Background: Little is known regarding the thermodynamics of binding of the broadly neutralizing anti-HIV-1 mAb 2F5 to its gp41 epitope.
Results: Isothermal titration calorimetry reveals strong differences between IgG and Fab.
Conclusion: Residues flanking the core epitope and the immunoglobulin Fc region contribute strongly to affinity by allosteric mechanisms.
Significance: The results may help to develop new therapeutics and/or vaccines against HIV and to understanding Ag-Ab recognition.
Keywords: Antibodies, HIV-1, Isothermal Titration Calorimetry, Peptides, Thermodynamics, MPER, gp41
Abstract
Immunotherapies and vaccines based on the induction of broadly neutralizing monoclonal antibodies (bNAbs) have become outstanding strategies against HIV-1. Diverse bNAbs recognizing different regions of the HIV-1 envelope have been identified and extensively studied. However, there is little information about the thermodynamics of binding of these bNAbs and their epitopes. We used isothermal titration calorimetry to characterize thermodynamically the interactions between bNAb 2F5 (in both the IgG and Fab forms) and its functional and core epitope peptides. We found that these interactions are enthalpically driven and opposed by a negative entropy change. The highest affinity was found for 2F5 IgG for its functional epitope, indicating that additional interactions involving residues flanking the core epitope contribute strongly to higher affinity. In addition, the strong influence of the Fc region on the binding affinity suggests long-range allosteric effects within IgG. Our results provide useful information for developing new therapeutics against HIV-1 and, in a broader scope, contribute to a better understanding of antigen-antibody recognition.
Introduction
Since 1983, when HIV was found to be the causal agent of AIDS, great efforts have been made to design an effective vaccine to contain the virus. Despite these efforts and even after encouraging results from the RV144 vaccine clinical trial (1), developing an HIV vaccine is still a daunting challenge. However, the field of HIV vaccine research has experimented a resurgence with the identification of antibodies that neutralize most circulating HIV-1 strains. Eliciting broadly neutralizing antibodies (bNAbs)2 provides a base for active and passive immunization strategies to prevent HIV infection (2). These antibodies can protect against infection and suppress established HIV infection in animal models. The finding that these antibodies develop in a fraction of infected individuals supports the idea that new approaches to vaccination might be developed by adapting the natural immune strategies or by structure-based design of immunogens (3). One of these bNAbs, 2F5, was discovered in a first generation of bNAbs against HIV-1 and has been extensively studied. bNAb 2F5 has strong neutralizing activity against a broad range of HIV-1 primary isolates (4–6). The core epitope of 2F5 was mapped onto the 662ELDKWAS668 linear sequence of gp41, which is located at the membrane-proximal external region (MPER) of the protein. A functional epitope including those residues whose substitution modifies the affinity for the corresponding antibody was later extended to the sequence 656NEQELLELDKWASLWN671 (7). The atomic interactions of the complex between the core epitope and antibody 2F5 have been described in detail (8–10). An additional complexity of the 2F5 epitope has been suggested, including other gp41 regions (11) and an influence of the proximal lipid bilayer (12, 13), which strongly affects its immunogenicity (14). Understanding the interaction of this antibody with its epitope requires not only a detailed knowledge of the structure but also knowing the physicochemical characteristics of the antigen (Ag)-antibody complex described by the kinetic rate constants, equilibrium constants, and thermodynamics of binding. Moreover, affinity and specificity are two of the most fundamental concepts of relevance to the molecular immunologist interested in understanding and characterizing Ag-Ab interactions. These concepts can be usefully defined in more than one way (15), but for some purposes, both terms can be specified precisely and quantitatively by reference to thermodynamics. However, very few data have been reported to date about the thermodynamics of binding of the interaction between bNAb 2F5 and its corresponding epitope peptide, and they account only for the Fab fraction (16). In this study, we have characterized thermodynamically the binding of bNAb 2F5 to peptides corresponding to both the core and functional epitopes by isothermal titration calorimetry (ITC). This technique is the only one capable of measuring not only the binding affinity but also its stoichiometry and the magnitude of the two thermodynamic terms that define the binding affinity: the enthalpy (ΔH) and entropy (ΔS) changes.
In contrast, numerous authors have reported that both variable (V) and constant (C) Ig regions contribute to the binding affinity and specificity of antibodies (11, 17–23). Most of these studies have focused on affinity differences between distinct Ig isotypes. For instance, the IgG isotype has been shown to affect the Ag-binding and HIV-neutralizing activity of monoclonal (24) and polyclonal (25) antibodies. Furthermore, 2F5 IgA2 and IgG1 display significantly different epitope specificity, antibody affinity, and functional activities (11). It remains unclear whether allosteric influence between C and V regions is limited to the Fab domain or extends further to the hinge and Fc regions of Ig. This may be clarified by a comparative analysis of epitope recognition between IgG and its Fab fragment. However, very few comparative thermodynamic studies of this type by ITC have been reported. This information would help to elucidate whether the flexibility of the IgG molecule, mostly around the hinge region between the Fc and Fab regions (26, 27), still allows for an effect on the Ab affinity by allosteric mechanisms. For this reason, we have completed this thermodynamic analysis by comparing the results of the binding of these peptides with 2F5 IgG and the 2F5 Fab fraction. Our results revealed higher binding affinities in the case of the functional epitope peptide, indicating that the presence of its flanking residues contributes to a strong affinity increase. Moreover, we also observed higher binding affinities for the complete IgG than for the Fab fraction, confirming that the immunoglobulin Fc region strongly influences bNAb 2F5 affinity.
EXPERIMENTAL PROCEDURES
Numbering of Residue Sequences
gp41 residues are numbered according to their positions in the full amino acid sequence of HXBc2 Env gp160 (http://www.hiv.lanl.gov/).
Materials
Antibody 2F5 (28) in the IgG1(κ) form was kindly provided by Dr. Dietmar Katinger (Polymun, Klosterneuburg, Austria). Two chemically synthesized peptides representing different fragments of the gp41 MPER region including the 2F5 epitope, N16N (656NEQELLELDKWASLWN671) and E7S (662ELDKWAS668), were purchased from CASLO Laboratory ApS. Both peptides were N-acetylated and C-amidated at their termini.
Fab Preparation
The 2F5 Fab fragments were generated via enzymatic digestion of IgG using a Fab preparation kit from Pierce Thermo Scientific. To maximize the purity, a size exclusion chromatography step with the pool of the 2F5 Fab fraction was carried out on a Superose 6 column (GE Healthcare) in PBS buffer (0.1 m sodium phosphate and 0.15 m sodium chloride; pH 7.2). Fab eluted from the column as a single peak at an elution volume corresponding to a monomer. Purity was also checked by SDS-PAGE.
Isothermal Titration Calorimetry
ITC experiments were performed with a high sensitivity VP-ITC microcalorimeter (MicroCal, Northampton, MA) at 25 °C. Antibody samples were extensively dialyzed against 50 mm sodium phosphate, pH 7.4, for 24 h at 4 °C prior to the experiments. Peptides were directly dissolved in the same buffer. Possible pH variations were corrected, and the concentrations were measured spectrophotometrically by UV absorption at 280 nm using extinction coefficients determined via the ProtParam algorithm based on the amino acid sequences. Protein and peptide solutions were degassed for 10 min before measurements.
Standard ITC Titrations
The antibody solutions (5–11 μm) in the calorimetric cell were titrated with the corresponding peptide (250–500 μm) in successive injections with variable volumes ranging from 5 to 15 μl. The corresponding heats of peptide dilution into buffer were measured in separate blank experiments and used to correct the binding heats. The experimental isotherms were analyzed according to a binding model of n independent and identical sites using Origin software (OriginLab, Northampton, MA). The fit of the binding curve yields the binding stoichiometry (n), the equilibrium binding constant (Kb), and the enthalpy change (ΔHb) of the binding reaction. The free energy (ΔGb) and entropy (ΔSb) changes of binding are determined by the basic thermodynamic expressions ΔGb = −RT ln Kb = ΔHb − TΔSb, where R and T are the gas constant and the absolute temperature, respectively.
Displacement Experiments
To accurately measure the extremely high binding affinity of 2F5 IgG for the N16N peptide, ITC displacement experiments were carried out. The protocol for this ITC displacement experiment requires two different titrations: (i) a standard titration with the E7S peptide (weak ligand) binding to 2F5 IgG and (ii) a displacement titration with the N16N peptide (strong ligand) of 2F5 IgG in the presence of the E7S peptide. Both titrations are performed following the same steps. The direct titration has been described above. For the displacement titration, 2F5 IgG (5 μm) was mixed with the weak ligand (E7S, 230 μm) in the cell, and the mixture was titrated with the strong ligand (N16N, 220 μm) in successive injections of 5 μl. The corresponding heats of dilution of the N16N peptide into the buffer were used to correct the data. The thermodynamic parameters for the high affinity binding were determined by fitting the binding isotherms according to the equations developed by Sigurskjold (29) using the binding parameters obtained for the direct titration of 2F5 IgG with the weak ligand as reference.
RESULTS
Thermodynamics of Binding of 2F5 IgG to Its Epitope
First, a 2F5 IgG solution was titrated with the E7S peptide corresponding to the core epitope of this bNAb. The ITC thermogram (Fig. 1A) shows a Kd value (1/Kb) in the micromolar range (Kd = 0.9 ± 0.1 μm) for this peptide (Table 1). The number of antibody-binding sites was found to be close to two, as expected. The binding enthalpy is large and negative (−10.3 ± 0.4 kcal·mol−1), and the binding entropy is unfavorable (T·ΔSb = −2.1 kcal·mol−1).
FIGURE 1.
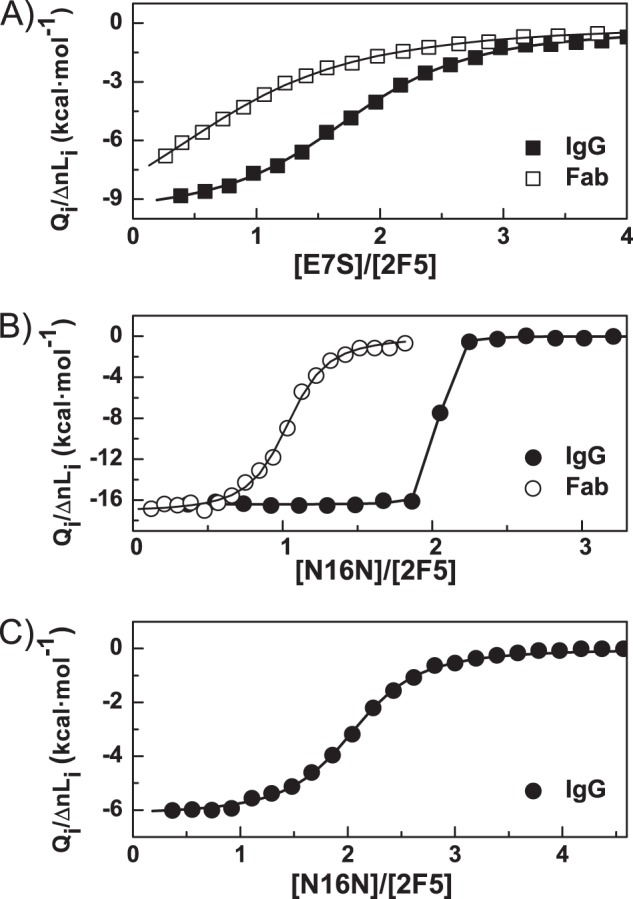
ITC isotherms for the binding of bNAb 2F5 to its core and functional epitope peptides. A and B, direct ITC isotherms for the binding of 2F5 (in both the IgG and Fab forms) to the E7S and N16N peptides, respectively. C, ITC displacement isotherm for the binding of 2F5 IgG to N16N using E7S as the weak ligand. In all panels, the symbols represent the experimental heats measured for each peptide injection normalized per mole of injected peptide. The curves correspond to the best fittings using a model of n identical sites for the direct titrations and a binding competition model for the displacement experiment.
TABLE 1.
Thermodynamic parameters of the association of the N16N and E7S peptides with 2F5 IgG and 2F5 Fab
Error calculations were based on 95% confidence limits of the fits.
| Complex | No. of binding sites | Kd | ΔHb | T·ΔSb | ΔGb |
|---|---|---|---|---|---|
| nm | kcal·mol−1 | kcal·mol−1 | kcal·mol−1 | ||
| N16N-2F5 IgG | 2.049 ± 0.002 | ≈3 | −16.4 ± 0.1 | ||
| N16N-2F5 IgGa | 1.72 ± 0.03 | 0.82 ± 0.03 | −16.4 ± 0.4 | −4.0 ± 0.4 | −12.37 ± 0.02 |
| E7S-2F5 IgG | 1.88 ± 0.04 | 885 ± 80 | −10.3 ± 0.4 | −2.1 ± 0.5 | −8.25 ± 0.08 |
| N16N-2F5 Fab | 1.05 ± 0.01 | 50 ± 10 | −17.2 ± 0.4 | −7.3 ± 0.5 | −9.9 ± 0.1 |
| E7S-2F5 Fab | 0.99 ± 0.03 | 5800 ± 900 | −12.0 ± 0.5 | −4.9 ± 0.5 | −7.14 ± 0.04 |
a Displacement ITC experiment.
Then, a 2F5 IgG solution was titrated with the N16N peptide corresponding to the functional epitope of this bNAb. The ITC thermogram (Fig. 1B) indicates an extremely high binding affinity of antibody 2F5 for the N16N peptide (7), which falls well above the proper range of the technique for an accurate direct determination. The Kd value estimated from this experiment is ∼3 nm, although this value has very high uncertainty. The number of antibody-binding sites derived from the isotherm is two, as expected for IgG. The binding enthalpy (ΔHb = −16.4 ± 0.1 kcal·mol−1) is negative and quite large for such a small peptide.
An accurate determination of very high binding affinities (at nanomolar levels or even higher) is quite difficult by direct ITC titration. To overcome such a drawback, ITC displacement experiments can extend the useful range for the association constant determination (29, 30).
For the ITC displacement experiment (Fig. 1C), we selected the E7S peptide. The resulting binding isotherm was fitted to a binding competition model. The binding parameters of the E7S peptide were fixed in the fitting using those previously determined by ITC. From this experiment, we could determine very accurately the binding affinity for the N16N peptide (Kd = 0.82 ± 0.03 nm). As expected, the number of binding sites in the antibody found with this model was again close to two. As expected, the binding enthalpy (ΔHb = −16.4 ± 0.4 kcal·mol−1) is fully coincident to that determined with the standard ITC titration, which already provided a very accurate value for this parameter (Table 1). These values result in ΔGb = −12.37 ± 0.02 kcal·mol−1 and T·ΔSb = −4.0 ± 0.4 kcal·mol−1, confirming our previous conclusions about the enthalpically driven binding of 2F5 IgG to its epitope. This is compensated by a relatively small loss of entropy, suggesting that the configurational entropy loss due to peptide immobilization dominates the binding entropy over the hydrophobic interaction.
If we compare the thermodynamic parameters obtained with the core or functional epitopes, we observe that the binding enthalpy is large and negative in both cases but significantly smaller in magnitude for the E7S peptide than for the N16N peptide. This result indicates less extensive interactions between the E7S peptide and the 2F5 paratope than those established with the N16N peptide. The binding entropy is also unfavorable in both cases but to a lower extent for E7S than for N16N, consistent with a lower entropy cost associated with peptide immobilization due the shorter peptide length.
In conclusion, the MPER epitope recognized by 2F5 IgG is not limited to the putative core sequence (ELDKWAS). Flanking residues along the MPER sequence are important to establish additional interactions with the antibody paratope, strongly enhancing the binding affinity (31–33).
Thermodynamics of Binding of 2F5 Fab to Its Epitope
2F5 Fab solutions were directly titrated with the peptides (N16N and E7S). The resulting binding isotherms were analyzed using a model of binding to n independent sites (Fig. 1, A and B). The number of binding sites of Fab resulting from the fits was close to one for both peptides, as expected. The binding enthalpies are ΔHb(N16N) = −17.2 ± 0.4 kcal·mol−1 and ΔHb(E7S) = −12.0 ± 0.5 kcal·mol−1, and the dissociation constants are Kd(N16N) = 50 ± 10 nm and Kd(E7S) = 5.8 ± 0.9 μm. The binding enthalpies for Fab are quite similar to those obtained for 2F5 IgG, indicating a similar interaction between the Ig paratopes and each of the epitopes, although the ΔHb values found for Fab are slightly more negative.
The binding affinities are much lower, however, for Fab than for IgG under the same conditions (Table 1). The affinity of 2F5 Fab for the N16N peptide is of the same order as that reported elsewhere for recombinant 2F5 Fab (Kd = 20 nm) (16), although the binding enthalpy is higher in our case, possibly due to the use of different buffer conditions. In fact, HEPES was used as the buffer at the same pH, so the ionization heat of this buffer may influence the observed binding enthalpy if proton exchange is coupled to the binding process. In our case, we used sodium phosphate buffer, which has a practically null ionization heat, therefore providing a binding enthalpy value unaffected by ionization effects. The strong decrease in affinity of 2F5 Fab for N16N and E7S can be fully ascribed to a higher binding entropy cost, partially opposed by a slightly more favorable binding enthalpy.
DISCUSSION
We have characterized by ITC the thermodynamics of binding between antibody 2F5 (in both the IgG and Fab forms) and its cognate peptide epitope. The binding of these complexes is driven by a large negative enthalpy change and opposed by a negative entropy change, showing typical enthalpy-entropy compensation. Our results confirm previous reports concluding that the putative core epitope ELDKWAS is not sufficient for high binding affinity but that additional flanking residues strongly increase this affinity by the establishment of additional interactions with the antibody. This is consistent with early observations by limited proteolysis extending the 2F5-recognized epitope to the N16N sequence (7). Moreover, there are also close and specific contacts with residues located N-terminal to the epitope core, and several residues C-terminal to the ELDKWAS core have been observed to adopt an ordered helical conformation in the crystal structures of the complexes between peptides and 2F5 Fab (8, 9). Our thermodynamic data indicate that interactions established by the flanking residues with the core contribute strongly to the favorable binding enthalpy with a relatively small entropy penalty and therefore suggest that these residues appear to establish interactions with the antibody paratope that may be stronger than anticipated by the reported crystal structures of 2F5 Fab with MPER peptides. Nevertheless, it is unlikely that the N16N peptide encompasses all of the antigenic elements of the 2F5 epitope. Additional hydrophobic contacts between the long CDRH3 loop of 2F5 and residues downstream into the MPER C terminus have been proposed to help stabilize the extended loop-bound conformation (16, 34). The CDRH3 loop has also been reported recently to assist in the extraction of the lipid-immersed MPER prior to tight binding (12–14). Because the MPER peptides adopt an α-helical conformation in a membrane environment (35), the conformational change needed for 2F5 binding should imply an additional energy cost. Other MPER-distant gp41 regions have been recently proposed to participate in stabilizing the antigenic conformation of the epitope in the lipid context (36). Calorimetric studies with MPER peptides in a model membrane environment would be needed to elucidate the thermodynamic determinants of these effects.
In addition, we have measured a much higher affinity of 2F5 IgG compared with 2F5 Fab for the binding of each peptide. The affinity for N16N is increased by ∼60 times, corresponding to a binding Gibbs energy decrease of about −2.4 kcal·mol−1, and in the case of E7S, this affinity increase is ∼6-fold, with a binding Gibbs energy decrease of −1.1 kcal·mol−1. Interestingly, the binding enthalpies are slightly more negative for Fab compared with IgG by −0.8 kcal·mol−1 for N16N and −1.7 kcal·mol−1 for E7S. These differences cannot be explained solely from the point of view of the Ab-epitope binding interface at the V domain and may be attributable to a higher structural flexibility of free Fab in comparison with IgG. Upon complex formation, a larger decrease in conformational flexibility and tightening of structural interactions within the Fab domain structure may account for the observed thermodynamic differences. This suggests that the presence of the Fc domain in the whole 2F5 IgG contributes strongly to conformationally stabilizing the binding-competent Fab domain by an allosteric mechanism.
There are very few thermodynamic studies comparing the Ag binding of whole Abs with that of the Fab or Fv fragments, and they generally show similar thermodynamic magnitudes (37–39). However, although traditionally the binding sites in the V regions of immunoglobulins have been considered functionally independent, significant binding-related allosteric effects between the V and C regions of antibodies have also become progressively evident. For example, Pritsch et al. (18) demonstrated that two human Ab isotypes sharing identical V regions bind tubulin with significantly different affinities. Putterman and co-workers (22) also demonstrated that anti-nuclear Abs with identical V regions but different C regions exhibit diverse binding affinities for histones. Their observations reflect differences in accessibility to binding sites depending on the flexibility of the various Fab fragments in association with different C regions and differences at the paratope level. Dam et al. (21) also used ITC to investigate the interactions between four monoclonal antibodies with identical V regions and a univalent peptide Ag, providing unambiguous thermodynamic evidence for the influence of the C region on the interaction of the V region with an Ag. Torres and Casadevall (23) have proposed that electrostatic and hydrophobic interactions resulting from differences in the microenvironment of constant heavy chain (CH) domains (e.g. pH and ionic strength) may affect the Ag-binding site. Additionally, the arrangement of the Fab C domains relative to the V domains and to each other may increase the probability of an appropriate relative orientation of the V heavy (H) and light (L) chains, which, in turn, can shape the Ag-binding site and affect affinity. The influence of the C region on V region affinity and/or specificity has important implications for the choice of isotype in therapeutic Abs and for the creation of more effective vaccines (20). Very recently, Casadevall and co-workers (40) studied four IgG isotypes with identical V regions by small angle x-ray scattering, and they provided additional evidence that the Ig V and C domains influence each other structurally and suggested that V region structure can have significant effects on the overall Ig structure. Also significantly, differences in affinity and epitope specificity have been described between 2F5 IgA and IgG isotypes (11), supporting a role of the CH1 domain in modulating antibody affinity as a consequence of differences in CH1 flexibility. Moreover, using bacterial proteins A and G as probes for CH binding, Sagawa et al. (41) demonstrated that hapten binding to the Fv region can induce long-range conformational changes in the Fc region of IgG. Recently, Ofran and co-workers (42) compared a large number of free and bound antibody crystal structures to explore possible allosteric effects associated with binding. They detected many structural changes occurring far from the Ag-binding site, including changes in the relative orientation of the H and L chains in both the V and C domains; changes in the elbow angle between the V and C domains; and quite consistently, conformational changes in an intrinsically disordered loop in the CH domain, which is implicated in the interaction between the H and L chains. This CH1 loop is also close in space to the hinge region connecting the CH1 domain to Fc in IgG, and its conformational changes may affect the relative orientation between the CH1 domain versus Fc, thereby influencing effector binding to Fc.
The results presented here provide direct evidence from a thermodynamic point of view of the effect of the Fc region on the immunoglobulin affinity. This and other previous evidence support that long-range cooperative motions in the 2F5 Fab domain may be more or less restricted by the presence or absence of Fc mediated by the CH1 domain, and this may in turn influence binding affinity.
In conclusion, the thermodynamic parameters determined here for binding of bNAb 2F5 to its cognate epitope and the results related to the involvement of the Fc region in Ab affinity are important for understanding the structural and thermodynamic determinants of Ag-Ab recognition and provide useful information for the design of new therapeutics and vaccines against HIV.
Acknowledgment
We thank Dr. Dietmar Katinger for kindly providing bNAb 2F5.
This work was supported by European Union Seventh Framework Programme (FP7/2007–2013) Grant 201038.
- bNAb
- broadly neutralizing antibody
- MPER
- membrane-proximal external region
- Ag
- antigen
- ITC
- isothermal titration calorimetry
- V region
- variable Ig region
- C region
- constant Ig region
- CH
- constant heavy chain
- H
- heavy
- L
- light.
REFERENCES
- 1. Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., Benenson M., Gurunathan S., Tartaglia J., McNeil J. G., Francis D. P., Stablein D., Birx D. L., Chunsuttiwat S., Khamboonruang C., Thongcharoen P., Robb M. L., Michael N. L., Kunasol P., Kim J. H. (2009) Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361, 2209–2220 [DOI] [PubMed] [Google Scholar]
- 2. Kwong P. D., Mascola J. R., Nabel G. J. (2013) Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13, 693–701 [DOI] [PubMed] [Google Scholar]
- 3. Klein F., Mouquet H., Dosenovic P., Scheid J. F., Scharf L., Nussenzweig M. C. (2013) Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conley A. J., Kessler J. A., 2nd, Boots L. J., Tung J. S., Arnold B. A., Keller P. M., Shaw A. R., Emini E. A. (1994) Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41–2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. U.S.A. 91, 3348–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muster T., Steindl F., Purtscher M., Trkola A., Klima A., Himmler G., Rüker F., Katinger H. (1993) A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67, 6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Purtscher M., Trkola A., Grassauer A., Schulz P. M., Klima A., Döpper S., Gruber G., Buchacher A., Muster T., Katinger H. (1996) Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10, 587–593 [DOI] [PubMed] [Google Scholar]
- 7. Parker C. E., Deterding L. J., Hager-Braun C., Binley J. M., Schülke N., Katinger H., Moore J. P., Tomer K. B. (2001) Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75, 10906–10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ofek G., Tang M., Sambor A., Katinger H., Mascola J. R., Wyatt R., Kwong P. D. (2004) Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78, 10724–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Julien J. P., Bryson S., Nieva J. L., Pai E. F. (2008) Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J. Mol. Biol. 384, 377–392 [DOI] [PubMed] [Google Scholar]
- 10. Bryson S., Julien J. P., Hynes R. C., Pai E. F. (2009) Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications. J. Virol. 83, 11862–11875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tudor D., Yu H., Maupetit J., Drillet A. S., Bouceba T., Schwartz-Cornil I., Lopalco L., Tuffery P., Bomsel M. (2012) Isotype modulates epitope specificity, affinity, and antiviral activities of anti-HIV-1 human broadly neutralizing 2F5 antibody. Proc. Natl. Acad. Sci. U.S.A. 109, 12680–12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song L., Sun Z. Y., Coleman K. E., Zwick M. B., Gach J. S., Wang J. H., Reinherz E. L., Wagner G., Kim M. (2009) Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc. Natl. Acad. Sci. U.S.A. 106, 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim M., Sun Z. Y., Rand K. D., Shi X., Song L., Cheng Y., Fahmy A. F., Majumdar S., Ofek G., Yang Y., Kwong P. D., Wang J. H., Engen J. R., Wagner G., Reinherz E. L. (2011) Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat. Struct. Mol. Biol. 18, 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim M., Song L., Moon J., Sun Z. Y., Bershteyn A., Hanson M., Cain D., Goka S., Kelsoe G., Wagner G., Irvine D., Reinherz E. L. (2013) Immunogenicity of membrane-bound HIV-1 gp41 membrane-proximal external region (MPER) segments is dominated by residue accessibility and modulated by stereochemistry. J. Biol. Chem. 288, 31888–31901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greenspan N. S. (2010) Cohen's Conjecture, Howard's Hypothesis, and Ptashne's Ptruth: an exploration of the relationship between affinity and specificity. Trends Immunol. 31, 138–143 [DOI] [PubMed] [Google Scholar]
- 16. Julien J. P., Huarte N., Maeso R., Taneva S. G., Cunningham A., Nieva J. L., Pai E. F. (2010) Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J. Virol. 84, 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Torres M., Fernández-Fuentes N., Fiser A., Casadevall A. (2007) The immunoglobulin heavy chain constant region affects kinetic and thermodynamic parameters of antibody variable region interactions with antigen. J. Biol. Chem. 282, 13917–13927 [DOI] [PubMed] [Google Scholar]
- 18. Pritsch O., Hudry-Clergeon G., Buckle M., Petillot Y., Bouvet J. P., Gagnon J., Dighiero G. (1996) Can immunoglobulin C(H)1 constant region domain modulate antigen binding affinity of antibodies? J. Clin. Invest. 98, 2235–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Janda A., Eryilmaz E., Nakouzi A., Cowburn D., Casadevall A. (2012) Variable region identical immunoglobulins differing in isotype express different paratopes. J. Biol. Chem. 287, 35409–35417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casadevall A., Janda A. (2012) Immunoglobulin isotype influences affinity and specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 12272–12273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dam T. K., Torres M., Brewer C. F., Casadevall A. (2008) Isothermal titration calorimetry reveals differential binding thermodynamics of variable region-identical antibodies differing in constant region for a univalent ligand. J. Biol. Chem. 283, 31366–31370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xia Y., Janda A., Eryilmaz E., Casadevall A., Putterman C. (2013) The constant region affects antigen binding of antibodies to DNA by altering secondary structure. Mol. Immunol. 56, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Torres M., Casadevall A. (2008) The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol. 29, 91–97 [DOI] [PubMed] [Google Scholar]
- 24. Cavacini L. A., Emes C. L., Power J., Desharnais F. D., Duval M., Montefiori D., Posner M. R. (1995) Influence of heavy chain constant regions on antigen binding and HIV-1 neutralization by a human monoclonal antibody. J. Immunol. 155, 3638–3644 [PubMed] [Google Scholar]
- 25. Scharf O., Golding H., King L. R., Eller N., Frazier D., Golding B., Scott D. E. (2001) Immunoglobulin G3 from polyclonal human immunodeficiency virus (HIV) immune globulin is more potent than other subclasses in neutralizing HIV type 1. J. Virol. 75, 6558–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris L. J., Skaletsky E., McPherson A. (1998) Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275, 861–872 [DOI] [PubMed] [Google Scholar]
- 27. Saphire E. O., Stanfield R. L., Crispin M. D., Parren P. W., Rudd P. M., Dwek R. A., Burton D. R., Wilson I. A. (2002) Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 319, 9–18 [DOI] [PubMed] [Google Scholar]
- 28. Kunert R., Rüker F., Katinger H. (1998) Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res. Hum. Retroviruses 14, 1115–1128 [DOI] [PubMed] [Google Scholar]
- 29. Sigurskjold B. W. (2000) Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal. Biochem. 277, 260–266 [DOI] [PubMed] [Google Scholar]
- 30. Velazquez-Campoy A., Freire E. (2006) Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat. Protoc. 1, 186–191 [DOI] [PubMed] [Google Scholar]
- 31. Biron Z., Khare S., Samson A. O., Hayek Y., Naider F., Anglister J. (2002) A monomeric 310-helix is formed in water by a 13-residue peptide representing the neutralizing determinant of HIV-1 on gp41. Biochemistry 41, 12687–12696 [DOI] [PubMed] [Google Scholar]
- 32. Liu J., Deng Y., Dey A. K., Moore J. P., Lu M. (2009) Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry 48, 2915–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu Z., Tan Y., Tong P., Yu Y., Chen Y. H. (2013) Possible explanations for the broadly neutralizing activity of HIV-1 gp41 specific monoclonal antibodies by recognition pattern based amino acid sequence analyses. Immunol. Lett. 150, 152–154 [DOI] [PubMed] [Google Scholar]
- 34. Guenaga J., Wyatt R. T. (2012) Structure-guided alterations of the gp41-directed HIV-1 broadly neutralizing antibody 2F5 reveal new properties regarding its neutralizing function. PLoS Pathog. 8, e1002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun Z. Y., Oh K. J., Kim M., Yu J., Brusic V., Song L., Qiao Z., Wang J. H., Wagner G., Reinherz E. L. (2008) HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28, 52–63 [DOI] [PubMed] [Google Scholar]
- 36. Huarte N., Araujo A., Arranz R., Lorizate M., Quendler H., Kunert R., Valpuesta J. M., Nieva J. L. (2012) Recognition of membrane-bound fusion-peptide/MPER complexes by the HIV-1 neutralizing 2F5 antibody: implications for anti-2F5 immunogenicity. PLoS ONE 7, e52740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwarz F. P., Tello D., Goldbaum F. A., Mariuzza R. A., Poljak R. J. (1995) Thermodynamics of antigen-antibody binding using specific anti-lysozyme antibodies. Eur. J. Biochem. 228, 388–394 [PubMed] [Google Scholar]
- 38. Wibbenmeyer J. A., Xavier K. A., Smith-Gill S. J., Willson R. C. (1999) Cloning, expression, and characterization of the Fab fragment of the anti-lysozyme antibody HyHEL-5. Biochim. Biophys. Acta 1430, 191–202 [DOI] [PubMed] [Google Scholar]
- 39. Welfle K., Misselwitz R., Höhne W., Welfle H. (2003) Interaction of epitope-related and -unrelated peptides with anti-p24 (HIV-1) monoclonal antibody CB4-1 and its Fab fragment. J. Mol. Recognit. 16, 54–62 [DOI] [PubMed] [Google Scholar]
- 40. Eryilmaz E., Janda A., Kim J., Cordero R. J., Cowburn D., Casadevall A. (2013) Global structures of IgG isotypes expressing identical variable regions. Mol. Immunol. 56, 588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sagawa T., Oda M., Morii H., Takizawa H., Kozono H., Azuma T. (2005) Conformational changes in the antibody constant domains upon hapten-binding. Mol. Immunol. 42, 9–18 [DOI] [PubMed] [Google Scholar]
- 42. Sela-Culang I., Alon S., Ofran Y. (2012) A systematic comparison of free and bound antibodies reveals binding-related conformational changes. J. Immunol. 189, 4890–4899 [DOI] [PubMed] [Google Scholar]


