confluent cells were serum-starved for 16 h and incubated for 40 min with either 10 μm PP2 or an equal amount of diluent. Cells were then activated for 15 min with 1 mm NaVO3 + 3 mm H2O2, lysed in radioimmuneprecipitation assay buffer + proteases inhibitors, resolved electrophoretically, and blotted for total and phospho-Src.
The experiments summarized in Figs. 8 and 9 argue in favor of the hypothesis that internalization of α1 is an outcome of interaction between ouabain and the pump, leading to a conformational change that triggers internalization. Moreover, according to Fig. 6 the process may also involve α ubiquitination. Ouabain is known to bind most tightly to and stabilize the E2P conformation of the pump. In a cellular situation stabilization of E2P can also be achieved by removing extracellular K+. Thus, we have tested the effect of removing extracellular K+ on internalization of α1. As seen in Fig. 10, incubating cells in K+-free medium did not itself induce significant internalization, nor did it alter the response to ouabain (Fig. 10). Thus, internalization does not appear to be directly linked to E2P stabilization per se, but rather it requires binding of the ouabain. Furthermore, because in the absence of external K+ the pump is largely inhibited, this experiment provides additional confirmation that the ouabain-induced internalization is not secondary to impaired cation balance or altered ATP/ADP ratio.
FIGURE 10.
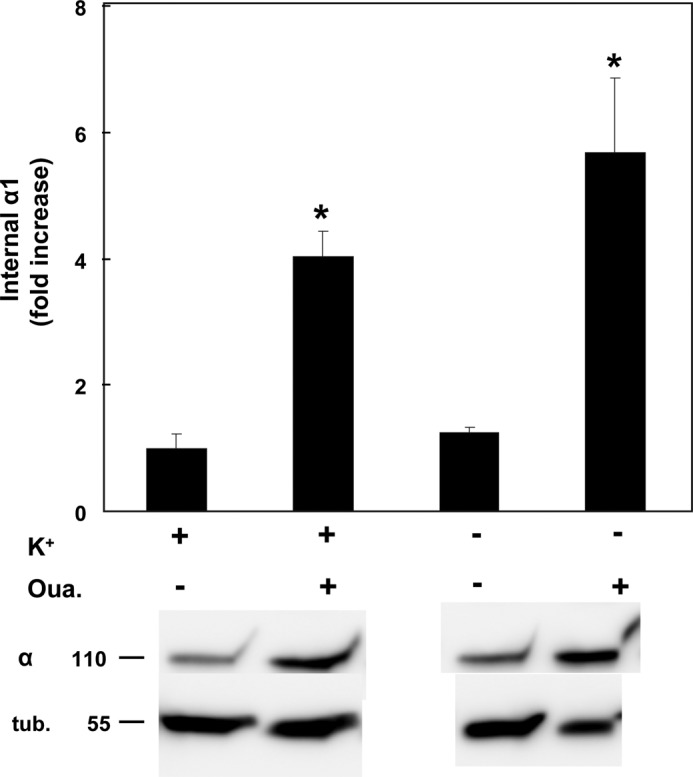
Effect of external K+ on ouabain-induced internalization. α1 internalized by incubation with 100 nm ouabain was measured as above in either regular or K+-free media. Means ± S.E. (error bars) of five experiments are depicted. *, p < 0.0001.
DISCUSSION
The Na+/K+-ATPase has been shown before to undergo internalization in response to a number of stimuli such as dopamine (13), parathyroid hormone (14), hypoxia and lung injury (15, 16), hypercapnia (17), and sepsis (18). The time course varies from minutes to hours, and the suggested mechanisms involve activation phosphorylation of α1 by protein kinases and in some cases its subsequent ubiquitination. A number of previous studies have also reported that external application of ouabain as well as other CGs results in internalization of both α and β subunits of the Na+/K+-ATPase. One role suggested for this process is acidification of endosomal pH (11, 30). Other studies related this process to the activation of caveolar Src and the subsequent ouabain-induced signaling cascade. The internalization observed in these studies was relatively rapid (1 h) and sensitive to Src and PI3K inhibitors (9, 31, 32). Such studies also identified Na+/K+-ATPase in the nucleus and suggested that it mediates altered gene expression (9, 33). Finally, it has been reported that in polarized epithelia ouabain can induce internalization of both the basolateral Na+/K+-ATPase and the apical Na+/H+ exchanger and thereby down-regulate transepithelial Na+ transport (32).
The current study characterizes ouabain-induced internalization of the α1 subunit of Na+/K+-ATPase in the human lung carcinoma cell line H1299. As discussed above, this cell line was selected due to the existence of a cell clone expressing YFP-tagged α1 from the normal genomic locus, enabling real-time detection of the location of endogenous protein. The ouabain-induced internalization identified and characterized in this cell line differ from those associated with the signaling cascade in the sense that it develops over a time scale of several hours, it is not affected by Src and PI3K inhibitors, and is associated with lysosomal degradation of the internalized pumps. Some of these differences may be due to the different cell line model used i.e. human carcinoma versus pig kidney epithelium.
A major question is the mechanism by which the association of ouabain with α1 induces its internalization. Two possibilities have been considered in this work. First, internalization may be an outcome of CG binding to a particular α1 molecule that triggers a conformational change of this pump. Second, Na+/K+-ATPase internalization might be secondary to the partial inhibition of Na+ and K+ transport that result in alterations in the intracellular Na+, K+, Ca2+, or H+ activities, as well as the ATP/ADP ratio, which then affect various intracellular pathways. One approach to discriminating between the two possibilities involved transfecting H1299 cells with rat α1, which is insensitive to nanomolar concentrations of ouabain. The result that expression of rat α1 did not inhibit internalization of human α1 argues against the possibility that Na+/K+-ATPase internalization is mediated by an altered cation balance. This observation also argues against the notion that internalization is the outcome of ouabain-induced signaling of MAPK/ERK or Akt/PI3K pathways. Because at sufficiently high concentration of ouabain both human and rat α1 are internalized, if these signaling pathways were involved, one would have expected that triggering the cascade by the binding of ouabain to human α1 at 100 nm would also internalize rat α1 expressed in the same cell.
In view of the arguments just presented, our best hypothesis is that internalization is a direct outcome of a ouabain-induced conformational change in the CG-bound molecule. One such obvious conformational change in a cell would be a shift of the steady-state E1/E2 distribution caused by tight binding of ouabain and stabilization of the E2P conformation (34). A similar accumulation of E2P should be achieved by removing extracellular K+, which greatly reduces the rate of hydrolysis of E2P. However, we found that maintaining cells in K+-free medium neither induces α1 internalization, nor does it augment the response to ouabain. This observation shows that it is not accumulation of E2P as such that is associated with internalization and that internalization is not secondary to the inhibition of Na+ and K+ transport. Recent evidence for a specific ouabain-induced conformational change that alters accessibility of cytoplasmic domains in α to tryptic digestion was provided in Ref. 35. These authors demonstrated a different ouabain-dependent pattern of the tryptic fragments of shark α stabilized in the E2·MgF42− state. In the absence of ouabain, trypsin cleaved the enzyme at Arg-269 in a region connecting the A domain with the M3 transmembrane helix. Ouabain protected this site and instead enabled a different split at Lys-161 located in a region connecting the M2 transmembrane helix with the A domain. In addition, it was shown many years ago that in the presence of ouabain, chymotrypsin specifically cleaves the cytoplasmic N domain of the protein into two fragments of 35 and 40 kDa (36). Thus, overall, it appears most likely that the ouabain-bound protein itself adopts a specific conformation that is susceptible to internalization, perhaps by altered interactions with other cellular proteins, for example of the cytoskeleton. In this respect, both ankyrin and adducin have been shown to interact specifically with the Na+/K+-ATPase (37, 38). It has also been demonstrated that mutations in α-adducin alter the rate of Na+/K+-ATPase internalization (39).
Another unresolved issue is the physiological role of the ouabain-induced internalization and lysosomal degradation characterized in this study. As discussed above it is clear today that CGs are synthesized in the adrenal gland and function as endogenous hormones (40). Although the exact roles and cellular functions of endogenous CGs are diverse and have not all been established, it is obvious that circulating CGs should bind the Na+/K+-ATPase in various tissues and block its function. Inhibition of the Na+/K+-ATPase activity is effectively irreversible because the dissociation rate of the ouabain-α complex is very slow (24, 34). Therefore, one can propose that internalization and degradation of the inhibited pumps, and their replacement by newly synthesized proteins, represent an efficient and rapid way for cells to recover from a transiently high level of circulating CG and remove endogenous ouabain from the circulation.
Acknowledgments
We thank Prof. Zvi Kam for useful discussions and Vladimir Kiss or help with the confocal microscopy measurements.
Footnotes
- CG
- cardiac glycoside
- CHX
- cycloheximide
- FOV
- field of view
- PP2
- 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine
- XTT
- 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide.
REFERENCES
- 1. Blanco G., Mercer R. W. (1998) Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 275, F633–650 [DOI] [PubMed] [Google Scholar]
- 2. Garty H., Karlish S. J. (2006) Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459 [DOI] [PubMed] [Google Scholar]
- 3. Schoner W., Scheiner-Bobis G. (2007) Endogenous and exogenous cardiac glycosides and their mechanisms of action. Am. J. Cardiovasc. Drugs 7, 173–189 [DOI] [PubMed] [Google Scholar]
- 4. Blaustein M. P. (1996) Endogenous ouabain: role in the pathogenesis of hypertension. Kidney Int. 49, 1748–1753 [DOI] [PubMed] [Google Scholar]
- 5. Silva E., Soares-da-Silva P. (2012) New insights into the regulation of Na+,K+-ATPase by ouabain. Int. Rev. Cell Mol. Biol. 294, 99–132 [DOI] [PubMed] [Google Scholar]
- 6. Liu J., Xie Z. J. (2010) The sodium pump and cardiotonic steroids-induced signal transduction protein kinases and calcium-signaling microdomain in regulation of transporter trafficking. Biochim. Biophys. Acta 1802, 1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang L., Zhang Z., Guo H., Wang Y. (2008) Na+/K+-ATPase-mediated signal transduction and Na+/K+-ATPase regulation. Fundam. Clin. Pharmacol. 22, 615–621 [DOI] [PubMed] [Google Scholar]
- 8. Kotova O., Al-Khalili L., Talia S., Hooke C., Fedorova O. V., Bagrov A. Y., Chibalin A. V. (2006) Cardiotonic steroids stimulate glycogen synthesis in human skeletal muscle cells via a Src- and ERK1/2-dependent mechanism. J. Biol. Chem. 281, 20085–20094 [DOI] [PubMed] [Google Scholar]
- 9. Liu J., Kesiry R., Periyasamy S. M., Malhotra D., Xie Z., Shapiro J. I. (2004) Ouabain induces endocytosis of plasmalemmal Na/K-ATPase in LLC-PK1 cells by a clathrin-dependent mechanism. Kidney Int. 66, 227–241 [DOI] [PubMed] [Google Scholar]
- 10. Liu J., Shapiro J. I. (2007) Regulation of sodium pump endocytosis by cardiotonic steroids: molecular mechanisms and physiological implications. Pathophysiology 14, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feldmann T., Glukmann V., Medvenev E., Shpolansky U., Galili D., Lichtstein D., Rosen H. (2007) Role of endosomal Na+-K+-ATPase and cardiac steroids in the regulation of endocytosis. Am. J. Physiol. Cell Physiol. 293, C885–896 [DOI] [PubMed] [Google Scholar]
- 12. Gupta S., Yan Y., Malhotra D., Liu J., Xie Z., Najjar S. M., Shapiro J. I. (2012) Ouabain and insulin induce sodium pump endocytosis in renal epithelium. Hypertension 59, 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chibalin A. V., Katz A. I., Berggren P. O., Bertorello A. M. (1997) Receptor-mediated inhibition of renal Na+-K+-ATPase is associated with endocytosis of its α- and β-subunits. Am. J. Physiol. 273, C1458–1465 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Norian J. M., Magyar C. E., Holstein-Rathlou N. H., Mircheff A. K., McDonough A. A. (1999) In vivo PTH provokes apical NHE3 and NaPi2 redistribution and Na-K-ATPase inhibition. Am. J. Physiol. 276, F711–719 [DOI] [PubMed] [Google Scholar]
- 15. Dada L. A., Chandel N. S., Ridge K. M., Pedemonte C., Bertorello A. M., Sznajder J. I. (2003) Hypoxia-induced endocytosis of Na,K-ATPase in alveolar epithelial cells is mediated by mitochondrial reactive oxygen species and PKC-ζ. J. Clin. Invest. 111, 1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lecuona E., Sun H., Vohwinkel C., Ciechanover A., Sznajder J. I. (2009) Ubiquitination participates in the lysosomal degradation of Na,K-ATPase in steady-state conditions. Am. J. Respir. Cell Mol. Biol. 41, 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Welch L. C., Lecuona E., Briva A., Trejo H. E., Dada L. A., Sznajder J. I. (2010) Extracellular signal-regulated kinase (ERK) participates in the hypercapnia-induced Na,K-ATPase down-regulation. FEBS Lett. 584, 3985–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berger G., Guetta J., Klorin G., Badarneh R., Braun E., Brod V., Saleh N. A., Katz A., Bitterman H., Azzam Z. S. (2011) Sepsis impairs alveolar epithelial function by down-regulating Na-K-ATPase pump. Am. J. Physiol. Lung Cell. Mol. Physiol. 301, L23–30 [DOI] [PubMed] [Google Scholar]
- 19. Sigal A., Danon T., Cohen A., Milo R., Geva-Zatorsky N., Lustig G., Liron Y., Alon U., Perzov N. (2007) Generation of a fluorescently labeled endogenous protein library in living human cells. Nat. Protoc. 2, 1515–1527 [DOI] [PubMed] [Google Scholar]
- 20. Cohen A. A., Geva-Zatorsky N., Eden E., Frenkel-Morgenstern M., Issaeva I., Sigal A., Milo R., Cohen-Saidon C., Liron Y., Kam Z., Cohen L., Danon T., Perzov N., Alon U. (2008) Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 [DOI] [PubMed] [Google Scholar]
- 21. Lindzen M., Gottschalk K. E., Füzesi M., Garty H., Karlish S. J. (2006) Structural interactions between FXYD proteins and Na,K-ATPase: α/β/FXYD subunit stoichiometry and cross-linking. J. Biol. Chem. 281, 5947–5955 [DOI] [PubMed] [Google Scholar]
- 22. Moshitzky S., Asher C., Garty H. (2012) Intracellular trafficking of FXYD1 (phospholemman) and FXYD7 proteins in Xenopus oocytes and mammalian cells. J. Biol. Chem. 287, 21130–21141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kometiani P., Liu L., Askari A. (2005) Digitalis-induced signaling by Na+/K+-ATPase in human breast cancer cells. Mol. Pharmacol. 67, 929–936 [DOI] [PubMed] [Google Scholar]
- 24. Crambert G., Hasler U., Beggah A. T., Yu C., Modyanov N. N., Horisberger J. D., Lelièvre L., Geering K. (2000) Transport and pharmacological properties of nine different human Na,K-ATPase isozymes. J. Biol. Chem. 275, 1976–1986 [DOI] [PubMed] [Google Scholar]
- 25. Katz A., Lifshitz Y., Bab-Dinitz E., Kapri-Pardes E., Goldshleger R., Tal D. M., Karlish S. J. (2010) Selectivity of digitalis glycosides for isoforms of human Na,K-ATPase. J. Biol. Chem. 285, 19582–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taguchi T. (2013) Emerging roles of recycling endosomes. J. Biochem. 153, 505–510 [DOI] [PubMed] [Google Scholar]
- 27. Xie Z., Askari A. (2002) Na+/K+-ATPase as a signal transducer. Eur. J. Biochem. 269, 2434–2439 [DOI] [PubMed] [Google Scholar]
- 28. Ferrandi M., Molinari I., Barassi P., Minotti E., Bianchi G., Ferrari P. (2004) Organ hypertrophic signaling within caveolae membrane subdomains triggered by ouabain and antagonized by PST 2238. J. Biol. Chem. 279, 33306–33314 [DOI] [PubMed] [Google Scholar]
- 29. Lifshitz Y., Petrovich E., Haviv H., Goldshleger R., Tal D. M., Garty H., Karlish S. J. (2007) Purification of the human α2 isoform of Na,K-ATPase expressed in Pichia pastoris: stabilization by lipids and FXYD1. Biochemistry 46, 14937–14950 [DOI] [PubMed] [Google Scholar]
- 30. Cain C. C., Sipe D. M., Murphy R. F. (1989) Regulation of endocytic pH by the Na+,K+-ATPase in living cells. Proc. Natl. Acad. Sci. U.S.A. 86, 544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu L., Abramowitz J., Askari A., Allen J. C. (2004) Role of caveolae in ouabain-induced proliferation of cultured vascular smooth muscle cells of the synthetic phenotype. Am. J. Physiol. Heart Circ. Physiol. 287, H2173–2182 [DOI] [PubMed] [Google Scholar]
- 32. Yan Y., Haller S., Shapiro A., Malhotra N., Tian J., Xie Z., Malhotra D., Shapiro J. I., Liu J. (2012) Ouabain-stimulated trafficking regulation of the Na/K-ATPase and NHE3 in renal proximal tubule cells. Mol. Cell. Biochem. 367, 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akimova O. A., Hamet P., Orlov S. N. (2008) [Na+]i/[K+]i-independent death of ouabain-treated renal epithelial cells is not mediated by Na+,K+-ATPase internalization and de novo gene expression. Pflugers Arch. 455, 711–719 [DOI] [PubMed] [Google Scholar]
- 34. Wallick E. T., Schwartz A. (1988) Interaction of cardiac glycosides with Na+,K+-ATPase. Methods Enzymol. 156, 201–213 [DOI] [PubMed] [Google Scholar]
- 35. Cornelius F., Mahmmoud Y. A. (2009) Interaction between cardiotonic steroids and Na,K-ATPase: effects of pH and ouabain-induced changes in enzyme conformation. Biochemistry 48, 10056–10065 [DOI] [PubMed] [Google Scholar]
- 36. Castro J., Farley R. A. (1979) Proteolytic fragmentation of the catalytic subunit of the sodium and potassium adenosine triphosphatase: alignment of tryptic and chymotryptic fragments and location of sites labeled with ATP and iodoacetate. J. Biol. Chem. 254, 2221–2228 [PubMed] [Google Scholar]
- 37. Ferrandi M., Salardi S., Tripodi G., Barassi P., Rivera R., Manunta P., Goldshleger R., Ferrari P., Bianchi G., Karlish S. J. (1999) Evidence for an interaction between adducin and Na+-K+-ATPase: relation to genetic hypertension. Am. J. Physiol. 277, H1338–1349 [DOI] [PubMed] [Google Scholar]
- 38. Devarajan P., Scaramuzzino D. A., Morrow J. S. (1994) Ankyrin binds to two distinct cytoplasmic domains of Na,K-ATPase α subunit. Proc. Natl. Acad. Sci. U.S.A. 91, 2965–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Torielli L., Tivodar S., Montella R. C., Iacone R., Padoani G., Tarsini P., Russo O., Sarnataro D., Strazzullo P., Ferrari P., Bianchi G., Zurzolo C. (2008) α-Adducin mutations increase Na/K pump activity in renal cells by affecting constitutive endocytosis: implications for tubular Na reabsorption. Am. J. Physiol. Renal Physiol. 295, F478–487 [DOI] [PubMed] [Google Scholar]
- 40. Schoner W., Scheiner-Bobis G. (2007) Endogenous and exogenous cardiac glycosides: their roles in hypertension, salt metabolism, and cell growth. Am. J. Physiol. Cell Physiol. 293, C509–536 [DOI] [PubMed] [Google Scholar]