Background: Older patients are more sensitive to drug-mediated development of liver disorders.
Results: Age and CCl4 treatments change expression of C/EBP proteins leading to repression of key regulators of liver biology.
Conclusion: The age-associated alterations of C/EBP proteins cause severe liver injury after CCl4 treatments.
Significance: Understanding of mechanisms of age-associated severe liver injury is important for development of therapeutic approaches.
Keywords: C/EBP Transcription Factor, Chromatin Remodeling, Epigenetics, Liver Injury, Proliferation
Abstract
The aged liver is more sensitive to the drug treatments and has a high probability of developing liver disorders such as fibrosis, cirrhosis, and cancer. Here we present mechanisms underlying age-associated severe liver injury and acceleration of liver proliferation after CCl4 treatments. We have examined liver response to CCl4 treatments using old WT mice and young C/EBPα-S193D knockin mice, which express an aged-like isoform of C/EBPα. Both animal models have altered chromatin structure as well as increased liver injury and proliferation after acute CCl4 treatments. We found that these age-related changes are associated with the repression of key regulators of liver biology: C/EBPα, Farnesoid X Receptor (FXR) and telomere reverse transcriptase (TERT). In quiescent livers of old WT and young S193D mice, the inhibition of TERT is mediated by HDAC1-C/EBPα complexes. After CCl4 treatments, TERT, C/EBPα and FXR are repressed by different mechanisms. These mechanisms include the increase of a dominant negative isoform, C/EBPβ-LIP, and subsequent repression of C/EBPα, FXR, and TERT promoters. C/EBPβ-LIP also disrupts Rb-E2F1 complexes in C/EBPα-S193D mice after CCl4 treatments. To examine if these alterations are involved in drug-mediated liver diseases, we performed chronic treatments of mice with CCl4. We found that C/EBPα-S193D mice developed fibrosis much more rapidly than WT mice. Thus, our data show that the age-associated alterations of C/EBP proteins create favorable conditions for the increased liver proliferation after CCl4 treatments and for development of drug-mediated liver diseases.
Introduction
Treatment of aged patients involves the use of drugs including those utilized for chemotherapy. The liver is the main organ of this drug and toxicant metabolism, which makes it the primary target of chemically induced injury. As the result of this injury, the liver starts proliferating, a characteristic that contributes to development of several diseases including fibrosis, cirrhosis, and hepatocellular carcinoma, HCC (1). Liver repair after toxicant exposure depends on age of the animals. Murali et al. have found that old animals initiate a much stronger proliferative response to CCl4 injury (2). These observations have been further confirmed by other groups (3). The mechanisms by which age increases sensitivity of liver to CCl4-mediated injury and the mechanisms responsible for the increased liver proliferation after CCl4 treatment are not known. In this report, we propose mechanisms which involve alterations of chromatin structure and subsequent alterations of key regulators of liver proliferation after CCl4 treatments.
A growing number of recent studies show a critical role for epigenetic changes in the regulation of liver functions (4–6). Epigenetic control is involved in development of alcoholic liver disease. It has been shown that alcohol causes acetylation of histone H3 at K9 in cultured primary hepatocytes (7). This epigenetic change is associated with the increase of the histone acetyl-transferase activity and with the reduction of histone deacetylase (HDAC)2 activity (7). In agreement with these observations, H3K9 acetylation has been observed in the liver after alcohol exposure (8). Meng et al. have recently shown that epigenetic changes in hepatocytes regulate expression of certain micro-RNAs through methylation of CpG islands in their promoters (9). It has been shown that the epigenetic switch of micro-RNA expression is mediated by the liver-specific transcription factor HNF4α and might link liver inflammation and tumorigenesis (10). A number of reports have demonstrated that epigenetic control is also involved in development of liver cancer (11, 12).
Two members of C/EBP family, C/EBPα and C/EBPβ, are expressed at high levels in the liver and are involved in the epigenetic control of liver proliferation and steatosis by interacting with p300 and HDAC1 (13–15). Studies with young and old mice suggested that the phosphorylation of C/EBPα at S193 is the major pathway of regulation of interactions of C/EBPα with HDAC1 (14). In agreement with these observations, generation of C/EBPα-S193D mice (further called S193D mice) showed that young S193D mice have increased amounts of HDAC1-C/EBPα complexes and have developed liver dysfunctions that are typically observed in old mice (6). These dysfunctions include altered chromatin structure; changes in liver morphology, accumulation of glycogen, development of hepatic steatosis, and accumulation of triglycerides in the blood (6, 15). Therefore, the S193D mice represent a powerful animal model for the investigations of the responses of livers of aged mice to drug treatments. Another member of C/EBP family, C/EBPβ, is similar to C/EBPα and is involved in the regulation of chromatin structure of hepatocytes through interactions with HDAC1 (16, 17). It has been shown that the C/EBPβ-HDAC1 complexes are abundant in livers of old mice and repress SIRT1 (18). The formation of C/EBPβ-HDAC1 complexes is mainly controlled by the RNA-binding protein CUGBP1 (14). CUGBP1 increases translation of HDAC1 and two isoforms of C/EBPβ: C/EBPβ-LAP and C/EBPβ-LIP (16, 17, 19). A truncated isoform of C/EBPβ-LIP lacks activation domains and functions as a dominant-negative molecule by inhibiting activities of full-length C/EBPα and C/EBPβ proteins (20, 21). In addition, C/EBPβ-LIP releases the negative control of liver proliferation through interactions with and disruption of E2F1-Rb complexes (21).
In this report, we show that an increase in repressive histone modifications in livers of old WT mice and in livers of young S193D mice changes the response of the liver to CCl4 injury. The main differences between young and old WT mice in the response to CCl4 are elevation of C/EBPβ-LIP in livers of old mice and subsequent repression of three key regulators of liver functions: TERT, C/EBPα, and FXR. These age-related alterations lead to an increase in liver injury and apoptosis and to an increase in liver proliferation after CCl4 treatments. We also found that chronic treatments with CCl4 cause a significantly higher level of fibrosis in S193D mice than in WT mice. This suggests that the molecular alterations observed in S193D mice may contribute to development of drug-mediated liver diseases in old patients.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies to C/EBPα (14AA), C/EBPβ (C-19), cdc2 (L-19), PCNA (FL-261), FXR (H-130), Rb (IF8), E2F1, HP1α, CUGBP1, elF2α, GSK-3β, p-GSK-3β (Tyr 216), CYP2E1, CYP2B1/2, and CYP3A were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies to C/EBPβ (A16) and trimethylated histone H3K9 were from Abcam (Cambridge, UK). Monoclonal antibody against to β-actin was from Sigma-Aldrich. Telomerase activity was measured using quantitative telomerase detection kit (Allied Biotech, Vallejo, CA).
Animal Work and CCl4-induced Liver Injury
Wild-type and S193D mice were maintained as previously described (22). In this report, we used young mice (2–3-month-old) and old mice (20–22-month-old) for all studies. For the simplicity of presentation, young mice are shown on figures as 2-month-old (2m) mice and old mice are shown as 22-month-old (22m).
Acute CCl4 Treatments
Young and old mice (males) were injected intraperitoneally with a single dose of 10% CCl4 in olive oil (5.0 ml/kg) or vehicle control as the zero time point.
Chronic CCl4 Treatments
Mice were treated for 4 weeks with 10% CCl4 in olive oil (5 ml/kg I.P; 3 times/week). BrdU was injected in mice 4 h before they were sacrificed. Liver damage and fibrosis were examined by staining with markers of fibrosis α-SMA and Sirius Red. All data in this report represent summaries of work with 5–6 mice of each age-group per each time point after CCl4 injections. Experiments with animals have been approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine (protocol AN-2503).
Histology
Mice were anesthetized and rapidly exsanguinated through the caudal vana cava. Liver pieces were quickly removed and fixed in 10% neutral buffered formalin, processed routinely and embedded in paraffin wax for hematoxylin and eosin (H&E) stain. BrdU was injected intraperitoneally 2 h before mice were sacrifice, and BrdU staining was performed using BrdU uptake assay kit from Invitrogen (Carlsbad, CA). Hepatocyte apoptosis was detected by using In situ Cell Death Detection (TUNEL) kit from Roche Applied Science (Mannheim, Germany). Sirius Red staining was performed using “Direct Red 80” (Sirius Red, Sigma-Aldrich) kit. α-SMA immunostaining was performed with “Actin, α-Smooth Muscle, Immunohistology kit” (Sigma).
Serum Analysis
Serum ALT and AST were measured at Baylor College of Medicine Facilities.
Protein Isolation and Western Blotting
Nuclear extracts were isolated from liver tissues and used for the Western blotting and for Co-IP as described in our previous publication (21).
Real-Time Quantitative Reverse-Transcriptase PCR
The Rneasy Plus minikit (Qiagen, Germantown, MD) was applied to extract total RNA, according to the manufacturer's protocol. The sequences of PCR primers were as follows: mC/EBPα: 5′-ATGGAGTCGGCCGACTTCTAC-3′ (forward) and 5′-CGTCTCGTGCTCGCAGATGCC-3′ (reverse); mCUGBP1: 5′-TCTGGGAAAACCTGTGGATA-3′ (forward) and 5′-CACAAAAAGCAGCTGGAAAT-3′(reverse); β-actin: 5′-TCTTGGGTATGGAATCCTGTGGCA-3′ (forward) and 5′-ACTCCTGCTTGCTGATCCACATCT-3′ (reverse); primers for mFXR and mTERT were the same as in references (18, 23).
Co-immunoprecipitation
Rb and CUGBP1 were immunoprecipitated from nuclear or cytoplasmic extracts correspondingly with monoclonal antibodies, and the presence of E2F1 and C/EBPβ in Rb IPs and eIF2α in CUGBP1 IPs was examined by Western blotting with polyclonal antibodies to the mentioned proteins.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay was performed as described in our previous articles, using the ChIP-IT Kit (Active Motif) according to the manufacturer's protocol. Briefly, the chromatin solutions were prepared from liver tissues, and DNA was sheared by sonication. C/EBPα, C/EBPβ (C-19 and A16), HDAC1, acetylated histone H3K9, and trimetylated histone H3K9 were immunoprecipitated from the solutions. DNA was isolated and used for PCRs with primers covering the C/EBP binding sites within C/EBPα, FXR and mTERT promoters. The sequences of the primers for the mTERT are as follows: 5′-AGGCGGCAGACCTCCTATAA-3′ (forward) and 5′-AGAGAGGGGCTGCTGAATTT-3′ (reverse). PCR mixtures were amplified, and PCR products were separated by 8% PAGE.
Statistical Analysis
All values are presented as means ± S.D. Statistical analyses were performed using the Student's t test. Statistical significance was assumed when *, p < 0.05.
RESULTS
Young S193D and Old WT Mice Have a Severe Liver Injury after CCl4 Injections
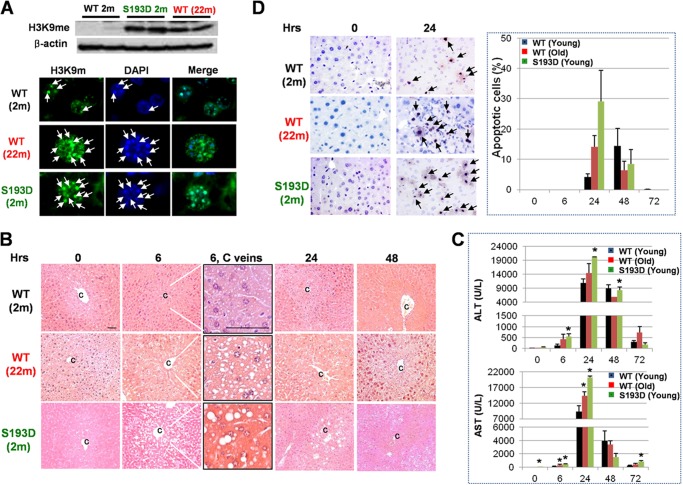
Liver functions decline with age, leading to development of age-associated diseases (11, 12). Previous studies with rats have shown that livers of old rats have an increased liver injury and accelerated liver proliferation after CCl4 treatments (2). Since C/EBPα is a critical regulator of liver functions in old mice, we have generated C/EBPα-S193D mice, which express an aged-like isoform of C/EBPα. These mice have altered chromatin structure and developed several aged-like dysfunctions of the liver at young age (6, 15, 22). Therefore, we examined if young S193D mice and old WT mice have similar responses to CCl4 treatments. In a prior investigation of effects of CCl4 on young S193D mice and old WT mice, we examined chromatin structure and expression of histone H3 trimethylation at K9 as a marker for heterochromatin in our experimental mice. Western blotting showed that amounts of H3K9-trimethyl are dramatically increased in livers of both old WT and young S193D mice (Fig. 1A). Consistent with this increase, immunostaining showed that chromatin structure is changed in both old WT mice and in young S193D mice (Fig. 1A). Therefore, given similarities in the change of chromatin structure, we used the S193D mouse model and old WT mice for the elucidation of mechanisms of age-mediated increase of liver proliferation after CCl4 injections. Mice were treated with CCl4 and investigated by several approaches. H&E staining showed that livers of old WT and young S193D mice have more fatty degenerated hepatocytes around central veins starting from 6 h after CCl4 treatments (Fig. 1B). Levels of ALT and AST are also higher in serum of old WT and young S193D mice than those in young WT mice at 6 and 24 h after CCl4 injections (Fig. 1C). Taken together, these studies showed that livers of old WT and young S193D mice undergo much higher damage after CCl4 treatment than livers of young WT mice.
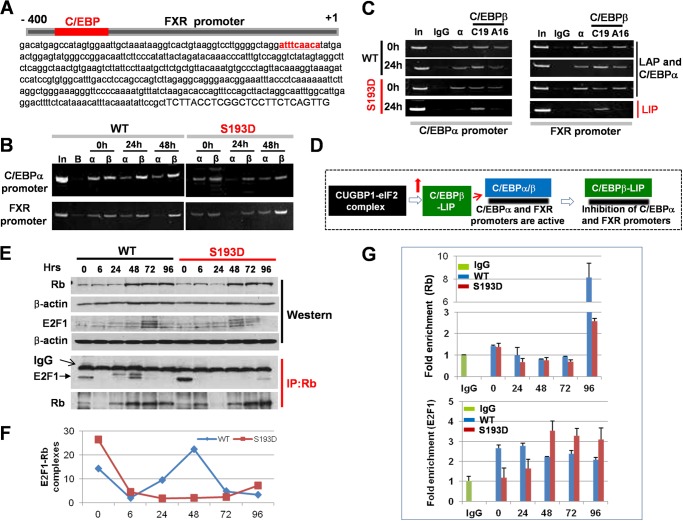
FIGURE 1.
Livers of old WT mice and young S193D mice have severe injury and increased apoptosis after CCl4 treatment. A, heterochromatin structure is abundant in livers of young S193D and old WT mice. Levels of H3K9me in livers of young and old WT and young S193D mice were detected by Western blotting with protein extracts. The membrane was re-probed with Abs to β-actin. Bottom image shows staining of hepatocytes with a marker of heterochromatin, histone H3-trimethylated at K9 (H3K9me), and with DAPI. The pictures were taken under original magnification 100× and enlarged two times. Arrows show H3K9me foci. B, H&E staining of the livers of 2 months (young), 20 months (old) old, and 2 months old (young) S193D mice. c, central vein; 6, C veins: the regions of central veins of 6 h-treated livers are enlarged. C, serum ALT and AST levels. p < 0.05 compared with young WT mice. D, TUNEL staining of the livers of young WT, old WT, and young S193D mice at 9 and 24 h after CCl4 treatments. Arrows indicate TUNEL-positive apoptotic cells. Bar graphs: the number of apoptotic cells was counted in >1,000 cells per each time point. Scale bar, 100 μm.
One of the important responses to CCl4 is the activation of apoptosis to remove the damaged cells. Therefore, we compared the number of apoptotic cells in livers of old WT, young S193D and young WT mice using TUNEL staining. In all three animal groups, TUNEL-positive cells were detected in the area of centrilobular necrosis at 24 h after CCl4 injections. However, the number of apoptotic cells is much higher in young S193D and old WT mice than that in young WT mice at 24 h (Fig. 1D). In livers of young WT mice, the number of TUNEL-positive cells was increased at later time points. Taken together, this study demonstrated that livers of young S193D and old WT mice are more susceptible to CCl4-induced liver injury and that apoptosis is dramatically activated in these mice at 24 h after CCl4 injection.
Liver Proliferation Is Accelerated in Old WT and Young S193D Mice after CCl4 Treatments
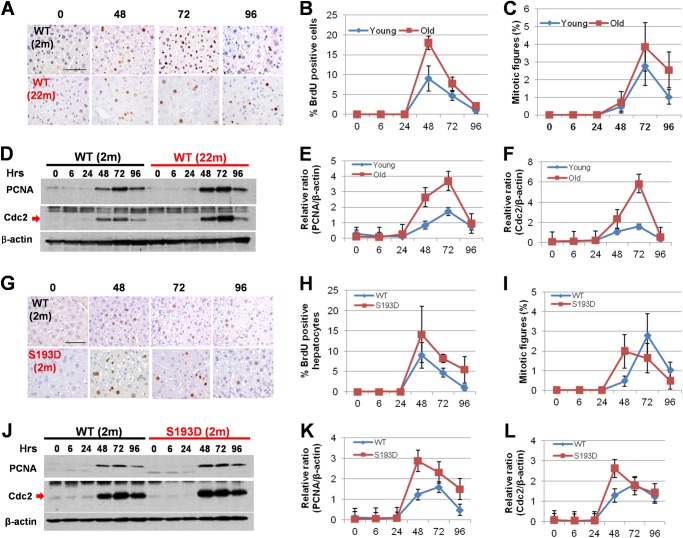
We next compared liver proliferation in young WT, old WT, and young S193D mice after CCl4 treatment. Since proliferative response to partial hepatectomy is reduced in old WT mice (11, 12), and because C/EBPα-S193D inhibits liver proliferation after partial hepatectomy (22), we expected that both old WT mice and young S193D mice might have a reduced proliferation of hepatocytes after CCl4 treatment. However, BrdU uptake showed that the number of proliferating hepatocytes is higher in livers of young S193D and old WT mice compared with that in young WT mice (Fig. 2, A, B, G, H). To confirm this observation, we applied two additional approaches: examination of mitosis and examination of cell cycle proteins. The examination of mitotic figures revealed that the number of hepatocytes with mitotic figures is higher in livers of old WT mice than that in livers of young WT mice (Fig. 2C). We also found that the mitosis occurs early in S193D mice with a broad peak between 48 and 72 h, while mitosis takes place at 72 h in young WT mice (Fig. 2I). Examination of expression of cell cycle proteins, PCNA and cdc2, by Western blotting showed that these proteins are elevated in all three groups of mice; however, the degree of elevation (as a ratio to β-actin) is higher in livers of old WT mice (Fig. 2, D–F) and in livers of young S193D mice (Fig. 2, J–L). Thus, three independent approaches showed that liver proliferation is accelerated in old WT and in young S193D mice treated with CCl4 compared with proliferation of the liver in young WT mice.
FIGURE 2.
Old WT mice and young S193D mice have increased hepatocyte proliferation after CCl4 treatment. A, examination of DNA replication in hepatocytes by BrdU uptake. B, number of BrdU-positive hepatocytes was counted and presented as percentage of total number of examined hepatocytes. C, examination of mitotic figures in young and old mice after CCl4 treatment. The number of mitotic figures was calculated as number per >1,000 hepatocytes. D, expression of PCNA and cdc2 was examined using Western blotting assay. E and F, bar graphs show the levels of these proteins as ratios to β-actin. G, examination of DNA replication in hepatocytes by BrdU uptake. Scale bar, 100 μm. H, number of BrdU-positive hepatocytes was calculated as percentage to the total number of hepatocytes. I, mitotic figures were calculated as number per >1,000 examined hepatocytes. J, expression of PCNA and cdc2 was examined using Western blotting. The filter was re-probed with Abs to β-actin. K and L, levels of PCNA and cdc2 were calculated as ratios to β-actin.
Levels of C/EBPα and FXR Are Reduced, but Levels of C/EBPβ-LIP Are Increased in Livers of Old WT and Young S193D Mice at Early Time Points after CCl4 Treatment
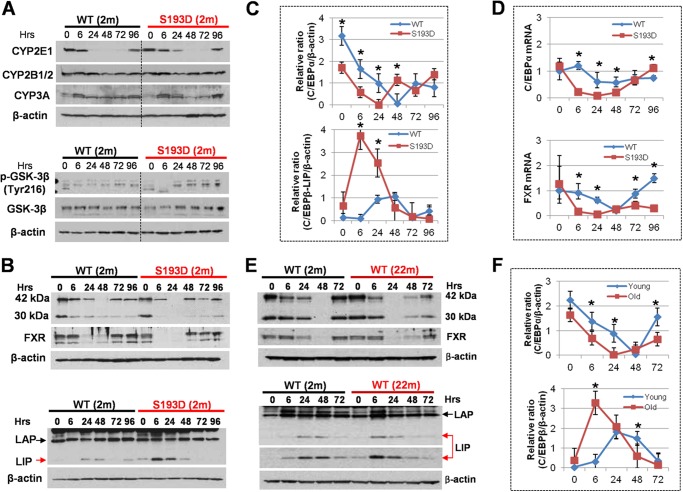
We further investigated several possibilities that could contribute to the molecular basis for the severe damage and for the increased liver proliferation in old WT and young S193D mice. First, we determined the expression of enzymes which convert CCl4 in the liver. We focused our studies on expression of cytochromes CYP2E1, CYP2B1/2, and CYP3A, which are major enzymes that contribute to execute the biotransformation of CCl4 (24–26). We did not observe significant differences for these proteins in the livers of WT and S193D mice (Fig. 3A) suggesting that S193D livers convert CCl4 as well as livers of young WT mice. Examination of the GSK3β pathway also showed no significant differences in the expression of total GSK3β in livers of young WT and S193D mice (Fig. 3A). We also examined expression of the active form, Tyr216-ph-GSK3β. These studies showed that the expression of Tyr216 ph-GSK3β is reduced at early time points in S193D mice, but it is slightly higher at later time points (Fig. 3A). Since these differences are minor, we think that it is unlikely that GSK3β is involved in acceleration of liver proliferation in S193D mice.
FIGURE 3.
Examination of key regulators of liver biology after CCl4 injection. A, upper: expression of isozymes of cytochrome P450 that catalyze CCl4 metabolism. Bottom: examination of GSK3β pathway by Western blotting with Abs to total GSK3β and to Tyr216-ph-GSK3β. B, expression of C/EBPα, FXR, and C/EBPβ in S193D mice after CCl4 treatment. Western blotting was performed with proteins isolated at different time points after CCl4 treatment. Each membrane was re-probed with β-actin. Positions of C/EBPα isoforms (42 kDa and 30 kDa) and C/EBPβ isoforms (LAP and LIP) are shown. C, levels of C/EBPα and C/EBPβ-LIP were calculated as ratios to β-actin. D, expression of C/EBPα and FXR mRNAs after CCl4 treatment was determined by Q-RT-PCR. E, expression of C/EBPα, FXR and C/EBPβ proteins in livers of old WT mice after CCl4 treatment. The experiment was performed as described above. Dark exposure for C/EBPβ-LIP is shown. F, levels of C/EBPα and C/EBPβ-LIP were calculated as ratios to β-actin.
Given our previous observations that both S193D-C/EBPα and ph-S193-C/EBPα inhibit liver proliferation (22), we next examined expression of C/EBPα in livers of young S193D and old WT mice after CCl4 treatments. We found that the levels of C/EBPα are reduced in all three groups; however, the reduction of C/EBPα in old WT and young S193D mice takes place early and to higher degree (Fig. 3, B–D). Previous reports have shown that farnesoid X receptor, FXR, is a critical regulator of liver biology and that the deletion of FXR affects liver repair after CCl4 treatment (27, 28). Therefore, we examined expression of FXR and found that, similar to C/EBPα, levels of FXR are dramatically reduced at 6 and 24 h after CCl4 treatment in livers of both old WT and young S193D mice while the reduction of FXR in livers of young WT mice is moderate and takes place at later time points after CCl4 injections (Fig. 3, B and E). Consistent with the patterns of expression of C/EBPα and FXR proteins, levels of C/EBPα and FXR mRNAs are also more greatly reduced in livers of young S193D and old WT mice (Fig. 3D). It is important to note that the significant differences in expression of C/EBPα and FXR are observed at 6 and 24 h, but do not differ significantly at later time points. Since these early time points determine DNA replication phase, we think that the early reduction of C/EBPα and FXR are involved in the acceleration of liver proliferation in young S193D and old WT mice.
Another member of C/EBP family, C/EBPβ, is expressed in the liver and regulates liver proliferation (29). A single C/EBPβ mRNA produces three isoforms: FL, LAP, and LIP, which have different functions in the liver. While C/EBPβ-LAP inhibits liver proliferation, C/EBPβ-LIP promotes liver proliferation by activation of promoters of cell cycle proteins (21). Therefore, we examined expression of C/EBPβ and found significant differences. The expression of C/EBPβ-LAP is not altered significantly after CCl4 injection in all three groups of mice; however, the levels of C/EBPβ-LIP are increased in livers of young S193D and old WT mice earlier and to a higher degree than in livers of young WT mice (Fig. 3, B and E). Note that the levels of C/EBPβ-LIP are higher in livers of old mice at 0 time point (see dark exposure in Fig. 3E). Densitometric calculations showed that levels of C/EBPβ-LIP are 40-fold and 10-fold higher at 6 h after CCl4 treatment in livers of young S193D and old WT mice, respectively (Fig. 3F). Taken together, these studies revealed that levels of growth inhibitory proteins C/EBPα and FXR are dramatically reduced in livers of old WT and young S193D mice; while levels of C/EBPβ-LIP are elevated in these two groups of mice at early time points after CCl4 injections.
CUGBP1-eIF2 Complex Increases Translation of C/EBPβ-LIP in Livers of S193D Mice at Early Time Points after CCl4 Injections
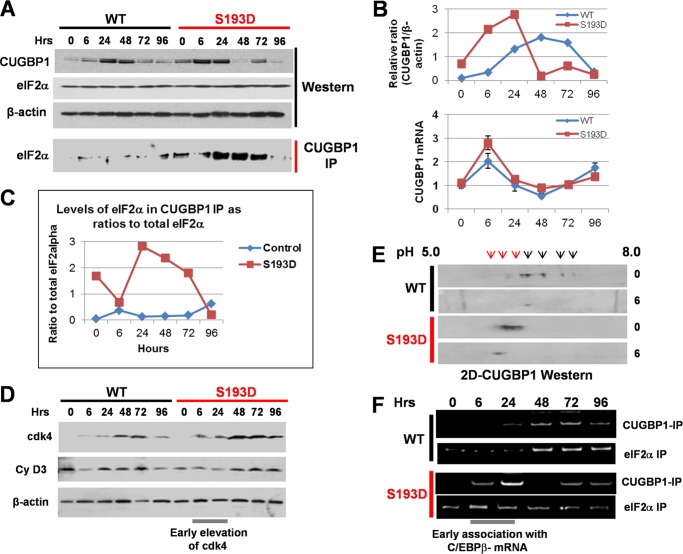
C/EBPβ-LIP is a strong activator of liver proliferation (17, 21). Therefore, we focused our further studies on mechanisms by which CCl4 increases C/EBPβ-LIP in S193D mice and on downstream targets of C/EBPβ-LIP. C/EBPβ-LIP is produced in the liver by the alternative translation from the third AUG codon of a single C/EBPβ mRNA (19, 20). The translation of C/EBPβ-LIP is regulated by a RNA-binding protein, CUGBP1, which interacts with eukaryotic translation initiation factor 2α, eIF2α and delivers C/EBPβ mRNA to polysomes (19). Therefore, we examined if CUGBP1 and the CUGBP1-eIF2 complex might be involved in the increase of translation of C/EBPβ-LIP in livers of S193D mice treated with CCl4. Western blotting revealed that, although CUGBP1 is elevated in both young WT and S193D mice, the elevation of CUGBP1 takes place much earlier in S193D livers and to higher degree than in livers of WT mice (Fig. 4, A and B). Levels of eIF2α are not changed by CCl4 treatment and do not differ in WT and S193D mice. Co-IP showed that the elevation of CUGBP1 in livers of S193D mice leads to a significant increase of the CUGBP1-eIF2 complex while this complex is only slightly increased in livers of WT mice (Fig. 4, A and C). Quantitative-RT-PCR with primers to CUGBP1 mRNA showed that the increase of CUGBP1 mRNA in S193D livers is higher than that in WT mice and correlates with the increase of CUGBP1 protein. On the later time points, however, CUGBP1 mRNA is reduced while protein levels remain at high levels, suggesting post-translational mechanisms of the regulation of CUGBP1 protein. Since the formation of CUGBP1-eIF2 complex also depends on the phosphorylation of CUGBP1 by cyclin D3-cdk4, we examined expression of cdk4 and cyclin D3 as well as the phosphorylation status of CUGBP1. Western blotting showed that levels of cyclin D3 are slightly increased in both WT and S193D livers; however, there is no difference between these two groups of animals. On the contrary, cdk4 is elevated in S193D mice at 6 and 24 h after CCl4 treatment while elevation of cdk4 takes place later in WT mice (Fig. 4D). Two-dimensional examination of CUGBP1 revealed that amounts of hyper-phosphorylated CUGBP1 (located in the acidic region of the two-dimensional gel) are much higher in livers of S193D mice before and after CCl4 injections (Fig. 4E). Interestingly, CUGBP1 is almost completely shifted to acidic region at 6 h after CCl4 treatments. We next examined if CUGBP1-eIF2 complex binds to the C/EBPβ mRNA and if this binding correlates with the translation of C/EBPβ-LIP. For this experiment, CUGBP1-eIF2α-RNA complexes were immunoprecipitated with Abs to CUGBP1 and to eIF2α and then RNA was isolated from these complexes and examined by RT-PCR with primers to C/EBPβ mRNA. Fig. 4F shows results of these studies. We found that amounts of C/EBPβ mRNA in CUGBP1 complexes are significantly increased in livers of S193D mice at 6–72 h after CCl4 injections, but the association of C/EBPβ mRNA with CUGBP1 is increased in WT mice at later time points (Fig. 4F). Examination of C/EBPβ mRNA in eIF2α IPs showed that eIF2α is associated with C/EBPβ mRNA in livers of both WT and S193D mice; however, amounts of C/EBPβ mRNA in eIF2α IPs from livers of S193D mice are higher than those in eIF2α IPs from livers of WT mice especially at early time points after CCl4 injections (Fig. 4F). Taken together, these studies showed that the CUGBP1-eIF2 complexes are elevated in livers of S193D mice and that CCl4 treatment causes further elevation of these complexes and subsequent interactions with C/EBPβ mRNA. Because the CUGBP1-eIF2α complex increases translation of C/EBPβ-LIP, these observations strongly suggest that the higher elevation of C/EBPβ-LIP in livers of young S193D mice is mediated by the CUGBP1-eIF2 complex.
FIGURE 4.
CUGBP1-eIF2 complexes increase translation of C/EBPβ-LIP in S193D mice. A, Western blotting assay shows expression of CUGBP1 and eIF2α in WT and in S193D livers after CCl4 treatment. Bottom part (CUGBP1 IP) shows Co-IP studies. B, levels of CUGBP1 protein were calculated as ratios to β-actin. Bottom image shows expression of CUGBP1 mRNA after treatments with CCl4. C, amounts of CUGBP1-eIF2α complexes were calculated as ratios of eIF2α in CUGBP1 IP to signals of total eIF2α. D, expression of cyclin D3 and cdk4 after CCl4 treatment was examined by Western blotting. E, examination of CUGBP1 phosphorylation by two-dimensional gel electrophoresis-Western blotting. Positions of highly phosphorylated forms of CUGBP1 are shown by red arrows. F, association of CUGBP1 and eIF2α with C/EBPβ-mRNA after CCl4 treatment. CUGBP1 and eIF2α were immunoprecipitated, RNA was isolated and the presence of C/EBPβ mRNA was determined by RT-PCR.
C/EBPβ-LIP Inhibits the C/EBPα and FXR Promoters in Young S193D Mice at Early Time Points after CCl4 Injections
The correlation of elevation of C/EBPβ-LIP and the down-regulation of C/EBPα and FXR suggested that C/EBPβ-LIP might inhibit promoters of these genes. It has been previously shown that the C/EBPα promoter contains a C/EBP site and that C/EBPβ-LIP inhibits the C/EBPα promoter after LPS treatments (20). Given the correlation between elevation of C/EBPβ-LIP and inhibition of FXR transcription, we searched the FXR promoter and found a high affinity C/EBP site in close proximity to the start of transcription (Fig. 5A). EMSA experiments showed that C/EBPα and C/EBPβ-LIP bind to this consensus (data not shown). We next examined if C/EBPβ-LIP binds to the C/EBPα and FXR promoters in livers of young S193D mice after CCl4 injections and if it inhibits these promoters at early time points by replacement of C/EBPα and C/EBPβ-LAP. In initial experiments, we used C19 (Santa Cruz) antibodies to C/EBPβ, which recognize both C/EBPβ-LAP and C/EBPβ-LIP proteins. ChIP assay showed that C/EBPα presence is reduced on the C/EBPα and FXR promoters in livers of WT mice. In livers of S193D mice; however, C/EBPα is not observed on the promoters at 24 h (Fig. 5B). We next performed ChIP with C19 antibodies and with A16 antibodies, which are generated to N terminus of C/EBPβ and recognize only C/EBPβ–LAP. These studies showed that only C/EBPβ-LIP occupies the FXR and C/EBPα promoters in livers of S193D mice at 24 h (Fig. 5C). These results show that C/EBPβ-LIP binds to the FXR and C/EBPα promoters and replaces C/EBPβ-LAP and C/EBPα from these promoters at 24 h after CCl4 treatment. Therefore, we conclude that the elevation of C/EBPβ-LIP in livers of young S193D mice after CCL4 injections leads to the repression of C/EBPα and FXR at early time points (Fig. 5D).
FIGURE 5.
C/EBPα-HDAC1 complexes and C/EBPβ-LIP repress promoters of C/EBPα and FXR; while C/EBPβ-LIP disrupts E2F1-Rb complexes in livers of S193D mice. A, FXR promoter contains a high affinity C/EBP site. The 400 nucleotide sequence of the FXR promoter is shown. Capital letters show the start of transcription. C/EBP consensus is shown in red. B, C/EBPα is not detected on the FXR and C/EBPα promoters in S193D mice at 24 h after CCl4 treatment. ChIP assay was performed with chromatin solutions from WT and S193D mice at 0, 24, and 48 h after CCl4 injection. In; 1/100 of input. C, C/EBPβ-LIP replaces C/EBPα and C/EBPβ-LAP from C/EBPα (left) and FXR (right) promoters at 24 h after CCl4 injection. ChIP assay was performed as described above. D, diagram summarizing results of the studies of the C/EBPα and FXR promoters. E, E2F1-Rb complexes are eliminated in S193D mice after CCl4 treatment. Upper image shows input (Western), bottom image shows Western blotting of Rb IPs with Abs to E2F1. Positions of E2F1 and heavy chains of IgGs are shown by arrows. Bottom panel shows Western of Rb IPs with antibodies to Rb. F, amounts of E2F1-Rb complexes were calculated as a ratio of E2F1 signals to Rb signals. G, semi-quantitative ChIP assay with cdc2 promoter using Abs to Rb (upper) and E2F1 (bottom). Data from three experiments are shown.
C/EBPβ-LIP Disrupts E2F1-Rb Complexes in S193D Mice after CCl4 Treatments
While down-regulation of C/EBPα and FXR eliminates negative control of liver proliferation, the Rb protein also inhibits liver proliferation and needs to be neutralized or eliminated to allow during liver proliferation. Because previous studies have shown that C/EBPβ-LIP disrupts Rb-E2F1 complexes, we suggested that the elevation of C/EBPβ-LIP might utilize this mechanism in S193D mice. To test this hypothesis, we examined E2F1-Rb complexes, which are downstream targets of C/EBPβ-LIP. We immunoprecipitated Rb from nuclear extracts of livers at each time point after CCl4 injection and examined E2F1 in these IPs. Fig. 5E shows that E2F1-Rb complexes are abundant in quiescent livers of both WT and S193D mice. Densitometric calculations showed that E2F1-Rb complex is elevated in WT mice starting at 24 h and with a peak at 48 h (Fig. 5F). The elevation of the E2F1-Rb complexes correlates with the end of proliferation in WT mice, suggesting that E2F1-Rb complexes are involved in the exit of hepatocytes from cell cycle division in WT mice at these time points. In contrast to WT mice, the E2F1-Rb complexes are reduced at a much higher degree in S193D mice and are only weakly elevated at 96 h after CCl4 treatment. Interestingly, the difference in the amounts of E2F1-Rb complexes correlates with the timing and degree of elevation of C/EBPβ-LIP suggesting that C/EBPβ-LIP might disrupt these complexes. To determine if the disruption of the E2F1-Rb complex changes occupation of promoters of cell cycle genes by these complexes, we performed quantitative ChIP assay with the cdc2 promoter. This promoter was selected because expression of this protein is higher in livers of young S193D mice than in WT mice. These studies demonstrated that Rb protein levels are dramatically increased on cdc2 promoter in WT mice at 96 h after CCl4 treatment; however, in S193D mice there is a minor elevation of Rb on cdc2 promoter (Fig. 5G). We also found that the amounts of E2F1 at 48–96 h are much higher on cdc2 promoter in S193D mice than in WT mice. Taken together, these data suggest that C/EBPβ-LIP disrupts E2F1-Rb complexes in S193D mice after CCl4 treatments.
C/EBPα-HDAC1 Complexes and C/EBPβ-LIP Inhibit Telomerase Reverse Transcriptase (TERT) in S193D Mice
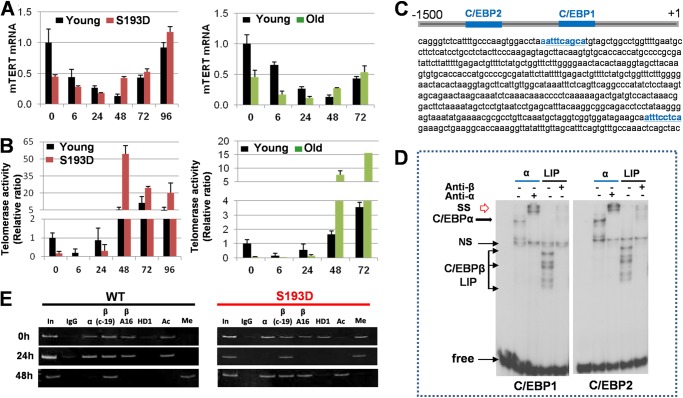
As it is shown in Fig. 1, old WT and young S193D mice have increased apoptosis after CCl4 treatment. It has been shown that telomerase knock-out mice have increased apoptosis and develop liver cirrhosis after CCl4 treatments much faster than WT mice and that the delivery of TERT in these mice inhibits apoptosis (23). Therefore, we suggested that the severe damage and increased apoptosis in S193D mice might be associated with low expression of TERT. We found that, in young WT mice, levels of TERT mRNA are high in quiescent livers and are gradually reduced from 0 to 48 h after CCl4 treatment (Fig. 6A). On the contrary, levels of TERT mRNA are significantly lower in livers of both young S193D and old WT mice before CCl4 injections. The CCl4 further down-regulates TERT to very low levels within the first 24 h. Examination of later time points showed that TERT mRNA is elevated at 48 h in all examined groups. Studies of telomerase activity confirmed that livers of WT mice have much higher levels of TERT activity at early time points after CCl4 injections (Fig. 6B). Thus, these observations showed a dramatic reduction of TERT activity in livers of young S193D and old WT mice within first 24 h after CCl4 treatments, which suggests that these differences might be responsible for the increased apoptosis in old WT and young S193D mice.
FIGURE 6.
The repression of TERT by C/EBPα-HDAC1 and C/EBPβ-LIP in livers of S193D mice. A, levels of TERT mRNA at different time points after CCl4 treatment were determined by quantitative RT-PCR in young S193D mice and in old WT mice. Data shown are mean ± S.D. p < 0.05. B, telomerase activity was examined in young S193D, old WT, and young WT mice after CCl4 injection. p < 0.05. C, nucleotide sequence of the mouse TERT promoter, which contains consensus for C/EBP proteins. The consensuses are shown in blue. D, C/EBPα and C/EBPβ-LIP bind to the TERT promoter. EMSA was performed with DNA probes covering C/EBP sites within the TERT promoter. E, ChIP assay was performed as described above.
We further determined mechanisms of down-regulation of TERT in S193D mice. We searched the promoter of mouse TERT and found two consensuses for C/EBP proteins that are located in close proximity to the start of transcription (Fig. 6C). EMSA analyses with probes covering these sites showed that overexpressed C/EBPα and C/EBPβ-LIP interact with both consensuses (Fig. 6D). We found that C/EBPα and C/EBPβ also interact with these sites in liver nuclear extracts (data not shown). Our previous reports and studies of C/EBPα and FXR promoters in this work showed that, in S193D mice, C/EBPα forms complexes with histone deacetylase 1 (HDAC1) and these complexes repress target promoters (17, 18). Given high affinity sites for C/EBPα and C/EBPβ-LIP within the TERT promoter, we examined the hypothesis that C/EBPα-HDAC1 complexes and C/EBPβ-LIP repress the TERT promoter in livers of S193D mice. The interactions of C/EBPα-HDAC1 complexes and C/EBPβ-LIP with the TERT promoter were examined by ChIP assay. To distinguish C/EBPβ-LIP and C/EBPβ-LAP, we used C19 antibodies (which recognize both proteins) and A16 antibodies (which recognize only LAP, but not LIP) as described above for studies of the C/EBPα and FXR promoters. In livers of young WT mice, C/EBPα and C/EBPβ are bound to the TERT promoter at 0 and 24 h after CCl4 injection; while HDAC1 is not detectable on the promoter at all examined time points (Fig. 6E). A quite different pattern of the interactions is observed in S193D mice. At 0 time point, C/EBPα-HDAC1 complexes occupy and repress the promoter because histone H3 is trimethylated at K9 on the promoter (Fig. 6E). At 24 h after CCl4 injections, C/EBPα, C/EBPβ-LAP (A16 Abs), and HDAC1 are not detectable on the promoter while C/EBP-LIP (C-19 Abs) is bound. Consistent with activation of TERT mRNA in S193D mice at 48 h, C/EBPα and C/EBPβ are bound to the promoter at 48 h after CCl4 injection. These patterns of binding confirm the hypothesis that the TERT promoter is activated in livers of WT mice by C/EBPα and C/EBPβ, but it is repressed in livers of S193D mice by C/EBPα-HDAC1 complexes at the zero time point and by C/EBPβ-LIP at 6–24 h after CCl4 injections.
Correction of TERT Levels in Livers of Old Mice Reduces Liver Proliferation after CCl4 Treatments
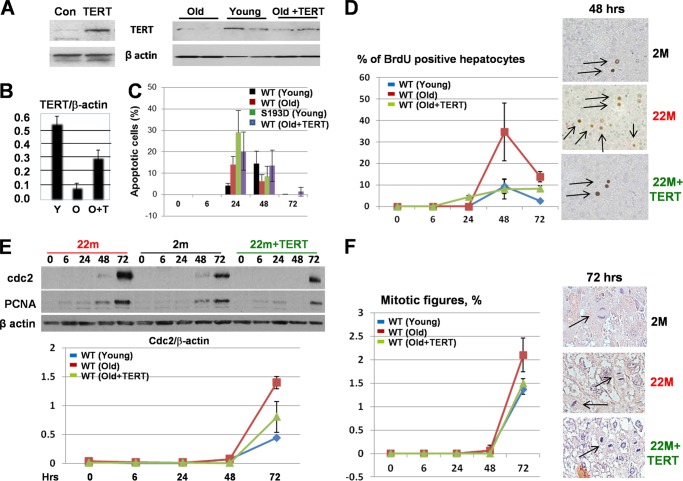
To determine if the reduction of TERT in livers of old mice is involved in severe liver injury and increased liver proliferation, we transfected old WT mice with a plasmid, which expresses mouse TERT using the “jetPei” protocol. This protocol delivers the plasmid mainly to liver and lung and, to a much lesser degree, other tissues (15). Examination of TERT by Western blotting showed that the levels of TERT in transfected old mice are close to the levels observed in livers of young mice (Fig. 7, A and B). However, studies of apoptosis revealed that the rescue of TERT does not reduce apoptosis in old mice treated with CCl4 (Fig. 7C). On the contrary, normalization of TERT levels in livers of old mice reduces DNA replication (Fig. 7D) and expression of cell cycle proteins cdc2 and PCNA after CCl4 treatments (Fig. 7E). The reduction of cdc2 by TERT-dependent normalization is more significant than that of PCNA. We also found that the normalization of TERT in livers of old mice leads to a reduction of mitotic cells after CCl4 injections close to the level observed in young mice (Fig. 7F). These data suggest that the reduction of TERT in livers of old mice is involved in the increase in liver proliferation after CCl4 treatments.
FIGURE 7.
Normalization of TERT levels in livers of old mice is not sufficient to reduce apoptosis; however, it reduces liver proliferation after treatments with CCl4. A, expression of TERT from the generated plasmid in HEK293 cells (left) and levels of TERT in livers of young, old, and old mice injected with TERT. Data with two mice of each group are shown. B, levels of TERT were calculated as ratios to β-actin. C, amounts of apoptotic cells in young WT and S193D mice and in old WT mice after injection of TERT were determined by TUNEL assay at different time points after CCl4 treatments. D, BrdU uptake in livers of WT mice treated with TERT. Bar graphs show summary of three independent experiments. Images on the right show typical pictures of BrdU staining at 48 h after CCl4 injections. Arrows show BrdU-positive hepatocytes. E, normalization of TERT reduces expression of cdc2 and PCNA in livers of old mice. Western blotting was performed with antibodies to cdc2 and PCNA. The filter was re-probed with β-actin. Bottom image: cdc2 levels were calculated as ratios to β-actin. Bar graphs show summary of three repeats. F, mitotic figures were examined in young and old WT mice and in old WT mice after TERT delivery at different time points after CCl4 treatments. Right images show typical pictures of mitotic figures observed at 72 h after CCl4 treatments.
The Aged-like Mutation of C/EBPα Makes Mice More Sensitive to the Development of Drug-associated Liver Fibrosis
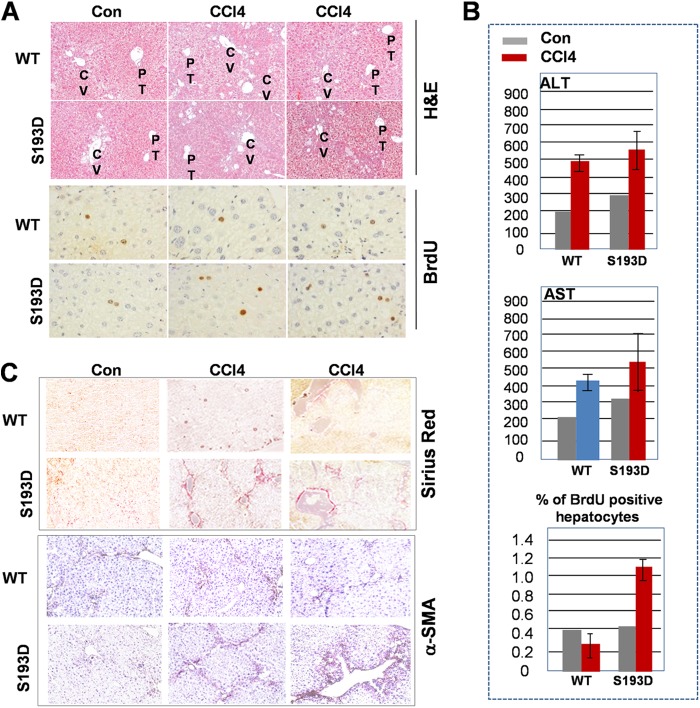
Our data with liver injury were obtained in acute treatments of mice with CCl4. To examine the role these alterations in liver disorders, we investigated development of liver fibrosis after chronic treatments of S193D mice with CCl4. The fibrosis is detectable within 2 weeks after initiation of treatments with CCl4 (30) and we examined if these early steps of development of liver disorders might be different in WT and C/EBPα-S193D mice. For this study, we treated mice for 4 weeks with 10% CCl4 in olive oil: 5 ml/kg I.P 3 times/week. Examination of liver damage by H&E staining and ALT/AST levels in the blood showed slightly higher levels of damage in S193D mice compared with that in WT mice (Fig. 8, A and B); however, these differences are minor. On the contrary, BrdU staining showed that livers of S193D mice have a significantly higher rate of proliferation after chronic treatments with CCl4 (Fig. 8B). We next examined the development of liver fibrosis by immunostaining of livers with a marker of fibrosis, Sirius Red. The Sirius Red staining revealed an increased distribution of collagen in S193D mice (Fig. 8C). Since the activation of hepatic stellate cells (HSC) plays critical role in the process of liver fibrosis, we next performed staining with α-SMA, a marker of activated HSC. In agreement with data obtained by Sirius Red staining, number of α-SMA-positive cells is significantly higher in livers of S193D mice (Fig. 8C). Taken together, these studies showed that S193D mice develop a more severe fibrosis than WT mice under conditions of chronic treatments with CCl4.
FIGURE 8.
Liver proliferation and liver fibrosis are increased in young S193D mice after chronic treatments with CCl4. A, animals were treated by CCl4 for 4 weeks as described under “Experimental Procedures.” Livers of WT and S193D mice were stained with H&E and with antibodies to BrdU. Reproducible pictures are shown. B, levels of ALT and AST in blood and calculations of BrdU-positive hepatocytes are shown in the bar graph pictures. C, livers were stained with α-SMA and Sirius Red.
DISCUSSION
Recovery of the liver after injury is a complex process which includes elimination of damaged cells and regeneration of hepatocytes to replace dead cells. This recovery is highly coordinated on different levels of gene expression. Although the severe liver damage in aged rodents after CCl4 injections has been previously reported, the underlying mechanisms are not known. In this paper, we present mechanisms by which age increases apoptosis and liver proliferation after CCl4 injury. Using old WT mice and an animal model of aged liver, C/EBPα-S193D knockin mice, we found significant alterations of the biochemical pathways in these mice after CCl4 injections. These alterations occur on different levels of the regulation of liver biology including signaling pathways leading to repression of promoters and activation of cell cycle regulators (Fig. 9). One of the critical and early steps of the cascade of alterations in young S193D and old WT mice is the elevation of a dominant negative isoform of C/EBPβ, C/EBPβ-LIP. In addition to elevation of C/EBPβ-LIP, livers of S193D mice contain high levels of C/EBPα-HDAC1 complexes that also repress C/EBP-dependent promoters (6, 18). As the result, livers of old WT and young S193D mice contain high levels of two repressors of the gene expression, C/EBPα-HDAC1 complexes and C/EBPβ-LIP. These repressors are key mediators of accelerated liver proliferation and severe liver injury. We found three critical targets of these repressors: FXR, C/EBPα, and TERT. Promoters of these three genes contain high affinity binding sites for C/EBP proteins. Using ChIP assay, we demonstrated that HDAC1-C/EBPα complexes and C/EBPβ-LIP repress promoters of these genes after CCl4 treatments. Because FXR and C/EBPα are strong inhibitors of liver proliferation, the rapid reduction of these proteins in livers of old WT and young S193D mice after CCl4 injections are likely to be involved in the acceleration of liver proliferation. C/EBPβ-LIP also promotes liver proliferation through additional mechanisms. The main targets of C/EBPβ-LIP inside the cell cycle machinery are the E2F1-Rb complexes. It has been shown that, after LPS treatment, C/EBPβ-LIP directly interacts with Rb and disrupts E2F1-Rb complexes; this leads to the release of promoters of S-phase genes from Rb-dependent repression (21). In agreement with these observations, C/EBPβ-LIP disrupts the E2F1-Rb complexes and activates E2F-dependent promoters in livers of S193D mice after CCl4 treatments.
FIGURE 9.
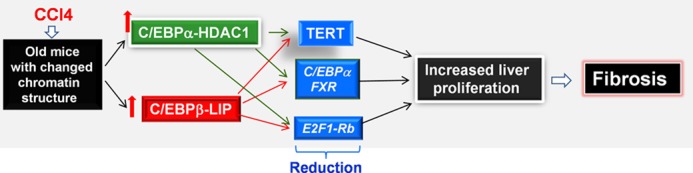
A hypothetical model for the age-associated increase of liver proliferation and early development of fibrosis after chronic treatments with CCl4.
One of the most important findings of our work is the observation that telomerase promoter is repressed in livers of old mice. This leads to very low levels of TERT and to low telomerase activity in both quiescent livers and in the livers within 24 h of CCl4 injections. In quiescent livers of S193D mice, this repression is mediated by C/EBPα-HDAC1 complexes, which are abundant in S193D mice. CCl4-mediated liver injury activates an additional mechanism of TERT repression in the livers of young S193D and old WT mice, which is the elevation a dominant negative C/EBPβ-LIP isoform. We found that C/EBPβ-LIP occupies and represses the TERT promoter after CCl4 treatment. Since telomerase activity is required to protect cells from apoptosis (23), we initially thought that ectopic expression of TERT might reduce liver injury and apoptosis, but we found that normalization of TERT in livers of old mice does not reduce liver injury and apoptosis. This suggests that other unknown mechanisms are involved in severe liver injury in old mice. These mechanisms remain to be elucidated. On the other hand, the normalization of levels of TERT in old mice is sufficient to reduce liver proliferation after CCl4 treatments. We found that DNA replication, mitosis and expression of cell cycle proteins are reduced by TERT in livers of old mice to the levels observed in livers of young WT mice. It is important to note that our data for TERT-mediated inhibition of liver proliferation in livers of old mice after CCl4 treatment do not support previous observations obtained in studies of liver regeneration after partial hepatectomy (PH). It has been shown that TERT activity was increased after PH in pig's livers (31) and that hTERT promoter is activated during liver proliferation in mouse hepatocytes (32). However, our data with CCl4-mediated liver proliferation in livers of old mice showed that the reduction of TERT is involved in the increased liver proliferation. We think that these different results reflect the differential environment mediated by age and the conditions of initiation of liver proliferation. In summary, our studies demonstrate that the age-associated changes of C/EBP family proteins caused severe liver injury, increased apoptosis, and accelerated proliferation of livers as well as the early development of liver fibrosis in old WT mice after chronic CCl4 treatment. The downstream events involve repression of key regulators of liver proliferation C/EBPα, TERT, and FXR (Fig. 9).
Acknowledgment
We thank Kelsey Jarrett for the discussions and editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM551888, AG039885, AG028865, and CA159942 (to N. T.) and R01AR044387 and R01AR052791 (to L. T.).
- HDAC
- histone deacetylase
- FXR
- farnesoid X receptor
- C/EBP
- CCAAT/enhancer-binding proteins
- C/EBPβ-LIP
- C/EBPβ liver inhibitory protein, a truncated isoform
- TERT
- telomerase reverse transcriptase, the catalytic component of telomerase
- CUGBP1
- CUG triplet repeat-binding protein 1
- ChIP
- chromatin immunoprecipitation assay
- CCl4
- carbon tetrachloride
- EMSA
- electrophoretic mobility shift assay.
REFERENCES
- 1. Fujii T., Fuchs B. C., Yamada S., Lauwers G. Y., Kulu Y., Goodwin J. M., Lanuti M., Tanabe K. K., (2010) Mouse model of carbon tetrachloride induced liver fibrosis: histopathological changes and expression of CD133 and epidermal growth factor. BMC Gastroenterology 10, 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murali B., Korrapati M. C., Warbritton A., Latendresse J. R., Mehendale H. M. (2004) Tolerance of aged Fisher 344 rats against chlordecone-amplified carbon tetrachloride toxicity. Mech. Aging Dev. 125, 421–435 [DOI] [PubMed] [Google Scholar]
- 3. López-Diazguerrero N. E., Luna-López A., Gutiérrez-Ruiz M. C., Zentella A., Königsberg M. (2005) Susceptibility of DNA to oxidative stressors in young and aging mice. Life Sci. 77, 2840–2854 [DOI] [PubMed] [Google Scholar]
- 4. Mandrekar P. (2011) Epigenetic regulation of alcoholic liver disease. World J. Gastroenterol. 17, 2456–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwabe R., Wang T. (2011) Targeting liver cancer: first steps toward a miracle. Cancer Cell 20, 698–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin J., Iakova P., Wang G-L., Shi X., Haefliger S., Finegold M., Timchenko N. A. (2010) Epigenetic changes play critical role in age-associated dysfunction of the liver. Aging Cell 9, 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park P. H., Lim R. W., Shukla S. D. (2005) Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: potential mechanism for gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G1124–G1136 [DOI] [PubMed] [Google Scholar]
- 8. Aroor A. R., James T. T., Jackson D. E., Shukla S. D. (2010) Differential changes in MAP kinases, histone modifications, and liver injury in rats acutely treated with ethanol. Alcohol Clin. Exp. Res. 34, 1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meng F., Glaser S. S., Francis H., Yang F., Han Y., Stokes A., Staloch D., McCarra J., Liu J., Venter J., Zhao H., Liu X., Francis T., Swendsen S., Liu C-G., Tsukamoto H., Alpini G. (2012) Epigenetic regulation of miR-34a in alcoholic liver injury. Am. J. Pathol. 181, 804–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cairo S., Buendia M. A. (2012) How transient becomes stable: An epigenetic switch linking liver inflammation and tumorigenesis. J. Hepatol. 57, 910–912 [DOI] [PubMed] [Google Scholar]
- 11. Timchenko N. A. (2011) in Molecular Pathology of the Liver Diseases (Monga S.P., ed), p. 279–290, Springer Science + Business Media, LLC, New York [Google Scholar]
- 12. Timchenko N. A. (2009) Aging and liver regeneration. Trends Endocrinol. Metabol. 20, 171–176 [DOI] [PubMed] [Google Scholar]
- 13. Iakova P., Timchenko L. T., Timchenko N. A. (2011) Intracellular signaling and hepatocellular carcinoma. Seminars in Cancer Biology 21, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones K., Timchenko L., Timchenko N. A. (2012) The role of CUGBP1 in age-dependent changes of liver functions. Ageing Research Reviews 11, 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin J., Iakova P., Breaux M., Sullivan E., Jawanmardi N., Chen D., Jiang Y., Medrano E. M., Timchenko N. A. (2013) Increased expression of enzymes of triglyceride synthesis is essential for the development of hepatic steatosis. Cell Reports 3, 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang G-L., Salisbury E., Shi X., Timchenko L. T., Medrano E. E., Timchenko N. A. (2008) HDAC1 cooperates with C/EBPα in the inhibition of liver proliferation in old mice. J. Biol. Chem. 283, 26169–26178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang G-L., Salisbury E., Shi X., Timchenko L. T., Medrano E. E., Timchenko N. A. (2008) HDAC1 promotes liver proliferation in young mice via interaction with C/EBPβ. J. Biol. Chem. 283, 26179–26187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin J., Iakova P., Jiang Y., Medrano E. E., Timchenko N. A. (2011) The reduction of SIRT1 in livers of old mice leads to impaired body homeostasis and to inhibition of liver proliferation. Hepatology 54, 989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Timchenko N. A., Wang G-L., Timchenko L. T. (2005) CUG triplet repeat binding protein, CUGBP1, increases translation of C/EBPβ isoform, LIP, by interacting with the α and β subunits of eIF2. J. Biol. Chem. 280, 20549–20557 [DOI] [PubMed] [Google Scholar]
- 20. Welm A. L., Mackey S. L., Timchenko L. T., Darlington G. J., Timchenko N. A. (2000) Translational induction of LIP during acute phase response leads to repression of C/EBP alpha mRNA. J. Biol. Chem. 275, 27406–27413 [DOI] [PubMed] [Google Scholar]
- 21. Orellana D., Liu X., Wang G-L., Jin J., Iakova P., Timchenko N. A. (2010) Calmodulin controls liver proliferation via interactions with C/EBPβ-LAP and C/EBPβ-LIP. J. Biol. Chem. 285, 23444–23456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang G-L., Shi X., Haefliger S., Jin J., Major A., Iakova P., Finegold M., Timchenko N. A. (2010) Elimination of C/EBPα through the ubiquitin-proteasome system promotes the development of liver cancer in mice. J. Clin. Invest. 120, 2549–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rudolph K. L., Chang S., Millard M., Schreiber-Agus N., DePinho R. A. (2000) Inhibition of experimental liver cirrhosis in mice by telomerase gene delivery. Science 287, 1253–1258 [DOI] [PubMed] [Google Scholar]
- 24. Zangar R. C., Benson J. M., Burnett V. L., Springer D. L. (2000) Cytochrome P450 is the primary enzyme responsible for low-dose carbon tetrachloride metabolism in human liver microsomes. Chem. Biol. Interact. 125, 233–243 [DOI] [PubMed] [Google Scholar]
- 25. Ibrahim Z. S., Ishizuka M., Soliman M., ElBohi K., Sobhy W., Muzandu K., Elkattawy A. M., Sakamoto K. Q., Fujita S. (2008) Protection by Nigella sativa against carbon tetrachloride-induced downregulation of hepatic cytochrome P450 isozymes in rats. Jpn. J. Vet. Res. 56, 119–128 [PubMed] [Google Scholar]
- 26. Akiyama E., Gonzalez F. J. (2003) Regulation of P450 genes by liver-enriched transcription factors and nuclear receptors. Biochim. Biophys. Acta 17, 223–234 [DOI] [PubMed] [Google Scholar]
- 27. Meng Z., Wang Y., Wang L., Jin W., Liu N., Pan H., Liu L., Wagman L., Forman B. M., Huang W. (2010) FXR regulates liver repair after CCl4-induced toxic injury. Mol. Endocrinol. 24, 886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee C. G., Kim Y. W., Kim E. H., Meng Z., Huang W., Hwang S. J., Kim S. G. (2012) Farnesoid X receptor protects hepatocytes from injury by repressing miR-199a-3p, which increases levels of LKB1. Gastroenterology 142, 1206–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greenbaum L. E., Li W., Cressman D. E., Peng Y., Ciliberto G., Poli V., Taub R. (1998) CCAAT enhancer- binding protein beta is required for normal hepatocyte proliferation in mice after partial hepatectomy. J. Clin. Invest. 102, 996–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Komatsu Y., Waku T., Iwasaki N., Ono W., Yamaguchi C., Yanagisawa J. (2012) Global analysis of DNA methylation in early-stage liver fibrosis. BMC Medical Genomics 5, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wege H., Müller A., Müller L., Petri S., Petersen J., Hillert C. (2007) Regeneration of pig livers by compensatory hyperplasia induces high levels of telomerase activity. Comp. Hepatology 6, 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sirma H., Kumar M., Meena J. K., Witt B. B., Weise J. M., Lechel A., Ande S., Sakk V., Guguen-Guillouzo C., Zender L., Rudolph K. L., Günes C. (2011) The promoter of human telomerase transcriptase is activated during liver regeneration and hepatocyte proliferation. Gastroenterology 141, 326–337 [DOI] [PubMed] [Google Scholar]