Background: BAF (BANF1) is a highly conserved essential protein that binds nuclear lamina proteins and DNA.
Results: Perturbing BAF phosphorylation results in nuclear envelope defects and impaired cell cycle progression.
Conclusion: Correct regulation of BAF phosphorylation is essential for its cellular function.
Significance: The phenotype resulting from perturbing BAF phosphorylation is strikingly similar to that of a progeroid syndrome resulting from a point mutation in BAF.
Keywords: Cell Cycle, Chromosomes, Mitosis, Nuclear Membrane, Phosphorylation, BAF, BAF BANF1, BAF BANF1 PP4C
Abstract
Barrier-to-autointegration factor (BAF or BANF1) is highly conserved in multicellular eukaryotes and was first identified for its role in retroviral DNA integration. Homozygous BAF mutants are lethal and depletion of BAF results in defects in chromatin segregation during mitosis and subsequent nuclear envelope assembly. BAF exists both in phosphorylated and unphosphorylated forms with phosphorylation sites Thr-2, Thr-3, and Ser-4, near the N terminus. Vaccinia-related kinase 1 is the major kinase responsible for phosphorylation of BAF. We have identified the major phosphatase responsible for dephosphorylation of Ser-4 to be protein phosphatase 4 catalytic subunit. By examining the cellular distribution of phosphorylated BAF (pBAF) and total BAF (tBAF) through the cell cycle, we found that pBAF is associated with the core region of telophase chromosomes. Depletion of BAF or perturbing its phosphorylation state results not only in nuclear envelope defects, including mislocalization of LEM domain proteins and extensive invaginations into the nuclear interior, but also impaired cell cycle progression. This phenotype is strikingly similar to that seen in cells from patients with progeroid syndrome resulting from a point mutation in BAF.
Introduction
Barrier-to-autointegration factor (BAF or BANF1)3 is a highly conserved protein in multicellular eukaryotes. It was first identified as a factor that protects Moloney murine leukemia virus from integrating into its own genome (1), a self-destructive pathway termed autointegration. BAF is a dimer (2, 3), and each monomer within the dimer binds double-stranded DNA non-specifically (4, 5). DNA bridging by BAF compacts the viral DNA making it unavailable as a target for integration (6). BAF also binds the LEM domain of the inner nuclear membrane protein family, which includes lamina-associated-polypeptide 2 (Lap2), emerin, and MAN1 (7–10). Direct interactions between BAF and the LEM domain has been confirmed by NMR (11).
BAF is an essential protein for the host cell. Homozygous BAF mutants are lethal in Drosophila at the larval-pupal transition (12) and exhibit abnormal nuclear organization, including clumping of chromatin and aberrant morphology. Knockdown of BAF by RNAi results in a defect in chromatin segregation during mitosis (4, 12, 13). Studies with a temperature-sensitive BAF mutant in Caenorhabditis elegans indicate that the nuclear envelope (NE) abnormality is independent of the chromatin segregation defect (14). Live cell images show that BAF assembles first at the core region of telophase chromosome and forms an immobile complex by directly binding with other core localizing NE proteins (15). BAF may provide a structural foothold to selectively recruit other NE proteins and allow assembly of the NE. BAF exists in both phosphorylated and unphosphorylated states. The major site of phosphorylation is Ser-4, with a minor population phosphorylated at Thr-2 and/or Thr-3 (16, 17). Phosphorylation is mediated by vaccinia-related kinase 1 (VRK1). Depletion of VRK1 results in mitotic defects, including impaired NE formation and BAF delocalization (14). Phosphorylation of BAF not only abrogates its DNA binding activity by introducing negative charge on the DNA interacting surface (6, 16, 17) but reduces the binding to LEM domain proteins and leads to mislocalization from the newly formed NE (5). It has recently been reported that LEM4 is required for the dephosphorylation of BAF by inhibiting activity of VRK1 (18).
We identified protein phosphatase 4 catalytic subunit (PP4C) as the major phosphatase responsible for dephosphorylation of the major phosphorylation site Ser-4 in BAF. By examining the cellular distribution of phosphorylated BAF (pBAF) and total BAF (tBAF) through the cell cycle, we find that BAF localizes to the core region of telophase chromosomes during mitosis in the phosphorylated form. Perturbing BAF phosphorylation has profound effects on the cellular localization of BAF and NE proteins and impairs cell cycle progression.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
BANF1 (M01) monoclonal antibody (Abnova) was used as tBAF antibody to detect tBAF at 5 μg/ml for Western blotting and 10 μg/ml for immunofluorescence. pBAF antibody was used at 1 μg/ml for Western blotting and 20 μg/ml for immunofluorescence. Other antibodies and their dilutions used in Western blotting were as follows: tubulin pAb (Abcam) 1/10,000 dilution; cyclin B1 mAb (GNS1) (Santa Cruz Biotechnology) 1/200; cyclin E mAb (Santa Cruz Biotechnology) 1/100; PP2A (G-4) mAb (Santa Cruz Bio.) 1/200; PP4C (PPX C-18) pAb (Santa Cruz Biotechnology) 1/500; R1 pAb (Bethyl Laboratories) 1/200; R2 pAb (Bethyl Laboratories) 1/500; emerin pAb (Abcam) 1/1000; and VRK1 pAb (Abcam) 1/500. Antibodies and their dilutions used in immunostaining were as follows: emerin mAb (Abcam) 1/50; lamin B1 pAb (Abcam) 1/500. Okadaic acid, calyculin A and propidium iodide (PI) were purchased from Sigma. All siRNAs were ordered from Thermo Dharmacon, including BAF siRNA pool (catalog no. L-011536-02-0005), VRK1 siRNA pool (catalog no. L-004683-00-0005), PP4C siRNA pool (catalog no. L-008486-00-0005), and PP4C individual siRNA set (catalog no. LQ-008486-00-0002).
Cell Culture and Transient Transfection
HEK293 and HeLa cells were obtained from ATCC, Inc. and maintained in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Plasmid DNA or siRNA were transiently introduced into cells by Lipofectamine LTX (Invitrogen) or Lipofectamine RNAiMAX (Invitrogen). Cells were harvested 48 h after transfection.
pBAF Antibody
pBAF polyclonal antibody was raised against the peptide N-MTTpSQKHRDFVAEPM by ProSci, Inc. (Poway, CA). Antibody that recognized phosphorylated BAF was separated from total antibody by a two-step immunoaffinity purification. Serum was first loaded on a column with immobilized non-phosphopeptide. The flow through was then loaded onto another column with phosphopeptide and eluted.
Western Blotting and Immunodepletion
Cells were lysed in TBS (pH 7.4) buffer containing 0.1% Triton X-100 with freshly added protease inhibitor and/or phosphatase inhibitor (Sigma). Cell lysates were separated on a 4–12% polyacrylamide gel and electrotransferred to PVDF membrane (Invitrogen). After blocking in 5% nonfat milk solution for 1 h, the membrane was incubated with the indicated primary antibody and secondary antibodies (KPL Laboratories). Signals were detected using a chemiluminescence reagent (Thermo) and exposed with x-ray film (Kodak).
Immunodepletion was carried out by incubating lysates with 2–3 μg of PP4C antibody and protein A-Sepharose CL-4B beads (GE Healthcare) for 4 h at 4 °C. The beads were pelleted by briefly centrifuging and discarded. The supernatant was kept for further assay.
Dephosphorylation of pBAF
pBAF was generated by incubating purified BAF protein from Escherichia coli with VRK1 protein for 1 h at 37 °C in 20 mm Tris-HCl buffer (pH 7.5), supplemented with 1 mm DTT, 1 mm ATP. Dephosphorylation reactions were performed by incubating pBAF with test samples at 30 °C for 0.5 h in 20 mm Tris-HCl buffer (pH 7.4), supplemented with 50 mm NaCl, 0.2 mm EDTA, and 0.2% 2-mercaptoethanol. Reaction mixtures were resolved on 4–12% SDS-PAGE gels, and the pBAF level was determined by Western blotting.
Co-immunoprecipitation of BAF WT and Mutants
A BAF-GFP expression construct in pCMV6-AC-GFP vector from Origene Technologies was used as the template for mutagenesis. Ser-4 was mutated to Asp or Ala to generate mutant BAF-S4D and BAF-S4A with a QuikChange site-directed mutagenesis kit (Stratagene) as described in the manual. Sequences were verified by ACGT, Inc. The resulting constructs were transfected into HEK 293 cells and lysed in 1×TBS buffer, supplemented with 1% Triton X-100 and protease inhibitor mixture (Roche Applied Science) and phosphatase inhibitor (Sigma). BAF-overexpressed cell lysates were incubated with GFP-Sepharose (Abcam) for 4 h at 4 °C. The Sepharose was washed with TBS buffer supplemented with 0.1% Triton X-100 three times and boiled in SDS loading buffer.
Immunopurification of PP4C
PP4C, PP4C-H56Q, and PP4C-R85A constructs with an N-terminal FLAG tag were gifts from Dr. Daniel Durocher (University of Toronto) (19). PP4C-overexpressed cell lysates were incubated with FLAG-M2 magnetic beads (Sigma) for 4 h at 4 °C. After three washes with TBS buffer supplemented with 0.1% Triton X-100, PP4C was eluted with 3×FLAG peptides (Sigma).
Immunostaining
HEK293 cells were grown on poly-d-lysine-coated glass coverslips (BD Biosciences) and stained as described (20). Briefly, cells were fixed with 4% formaldehyde in PBS for 20 min at room temperature, permeabilized by incubation in 50 mm NH4Cl/PBS and 1% Triton X-100/0.1% SDS for 5 min each and blocked in 0.2% gelatin/PBS for 30 min. Primary and secondary fluorescent antibodies were applied in gelatin/PBS for 1 h each at room temperature. Samples were embedded in Prolong Gold anti-fade mounting reagent with DAPI (Invitrogen) and viewed with a Zeiss confocal microscope. Signal densities were measured with ImageJ software and analyzed for significant differences by t test.
Cell Synchronization
HeLa cells from ATCC, Inc. were synchronized to M phase by treatment with 0.5 μm nocodazole or early S phase by double treatments with 2 mm thymidine. The cells were released by changing to fresh medium.
Flow Cytometry
HEK293 cells were fixed with 70% ethanol and stained with PI to analyze cell cycle stages. Cells were incubated with 10 mm 5-ethynyl-2′-deoxyuridine 2 h before harvesting to label the S phase cell population and stained using a Click-iT 5-ethynyl-2′-deoxyuridine flow cytometry assay kit (Invitrogen). Cell cycle stages were analyzed on a MoFlo Astrios cell sorter (Beckman Coulter). Kaluza software (version 1.2; Beckman Coulter) was used to plot and quantitate the cell cycle data.
RESULTS
BAF Is Dephosphorylated by Phosphatase PP4C
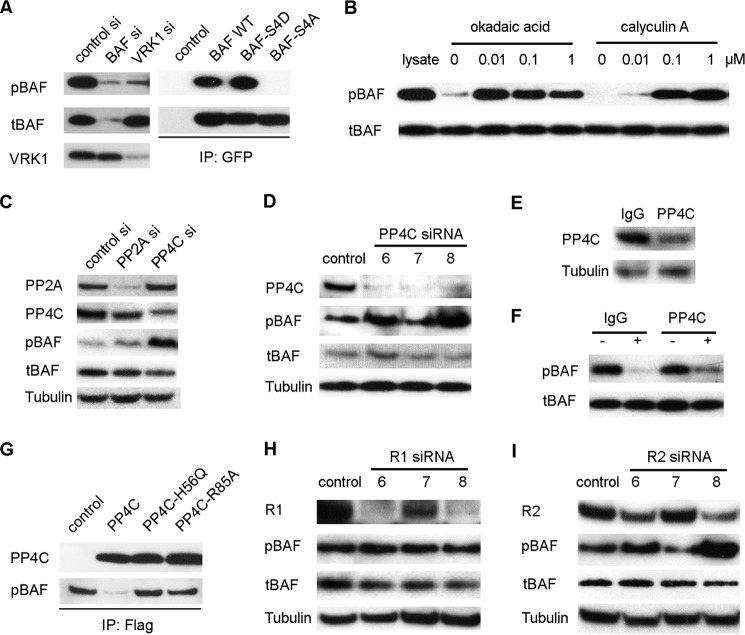
Commercial monoclonal BAF antibody M01 (Abnova) was used to detect tBAF, which includes BAF in both the phosphorylated and unphosphorylated forms. The antibody for pBAF was raised against residues 1–15 of BAF phosphorylated at Ser 4. To check the specificity of the pBAF antibody, lysates of cells transfected with BAF siRNA or VRK1 siRNA were immunoblotted with both of pBAF and tBAF antibodies. As a consequence of BAF knockdown, both pBAF and tBAF significantly decreased in BAF siRNA-transfected samples. VRK1 knockdown did not affect the level of tBAF but dramatically reduced pBAF (Fig. 1A, left panel). In addition, the specificity of the pBAF antibody was further verified with phospho-mimic mutants. GFP-tagged BAF WT construct was used as the template to generate the BAF constitutive phosphorylated mutant BAF-S4D and constitutive unphosphorylated mutant BAF-S4A by replacing Ser-4 with Asp or Ala as reported by Nichols et al. (17) (named BAF MTTDQ and BAFMTTAQ). To further confirm the specificity of the pBAF antibody, BAF WT or mutants were transiently transfected into HEK293 cells separately and immunoprecipitated with GFP antibody. tBAF was detected in all the immunoprecipitates, but no pBAF was found in the BAF-S4A immunoprecipates (Fig. 1A, right panel), confirming that pBAF antibody selectively detects BAF phosphorylated at Ser-4.
FIGURE 1.
pBAF is dephosphorylated by the PP4C complex. A, specificity of the pBAF antibody. Left panel, HEK293 cells were transfected with BAF siRNA or VRK1 siRNA, and the resulting lysates were separated on a 4–12% SDS-PAGE gel. The gel was transferred to PDVF membrane and blotted with pBAF antibody, tBAF antibody, or VRK1 antibody. Right panel, empty vector, BAF WT, BAF-S4D, or BAF-S4A was overexpressed in HEK293 cells. The resulting lysates were immunoprecipitated by GFP antibody and immunoblotted with both of pBAF or tBAF antibody. B, whole HEK293 cell lysates were incubated with increasing amounts of the indicated phosphatase inhibitor at 30 °C for 0.5 h. The lysates were then immunoblotted with pBAF antibody to check the effect of the phosphatase inhibitor on pBAF dephosphorylation by endogenous phosphatases present in the extract, with tBAF antibody as the control. Lane 1 shows the lysate control without incubation. C, HEK293 cells were transfected with PP2A siRNA pool or PP4C siRNA pool. The resulting cell lysates were blotted with the indicated antibodies. D, individual PP4C siRNA 6, 7, and 8 were transfected into HEK293 cells, and the effect on BAF phosphorylation was evaluated by immunoblotting. E, whole HEK293 cell lysate was immunodepleted with PP4C antibody and IgG (used as control). The level of remaining PP4C was evaluated by Western blot. F, immunodepleted cell lysate was incubated at 30 °C for 0.5 h, and cell lysate depleted with IgG was used as control. pBAF and tBAF were detected by immunoblotting. G, PP4C wild type and its inactive mutants PP4C-H56Q and PP4C-R85A were transfected into HEK293 cells separately, immunopurified by FLAG beads, and eluted with 3× FLAG peptides. Eluted PP4C or mutants was incubated with pBAF for 30 min and immunoblotted with PP4C or pBAF antibody. H and I, HEK 293 cells were transfected with individual R1 siRNA 6, 7, and 8 (H) or R2 siRNA 6, 7, and 8 (I). The cell lysates were immunoblotted with the indicated antibody.
Reversible phosphorylation of proteins is an important regulatory mechanism that occurs in both prokaryotic and eukaryotic organisms (21, 22). VRK1 is the major kinase responsible for phosphorylation of BAF, but the identity of the phosphatase responsible for dephosphorylation is not clear. We therefore performed in vitro phosphatase assays to assess the ability of known phosphatase families to dephosphorylate BAF. Phosphatase assays were performed in the presence of the Ser/Thr protein phosphatase inhibitors okadaic acid or calyculin A (Fig. 1B). Okadaic acid and calyculin A are selective for the two major families of mammalian phosphatases, PP1 and PP2A. Okadaic acid has higher inhibitory specificity for PP2A, and the closely related PP4 (IC50 = 0.1 nm) than PP1 (IC50 = 50 nm), whereas calyculin A shows a slightly lower IC50 for PP1 (0.25 nm) than for PP2A and PP4 (0.4 nm) (23). After 30 min of incubation, pBAF in the cell lysate without inhibitor was completely dephosphorylated by endogeneous phosphatase. However, the dephosphorylation of pBAF was totally blocked by addition of 0.01 μm okadaic acid, whereas tBAF remained constant. Calyculin A showed a weaker effect compared with okadaic acid and partly inhibited BAF dephosphorylation at 0.01 μm (Fig. 1B, right panel). The fact that okadaic acid had a stronger effect than calyculin suggested that the PP2A family composed of PP2A and PP4, but not the PP1 family, was responsible for BAF dephosphorylation.
To investigate a possible role of PP2A or PP4 in regulating BAF dephosphorylation, small interfering RNAs were employed to specifically down-regulate the catalytic subunits of PP2A or PP4C expression. HEK 293 cells were harvested 48 h after transient transfection with an siRNA pool targeting PP2A (gene ID 5515) or PP4C (gene ID 5531). As shown in Fig. 1C, both PP2A and PP4C were significantly reduced by their own specific siRNA but not by the other siRNA. A remarkable increase in pBAF was detected in the cells with PP4C knockdown but not in the cells with PP2A knockdown. In both cases, tBAF was unchanged (Fig. 1C). Individual PP4C siRNAs 6, 7, and 8 were used to further test the effect of the PP4C knockdown on pBAF abundance. PP4C was efficiently depleted by all of the three siRNAs and siRNA 6 and 8 led to an increase in pBAF (Fig. 1D).
The role of PP4C in BAF dephosphorylation was further confirmed by in vitro immunodepletion. About 70% of PP4C was depleted from the whole cell lysate by incubation with PP4C antibody, compared with lysate treated with IgG (Fig. 1E). The ability to dephosphorylate BAF was reduced in the lysate depleted of PP4C (Fig. 1F). PP4C or the inactive mutants PP4C-H56Q and PP4C-R85A (19) were immunopurified from overexpressing HEK293 cells by FLAG antibody and incubated with pBAF for 30 min. pBAF can be dephosphorylated by PP4C but not the inactive mutants PP4C-H56Q and PP4C-R85A (Fig. 1G). The results confirmed that PP4C is responsible for the dephosphorylation of BAF at Ser-4.
R2 Is a Component of PP4C Complex for BAF Dephosphorylation
Unlike kinases, Ser/Thr phosphatases are multimeric enzymes that are comprised of a small number of catalytic subunits and a large number of variable regulatory subunits (24). PP4 complexes comprise the catalytic subunit with a core regulatory subunit (R1 or R2) and a third variable regulatory subunit (25). In an attempt to identify the core regulatory subunit for BAF dephosphorylation, individual siRNAs against either R1 or R2 were used. Nearly 90% of R1 was depleted by the two R1 specific siRNAs 6 and 8, and 50% by siRNA 7. However, none of these siRNAs had any effect on the level of pBAF (Fig. 1H). R2-specific siRNA 6 and 8 reduced R2 to <50%, whereas siRNA 7 had no knockdown effect. The pBAF level was significantly increased in the cell lysates transfected with R2 siRNA 6 and 8, but not in the sample with siRNA 7 (Fig. 1I). Therefore, R2 may serve as the core regulatory subunit of PP4C complex that is responsible for BAF dephosphorylation. We also tried to isolate the third regulatory subunit in BAF dephosphorylation complex by fractionation of whole lysates, but the results were inconclusive (data not shown).
Phosphorylated and Unphosphorylated BAF Show Distinct Localization during Mitosis
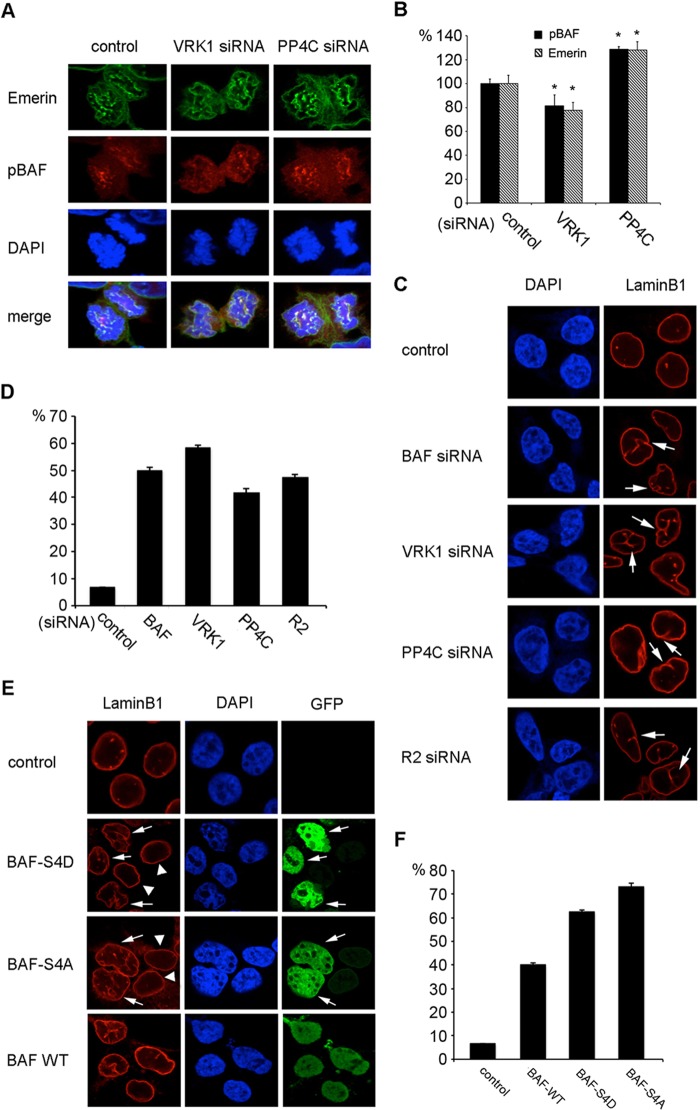
The localization of pBAF and tBAF during interphase and cell mitosis was studied by direct immunostaining. In interphase, pBAF and tBAF were present in both the cytoplasm and nucleus, although pBAF was more abundant in the nucleus with a punctate signal (Fig. 2A). However, pBAF and tBAF exhibited very different localization during mitosis.
FIGURE 2.
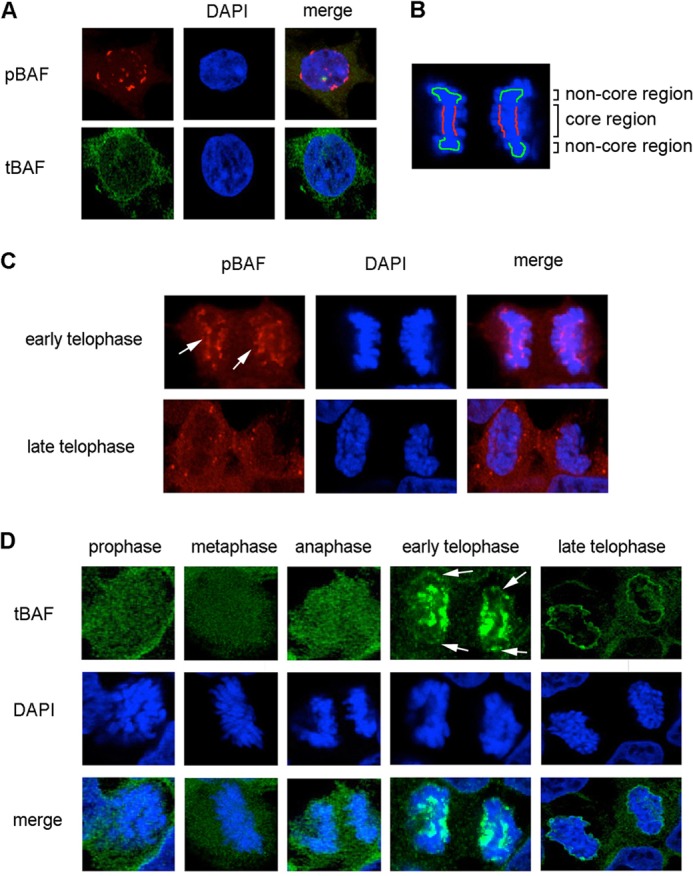
Localization of pBAF and tBAF through cell mitosis. A, HEK293 cells at interphase were immunostained with pBAF and tBAF antibodies. DAPI was used to stain the nucleus. B, schematic diagram of the core region and non-core region of early telophase chromosomes. The core region is indicated in red, and the non-core region is colored green. C, HEK293 cells at telophase were fixed and stained with the pBAF antibody and DAPI (DNA). Arrows indicate the signal at the core region. D, HEK293 cells during mitosis were stained with tBAF and DAPI. Arrows indicate the signal at the non-core region. Images were taken with a confocal microscope.
During early telophase, pBAF consistently accumulated at the central region of telophase chromosomes, which is designated as the core region (15) but not at non-core regions (Fig. 2B, arrows in C). In late telophase, most of pBAF dispersed to the cytoplasm with some punctate signals (Fig. 2C). To further confirm the localization pattern of pBAF at telophase, VRK1 siRNA or PP4C/R2 siRNA were used to modulate the phosphorylation of BAF. The pBAF signal at the core region was reduced to 60% by VRK1 siRNA transfection, but was increased by PP4C/R2 siRNA (Fig. 3, A and C). We conclude that the core-localized BAF at early telophase is in the phosphorylated form.
FIGURE 3.
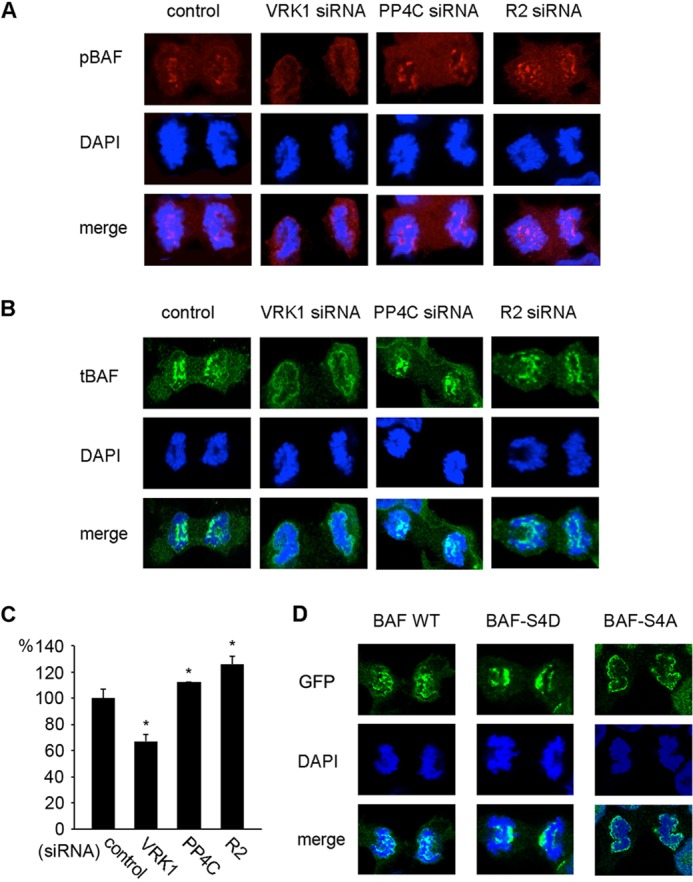
Effects of phosphorylation on pBAF and tBAF localization during telophase. A and B, HEK293 cells transfected with VRK1, PP4C, or R2 siRNA were immunostained with pBAF (A) or tBAF (B) antibody and DAPI at early telophase. C, fluorescence intensity of pBAF at the core region (B) was measured and graphed. Fluorescence intensity was normalized to the signals of the control (>20 separate cells were measured) and analyzed by t test (*, p < 0.05 versus control). D, GFP signals in phospho-mimic mutants overexpressing cells at early telophase were observed by confocal microscopy.
In early mitosis before telophase, tBAF was equally distributed throughout the cytoplasm and nucleus (Fig. 2D). At early telophase, when sister chromosomes separate to each pole of the cell, tBAF was strongly associated with the core region of telophase chromosomes and to a lesser extent with the non-core region at the ends of chromosomes (Fig. 2, B and D). When chromosomes decondensed in late telophase, tBAF translocated to the periphery of chromosomes (Fig. 2D). In the cells depleted of pBAF by VRK1 siRNA, tBAF mainly in the unphosphorylated form, located to the periphery of chromatin throughout telophase (Fig. 3B). The results show that unphosphorylated BAF is mainly localized at the periphery of chromosomes through telophase, where the new NE is formed. The localization of pBAF and unphosphorylated BAF was recapitulated with the BAF phospho-mimic mutants with a GFP tag. BAF-S4D strongly associated with the core region of telophase chromosomes, whereas BAF-S4A localized at the periphery of chromosomes (Fig. 3D).
Phosphorylation of BAF Mediates NE Formation
The best characterized binding partners of BAF are LEM domain proteins, which are integral to the inner NE. It has been reported that emerin localizes to the core region of chromosomes in a BAF-dependent manner (15). In an attempt to evaluate the role of BAF phosphorylation in the core localization of emerin, the localization pattern of emerin at telophase was studied in cells transfected with VRK1 siRNA or PP4C siRNA (Fig. 4A). Signal intensity quantitation of the results showed that the core region localization of emerin at early telophase is disrupted by knockdown of pBAF by VRK1 siRNA and enhanced by knockdown of unphosphorylated BAF by PP4C siRNA (Fig. 4B). No significant change in emerin at the periphery of chromosome at late telophase was observed in cells depleted of VRK1 or PP4C (data not shown). We conclude that BAF phosphorylation controls the recruitment of emerin to the core region of telophase chromosomes, which is essential for new NE assembly.
FIGURE 4.
BAF phosphorylation affects the core localization of emerin and NE formation. A and B, HEK293 cells transfected with VRK1 siRNA or PP4C siRNA were fixed and co-stained with pBAF antibody, emerin antibody, and DAPI. Cells at early telophase were photographed (A). Fluorescence intensities of pBAF and emerin at the core region were measured and normalized to % control (B) (>20 separate cells were measured; *, p < 0.05 versus control). C and D, HEK293 cells transfected with the indicated siRNAs were fixed and stained with lamin B1 antibody and DAPI (C). NE invaginations are indicated by arrows. The percentage of cells with NE invaginations was graphed (D). E and F, HEK293 cells transfected with BAF-S4D, BAF-S4A, or BAF WT GFP fusion proteins were fixed and stained with lamin B1 antibody and DAPI (E). Arrows indicate transfected cells, and arrowheads indicate non-transfected cells. The percentage of cells with NE invaginations was graphed (F).
To further understand the effect of BAF phosphorylation on NE formation, nuclear shape was observed by DAPI staining, and the NE was visualized by labeling lamin B1. About 50% of cells transfected with BAF siRNA showed an irregular shape and wrinkled NE with many invaginations into the nucleus compared with the round appearance of control nuclei (Fig. 4, C and D). This result supports a role for BAF in NE assembly. Knockdown of VRK1 or PP4C/R2 led to the same abnormal nuclear defects as BAF depletion, and 40–60% cells displayed NE invaginations (Fig. 4, C and D). The normal phosphorylation of BAF appears to be indispensible for normal nuclear morphology.
Because PP4 has many substrates involved in organelle assembly (26), the effect of PP4C depletion could be mediated through proteins other than BAF. We therefore overexpressed BAF-S4D and BAF-S4A to investigate their effects on nuclear morphology. Overexpression of either BAF-S4D or BAF-S4A led to an increased frequency of NE invaginations and more nuclear shape distortions (Fig. 4, E and F), whereas untransfected cells showed normal NE (arrowheads in Fig. 4E). It is interesting to note that similar nuclear abnormalities have been reported in a progeroid syndrome that results from a homozygous mutation in BAF (27).
BAF Phosphorylation Is Critical to Cell Cycle Progression
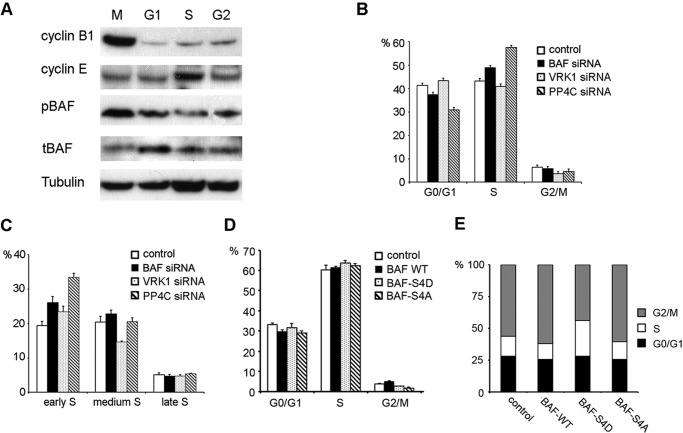
The cell cycle dependence of BAF phosphorylation was studied by Western blot. Cells were synchronized by nocodazole or double blocked by thymidine and released to corresponding cell cycle stages. The Western blot results showed that pBAF was more abundant at M phase (Fig. 5A).
FIGURE 5.
BAF phosphorylation perturbs normal cell cycle progression. A, synchronized HeLa cells at different cell cycle stages (M, G1, S, and G2 stage) were lysed and analyzed by immunoblotting with the indicated antibodies. Cyclin B1 and cyclin E, which peak at M phase and S phase, respectively, were immunoblotted to confirm cell cycle stages. B and C, HEK293 cells transfected with the indicated siRNA were labeled with 5-ethynyl-2′-deoxyuridine and stained with PI. Percentages of cells in G0/G1, S, and G2/M phase were analyzed by flow cytometry (B). S phase cells were further resolved into early, middle, and late stages using additional thresholds of PI staining intensity (C). Values are presented as the percentage of total population of triplicate measurements. Two separate experiments were repeated. D, HEK293 cells transfected with BAF WT or indicated BAF mutant constructs were stained with PI and analyzed cell cycle by flow cytometry. Values are presented as percentage of total cell population of triplicate measurements from two separate experiments. E, HeLa cells overexpressing BAF WT or indicated BAF mutants were synchronized by double thymidine blocks and released for 8 h. Cells were stained by PI and analyzed by flow cytometry. Results represent the percentage of the total population from triplicate samples.
Knockdown of BAF by RNAi caused a delay in S phase progression and a correspondingly longer doubling time (12, 28). We examined the effect of phosphorylation of BAF on cell cycle progression by flow cytometry. An increase in S phase cell population was found in BAF or PP4C knockdown cells, but no change was observed in VRK1 siRNA-transfected cells (Fig. 5B). To determine whether the defect lies in entry into mitosis or S phase progression, S phase cells were further resolved into early, middle, and late stages using additional thresholds of PI staining intensity, classifying them as 5-ethynyl-2′-deoxyuridine+ cells with diploid, hypo-tetraploid, and tetraploid DNA content, respectively. The population of early S phase cells was significantly increased in PP4C knockdown cells (Fig. 5C), suggesting that inhibition of BAF dephosphorylation impairs the progression from early S phase to middle S phase. Knockdown of either VRK1 or PP4C by siRNA also resulted in a drop in the G2/M phase population (Fig. 5B).
BAF WT and mutants were overexpressed to confirm the effect of BAF phosphorylation on the cell cycle. Overexpression of both BAF WT and BAF-S4A mutants showed no significant effect on the S phase cell population. BAF-S4D overexpression resulted in an ∼3% increase in S phase cells, which was consistent with the PP4C knockdown results, although the increase was less dramatic. Overexpression of BAF-S4D or BAF-S4A reduced the G2/M phase population, but BAF WT resulted in a small increase (Fig. 5D). To further confirm the result, Hela cells were arrested at early S phase entry by double thymidine and released for 8 h to allow the cells to exit S phase. Overexpression of BAF-S4D almost doubled the S phase population compared with control (Fig. 5E). The results suggest that inhibition of BAF dephosphorylation prolongs S phase progression and results in a reduced G2/M population.
DISCUSSION
Role of PP4C-R2 complex in BAF dephosphorylation
Genetic, biochemical and functional studies have implicated phosphatases, that include PP1, PP2A, PP4, Cdc25 and Cdc14, in the regulation of mitosis (29). These phosphatases exist as complexes with diverse regulatory subunits that control dephosphorylation activity and cellular localization. We identified the PP4C-R2 complex as a major phosphatase responsible for dephosphorylation of BAF. Knockdown of PP4C or R2 resulted in a major increase in pBAF. It has been reported that PP2A can also dephosphorylate BAF and influence the recruitment of BAF to chromatin (18). However, our results do not support the conclusion that knockdown of PP2A enhances the phosphorylation of BAF at Ser-4.
The interplay with different regulatory subunits is likely central to PP4C action. R2 is one of the structurally distinct regulatory subunits of the PP4C complex that does not interact with PP2A (26). In addition to a typical centrosome location, PP4C and R2 are also found in the nucleus and at low levels in the cytoplasm (18). In mammalian cells, a proteomic approach identified PP4C-R2 as interacting with gemins 3 and 4 to regulate the motor neuron SMN complex (30). PP4C-R2 also forms a complex with R3 to dephosphorylate histone γ-H2AX to maintain the efficiency of DNA repair (19, 31). Although we failed to isolate the third regulatory subunit in BAF dephosphorylation complex, we did notice that PP4C and R2 in eluted fractions from a gel filtration column formed a broader peak than the phosphatase activity for BAF dephosphorylation, suggesting the possible involvement of additional unidentified regulatory subunits.
Cellular Localization of pBAF and Unphosphorylated BAF
The localization of pBAF and unphosphorylated BAF was studied by indirect immunostaining with antibodies against pBAF and tBAF and further confirmed by observing the changes in the BAF distribution patterns in response to VRK1 or PP4C/R2 knockdown. The distribution of endogenous tBAF detected by indirect immunostaining is largely similar to that of exogenously expressed BAF fused with fluorescent protein (20). Unphosphorylated BAF was located throughout cells with no distinct pattern during early mitosis but abundantly colocalized with newly synthesized NE at late telophase. pBAF accumulated at the core region at early telophase and redistributed to the cytoplasm at late telophase.
The core localization of pBAF is consistent with the finding that depletion of VRK-1 abolishes core localization of BAF in C. elegans (14). However, the interactions responsible for localization of pBAF to the core region remain to be determined. Haraguchi et al. (15) found that core localization is also dependent on spindle MTs. It can be hypothesized that phosphorylated BAF localizes to the core region by directly binding the spindle MTs. However, the possibility that BAF has other as yet unidentified partners cannot be ruled out. Attempts to fish out interacting partners of BAF from cell extracts by biochemical means result in a very large number of candidates (32),4 making it hard to discriminate partners that may be biologically relevant. This is not surprising because any DNA binding protein is likely to be fished out through indirect interactions through DNA. In fact, some of the previously reported interacting partners of BAF only “interact” in the presence of DNA (33). It may be fruitful to repeat such screens with phosphorylated BAF where DNA binding would not be a problem and direct interacting partners of the phosphorylated form may be identified.
Involvement of BAF Phosphorylation in NE Morphology and Cell Cycle Progression
BAF has an important function in NE assembly (13, 34, 35). The core localization of the NE protein emerin largely depends on BAF phosphorylation. Modifying the BAF phosphorylation state by VRK1 siRNA or PP4C/R2 siRNA resulted in an abnormal NE phenotype in which there were extensive invaginations of the NE into the nucleus and mislocalization of NE proteins. This phenotype is strikingly similar to that observed in fibroblasts of individuals with a hereditary progeroid syndrome (27). The syndrome results from an A12T mutation in BAF, which likely destabilizes BAF and results in a reduced intracellular concentration. The abnormal NE structures can arise from imperfect synchrony between chromosome decondensation, fusion of NE fragments during NE assembly after mitosis, and extensive growth of NE (36, 37). It is noteworthy that mutations in a number of other genes encoding proteins associated with NE can also result in abnormalities in the NE and progeroid syndromes (38). Hutchinson-Gilford progeria syndrome, resulting from a point mutation of lamin A gene, causes similar changes in nuclear shape (39).
Knockdown of BAF by RNAi leads to a defect in S phase progression (12, 28). Our results suggest that phosphorylation and dephosphorylation of BAF play an essential role in maintaining normal cell cycle progression. Knockdown of BAF dephosphorylation led to impaired S phase progression and resulted in a reduced G2/M phase cell population. Although it is clear that BAF plays a major role in the assembly of the NE after mitosis, and mutations that affect the phosphorylation state of BAF have profound effects on NE shape integrity and cell cycle progression, the detailed mechanisms remain to be elucidated.
Acknowledgments
We are grateful to Dr. Daniel Durocher for generously providing the constructs PP4C-H56Q and PP4C-R85A. We thank Dr. Yihong Ye for critically reading the manuscript and giving comments.
This work was supported, in whole or in part, by National Institutes of Health Intramural Project DK036171. This work was supported by the Intramural Research Program of NIDDK, National Institutes of Health.
X. Zhuang, E. Semenova, and R. Craigie, unpublished data.
- BAF
- barrier-to-autointegration factor
- VRK1
- vaccinia-related kinase 1
- tBAF
- total BAF
- NE
- nuclear envelope
- PP4C
- protein phosphatase 4 catalytic subunit
- PI
- propidium iodide.
REFERENCES
- 1. Lee M. S., Craigie R. (1998) A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. U.S.A. 95, 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Umland T. C., Wei S. Q., Craigie R., Davies D. R. (2000) Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry 39, 9130–9138 [DOI] [PubMed] [Google Scholar]
- 3. Cai M., Huang Y., Zheng R., Wei S. Q., Ghirlando R., Lee M. S., Craigie R., Gronenborn A. M., Clore G. M. (1998) Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat. Struct. Biol. 5, 903–909 [DOI] [PubMed] [Google Scholar]
- 4. Zheng R., Ghirlando R., Lee M. S., Mizuuchi K., Krause M., Craigie R. (2000) Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 97, 8997–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradley C. M., Ronning D. R., Ghirlando R., Craigie R., Dyda F. (2005) Structural basis for DNA bridging by barrier-to-autointegration factor. Nat. Struct. Mol. Biol. 12, 935–936 [DOI] [PubMed] [Google Scholar]
- 6. Skoko D., Li M., Huang Y., Mizuuchi M., Cai M., Bradley C. M., Pease P. J., Xiao B., Marko J. F., Craigie R., Mizuuchi K. (2009) Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc. Natl. Acad. Sci. U.S.A. 106, 16610–16615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furukawa K. (1999) LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112, 2485–2492 [DOI] [PubMed] [Google Scholar]
- 8. Lee K. K., Haraguchi T., Lee R. S., Koujin T., Hiraoka Y., Wilson K. L. (2001) Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114, 4567–4573 [DOI] [PubMed] [Google Scholar]
- 9. Shumaker D. K., Lee K. K., Tanhehco Y. C., Craigie R., Wilson K. L. (2001) LAP2 binds to BAF center dot DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 20, 1754–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mansharamani M., Wilson K. L. (2005) Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J. Biol. Chem. 280, 13863–13870 [DOI] [PubMed] [Google Scholar]
- 11. Cai M., Huang Y., Suh J. Y., Louis J. M., Ghirlando R., Craigie R., Clore G. M. (2007) Solution NMR structure of the barrier-to-autointegration factor-emerin complex. J. Biol. Chem. 282, 14525–14535 [DOI] [PubMed] [Google Scholar]
- 12. Furukawa K., Sugiyama S., Osouda S., Goto H., Inagaki M., Horigome T., Omata S., McConnell M., Fisher P. A., Nishida Y. (2003) Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Sci. 116, 3811–3823 [DOI] [PubMed] [Google Scholar]
- 13. Margalit A., Segura-Totten M., Gruenbaum Y., Wilson K. L. (2005) Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc. Natl. Acad. Sci. U.S.A. 102, 3290–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gorjánácz M., Klerkx E. P., Galy V., Santarella R., López-Iglesias C., Askjaer P., Mattaj I. W. (2007) Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 26, 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haraguchi T., Kojidani T., Koujin T., Shimi T., Osakada H., Mori C., Yamamoto A., Hiraoka Y. (2008) Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 121, 2540–2554 [DOI] [PubMed] [Google Scholar]
- 16. Bengtsson L., Wilson K. L. (2006) Barrier-to-autointegration factor phosphorylation on Ser-4 regulates emerin binding to lamin A in vitro and emerin localization in vivo. Mol. Biol. Cell 17, 1154–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nichols R. J., Wiebe M. S., Traktman P. (2006) The vaccinia-related kinases phosphorylate the N terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 17, 2451–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Asencio C., Davidson I. F., Santarella-Mellwig R., Ly-Hartig T. B., Mall M., Wallenfang M. R., Mattaj I. W., Gorjánácz M. (2012) Coordination of kinase and phosphatase activities by Lem4 enables nuclearenvelope reassembly during mitosis. Cell 150, 122–135 [DOI] [PubMed] [Google Scholar]
- 19. Nakada S., Chen G. I., Gingras A. C., Durocher D. (2008) PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 9, 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dechat T., Gajewski A., Korbei B., Gerlich D., Daigle N., Haraguchi T., Furukawa K., Ellenberg J., Foisner R. (2004) LAP2 α and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 117, 6117–6128 [DOI] [PubMed] [Google Scholar]
- 21. Cozzone A. J. (1988) Protein-phosphorylation in prokaryotes. Ann. Rev. Microbiol. 42, 97–125 [DOI] [PubMed] [Google Scholar]
- 22. Barford D., Das A. K., Egloff M. P. (1998) The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 [DOI] [PubMed] [Google Scholar]
- 23. Honkanen R. E., Golden T. (2002) Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr. Med. Chem. 9, 2055–2075 [DOI] [PubMed] [Google Scholar]
- 24. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 25. Cohen P. T. (2005) Protein phosphatase 4: a multifunctional protein serine/threonine phosphatase sensitive to antitumour drugs. FEBS J. 272, 303–303 [Google Scholar]
- 26. Cohen P. T., Philp A., Vázquez-Martin C. (2005) Protein phosphatase 4 - from obscurity to vital functions. FEBS Lett. 579, 3278–3286 [DOI] [PubMed] [Google Scholar]
- 27. Puente X. S., Quesada V., Osorio F. G., Cabanillas R., Cadiñanos J., Fraile J. M., Ordóñez G. R., Puente D. A., Gutiérrez-Fernández A., Fanjul-Fernández M., Lévy N., Freije J. M., López-Otín C. (2011) Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am. J. Hum. Genet. 88, 650–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haraguchi T., Koujin T., Osakada H., Kojidani T., Mori C., Masuda H., Hiraoka Y. (2007) Nuclear localization of barrier-to-autointegration factor is correlated with progression of S phase in human cells. J. Cell Sci. 120, 1967–1977 [DOI] [PubMed] [Google Scholar]
- 29. Bollen M., Gerlich D. W., Lesage B. (2009) Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 19, 531–541 [DOI] [PubMed] [Google Scholar]
- 30. Carnegie G. K., Sleeman J. E., Morrice N., Hastie C. J., Peggie M. W., Philp A., Lamond A. I., Cohen P. T. (2003) Protein phosphatase 4 interacts with the Survival of Motor Neurons complex and enhances the temporal localisation of snRNPs. J. Cell Sci. 116, 1905–1913 [DOI] [PubMed] [Google Scholar]
- 31. Chowdhury D., Xu X., Zhong X., Ahmed F., Zhong J., Liao J., Dykxhoorn D. M., Weinstock D. M., Pfeifer G. P., Lieberman J. (2008) A PP4-phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Mol. Cell 31, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montes de Oca R., Shoemaker C. J., Gucek M., Cole R. N., Wilson K. L. (2009) Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One 4, e7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang Y., Cai M., Clore G. M., Craigie R. (2011) No interaction of barrier-to-autointegration factor (BAF) with HIV-1 MA, cone-rod homeobox (Crx) or MAN1-C in the absence of DNA. PLoS One 6, e25123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haraguchi T., Koujin T., Hayakawa T., Kaneda T., Tsutsumi C., Imamoto N., Akazawa C., Sukegawa J., Yoneda Y., Hiraoka Y. (2000) Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113, 779–794 [DOI] [PubMed] [Google Scholar]
- 35. Segura-Totten M., Kowalski A. K., Craigie R., Wilson K. L. (2002) Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 158, 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brandt A., Papagiannouli F., Wagner N., Wilsch-Bräuninger M., Braun M., Furlong E. E., Loserth S., Wenzl C., Pilot F., Vogt N., Lecuit T., Krohne G., Grosshans J. (2006) Developmental control of nuclear size and shape by kugelkern and kurzkern. Curr. Biol. 16, 543–552 [DOI] [PubMed] [Google Scholar]
- 37. Gupton S. L., Collings D. A., Allen N. S. (2006) Endoplasmic reticulum targeted GFP reveals ER organization in tobacco NT-1 cells during cell division. Plant Physiol. Biochem. 44, 95–105 [DOI] [PubMed] [Google Scholar]
- 38. Gruenbaum Y., Margalit A., Goldman R. D., Shumaker D. K., Wilson K. L. (2005) The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6, 21–31 [DOI] [PubMed] [Google Scholar]
- 39. Goldman R. D., Shumaker D. K., Erdos M. R., Eriksson M., Goldman A. E., Gordon L. B., Gruenbaum Y., Khuon S., Mendez M., Varga R., Collins F. S. (2004) Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 101, 8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]