Background: The role of S100A14 in tumorigenesis and the underlying mechanisms have not been fully understood.
Results: S100A14 binds HER2 (human epidermal growth factor receptor 2) and modulates HER2 phosphorylation and HER2-stimulated cell proliferation.
Conclusion: S100A14 acts as a functional partner of HER2.
Significance: These findings provide mechanistic evidence for S100A14 in breast cancer progression.
Keywords: Breast Cancer, Cell Proliferation, Protein Phosphorylation, Protein-Protein Interactions, S100 Proteins
Abstract
HER2 is overexpressed in 20–25% of breast cancers. Overexpression of HER2 is an adverse prognostic factor and correlates with decreased patient survival. HER2 stimulates breast tumorigenesis via a number of intracellular signaling molecules, including PI3K/AKT and MAPK/ERK. S100A14, one member of the S100 protein family, is significantly associated with outcome of breast cancer patients. Here, for the first time, we show that S100A14 and HER2 are coexpressed in invasive breast cancer specimens, and there is a significant correlation between the expression levels of the two proteins by immunohistochemistry. S100A14 and HER2 are colocalized in plasma membrane of breast cancer tissue cells and breast cancer cell lines BT474 and SK-BR3. We demonstrate that S100A14 binds directly to HER2 by co-immunoprecipitation and pull-down assays. Further study shows that residues 956–1154 of the HER2 intracellular domain and residue 83 of S100A14 are essential for the two proteins binding. Moreover, we observe a decrease of HER2 phosphorylation, downstream signaling, and HER2-stimulated cell proliferation in S100A14-silenced MCF-7, BT474, and SK-BR3 cells. Our findings suggest that S100A14 functions as a modulator of HER2 signaling and provide mechanistic evidence for its role in breast cancer progression.
Introduction
HER2 (human epidermal growth factor receptor 2), also known as Neu, ErbB2, or p185, is a transmembrane protein that is encoded by the ERBB2 gene in humans. It belongs to the epidermal growth factor receptor (EGFR3/ErbB) family, which contains four family members: EGFR (HER1, ErbB1), HER2 (ErbB2, HER2/neu), HER3 (ErbB3), and HER4 (ErbB4). The EGFR family is involved in regulating cell proliferation, survival, and differentiation through interlinked signal transduction involving hyperactivation of the PI3K/AKT and MAPK/ERK pathways (1). Amplification or overexpression of the HER2 gene occurs in ∼20–25% of breast cancers and is associated with more aggressive disease and a worse outcome (2). HER2 is a typical receptor tyrosine kinase, comprising an extracellular domain (ECD), a single transmembrane region, and an intracellular domain (ICD) (3). HER2 is unique among the ErbB receptors in that it does not bind a ligand directly but is preferentially recruited as a binding partner into heterodimers (4).
S100 is a family of low molecular weight proteins that contains more than 20 family members and comprises the largest subfamily of EF-hand Ca2+-binding proteins (5). S100 proteins are composed of two EF-hand calcium-binding domains: the N-terminal domain (also known as the S100 hand) and the C-terminal domain (also known as the canonical EF-hand) (6). S100 proteins interact with several targets, such as RAGE, p53, CacyBP/BP, Jab-1, and matrix metalloproteinases, and regulate Ca2+ homeostasis, protein phosphorylation, and degradation, thereby affecting cell proliferation and metastasis and many other biological events (5, 7).
S100A14 is a recently identified member of the S100 protein family. Differential expression of S100A14 has been reported in a variety of cell types and is overexpressed in certain types of tumors, such as lung, breast, and uterus, but underexpressed in some other tumors like colon, kidney, and rectal tumors (8). The heterogenic expression of S100A14 may indicate different and potentially tissue-specific roles. Down-regulated S100A14 expression was correlated with poor differentiation and simultaneous S100A14 underexpression, and S100A4 overexpression was correlated with high colorectal cancer metastatic potential (9). S100A14 was identified as a potential novel marker of breast cancer cells capable of predicting distant metastasis (10) and was found to be useful for detection and characterization of circulating tumor cells in peripheral blood from patients with colorectal, prostate, and breast cancers (11). Moreover, S100A14 was significantly associated with clinical outcome of breast cancer patients. Our previous studies showed that S100A14 requires functional p53 to affect MMP2 transcription (12). We also found that S100A14 could be secreted from stably overexpressing S100A14 of EC9706 cells, and low doses of extracellular S100A14 stimulate cell proliferation and promote survival in KYSE180 cells, but a high dose of S100A14 causes apoptosis via the mitochondrial pathway (13).
We have previously shown that S100A14 is a new target for p53 and could affect p53 transactivity and stability, and S100A14 affects cell invasiveness by regulating MMP2 transcription in a p53-dependent manner (12). In the present study, we demonstrate for the first time that there is a strong correlation between S100A14 and HER2 expression in breast cancer tissues, and S100A14 can interact with HER2 by co-immunoprecipitation and pull-down assays. Further study revealed that residues 956–1154 of the HER2 intracellular domain and residue 83 of S100A14 are essential for the two proteins binding. Furthermore, we found that S100A14 plays an important role in the HER2-induced cell proliferation of MCF-7, BT474, and SK-BR3 cells through interaction with HER2 and regulation of pERK and pAKT. This study provides further insights into the HER2 signaling pathway.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
HEK 293T/17 and MCF-7 cells were obtained from the American Type Culture Collection (Manassas, VA). SK-BR3, MDA-MB-453, and BT474 cells were obtained from the National Platform of Experimental Cell Resources for Sci-Tech (Beijing, China). HEK 293T/17 and MCF-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/ml streptomycin, and 100 units/ml penicillin. SK-BR3 and BT474 cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 units/ml streptomycin, and 100 units/ml penicillin. MDA-MB-453 cells were maintained in L15 supplemented with 10% fetal bovine serum, 100 units/ml streptomycin, and 100 units/ml penicillin. The medium was changed at alternate days, and cells were split before they reached confluence.
Patient Tissue Samples
Tissue specimens from 74 patients with invasive ductal carcinoma were analyzed. Patients were consecutively recruited at the Chinese Academy of Medical Sciences Cancer Hospital (Beijing). At recruitment, informed consent was obtained from each subject. This study was approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Institute and Hospital.
Immunohistochemistry
Immunohistochemistry staining was performed as described previously (13). Briefly, the tissues embedded in paraffin were sectioned and mounted on slides, deparaffinized in xylene, and rehydrated through a series of ethanol-water solutions. Antigen retrieval was carried out by immersing the sections in citrate acid buffer and heating using a pressure cooker for 2 min at full pressure, and then nonspecific activity was blocked sequentially with 3% hydrogen peroxide. Sections were incubated overnight at 4 °C with antibodies specific to S100A14 (1:100) (gift from Dr. Iver Petersen) and HER2 (ZA-0023, Beijing Golden Bridge Biotechnology Company, Beijing, China). Immunohistochemical analysis was performed using the PV-9000 polymer detection system for immunohistochemical staining kit (Beijing Golden Bridge Biotechnology Company, Beijing, China). 3,3′-Diaminobenzidine was used to visualize the reaction, followed by counterstaining with hematoxylin. Images were visualized and annotated with the ImageScope software (Aperio Technology), and the number of positive cells at ×200 magnification was quantified. A total of 4–6 fields/slide were selected, counted, and averaged. Expression score was determined by staining intensity and immunoreactive cell percentage. No staining was taken as 0, a weak positive staining as 1, a positive staining as 2, and a strong positive staining as 3. Two groups were divided according to the ratio of matched cancer/normal cells (a ratio of ≤1 was taken as the non-overexpressed group, and a ratio of >1 was taken as the overexpressed group). Representative areas of each section were selected.
Plasmids and Transfection
To construct FLAG-tagged ECD, ICD of HER2, and HER2-ICD-1 to -4, PCR was performed using plasmid pcDNA3-Zeo(+)-ErbB2 as a template which was a generous gift plasmid from Dr. Espen Stang (Institute of Pathology, Faculty of Medicine, University of Oslo, Norway). The ECD of HER-2, corresponding to amino acids 1–636, and the ICD of HER-2, corresponding to amino acids 676–1255, were subcloned into pcDNA3 FLAG-2Ab. The HER2-ICD-1 to -4 were subcloned into p3×FLAG-CMV-14 (Sigma-Aldrich). The S100A14 was subcloned into pEBG. The resulting construct was verified by direct sequencing. The plasmid pQE30-Myo117, with a size similar to that of S100A14, was a generous gift from Dr. Mariam Grigorian (Institute of Cancer Biology, Copenhagen, Denmark). Point mutation of S100A14 was made by a site-directed mutation kit (Saibaisheng, Shanghai, China). Primers used are listed in Table 1.
TABLE 1.
Primers and siRNAs used in this study
| Primer/siRNA | Sequence |
|---|---|
| Primers for plasmid construction | |
| FLAG-HER2-F | ttaagcttgccaccatgATGGAGCTGGCGGCCTTGTG |
| FLAG-HER2-R | ttgcggccgcCACTGGCACGTCCAGACCCA |
| HER2-ECD-F | ttaagcttATGGAGCTGGCGGCCTTGTG |
| HER2-ECD-R | ttgcggccgcGTCCACACAGGAGTGGGT |
| HER2-ICD-F | ttaagcttAAGCGACGGCAGCAGAA |
| HER2-ICD-R | ttgcggccgcCACTGGCACGTCCAGAC |
| HER2-ICD-1-F | ttaagcttgccaccatgAAGCGACGGCAGCAGAAGAT |
| HER2-ICD-1-R | ttgcggccgcGTCTGTAATTTTGACATGGT |
| HER2-ICD-2-F | ttaagcttgccaccatgGAAGCATACGTGATGGCTGG |
| HER2-ICD-2-R | ttgcggccgcTTTGACCAT GATCATGTAG |
| HER2-ICD-3-F | ttaagcttgccaccatgGTCAAATGTTGGATGATT |
| HER2-ICD-3-R | ttgcggccgcGGGCGAAGGGGGCTGGGGCCG |
| HER2-ICD-4-F | ttaagcttgccaccatgCGAGAGGGCCCTCTG |
| HER2-ICD-4-R | ttgcggccgcCACTGGCACGTCCAGACCCA |
| GST-S100A14-F | ttggatccATGGGACAGTGTCGGTCAGC |
| GST-S100A14-R | ataagatgcggccgcTCAGTGCCCCCGGACAGGCC |
| S100A14-Δ1-F | ttggatccATGGGACAGTGTCGGTCAGC |
| S100A14-Δ1-R | ataagatgcggccgcTCAGAAACTCCTGAACTCCAGTT |
| S100A14-Δ2-F | ttggatccCCTTCTGAGCTACGGGACCTGG |
| S100A14-Δ2-R | ataagatgcggccgcTCAGTGCCCCCGGACAGGCC |
| S100A14-Δ3-F | TTTCACCAGTACCCTTCTGAGCTA |
| S100A14-Δ3-R | TAGCTCAGAAGGGTACTGGTGAAA |
| S100A14-Δ4-F | ATTGCCAACCTGAGGAGTTTCTGG |
| S100A14-Δ4-R | CCAGAAACTCCTCAGGTTGGCAAT |
| D76A-F | CAGCTGCAATGCCTCTAAACTGG |
| D76A-R | CCAGTTTAGAGGCATTGCAGCTG |
| F81A-F | TAAACTGGAGGCCAGGAGTTTCT |
| F81A-R | AGAAACTCCTGGCCTCCAGT TTA |
| S83R-F | GGAGTTCAGGAGATTCTGGGAGC |
| S83R-R | GCTCCCAGAATCTCCTGAACTCC |
| F81AS83R-F | GGAGGCCAGGAGATTCTGGGAGC |
| F81AS83R-R | GCTCCCAGAATCTCCTGGCCTCC |
| siRNAs | |
| siControla | |
| siS100A14b | 1) GCUGACCCCUUCUGAGCUA |
| 2) CAGAGGAUGCUCAGGAAUU | |
| 3) GAGAACUCCCUCUGGAAUU | |
| 4) UCAAGAACUUUCACCAGUA | |
a Dharmacon ONTARGETplus non-targeting siRNA (D-001810-10-20).
b Dharmacon Non-TARGETplus siRNA.
Plasmid transfection was performed as described previously (10). ON-TARGETplus pool siRNAs (L-010723-00-0005, Dharmacon) (25 nm) were transfected by DharmaFECT 1 (T-2001-02, Dharmacon) following the manufacturers' protocol. The sequences for siRNAs were listed in Table 1.
Protein Expression and Purification
Expression and purification of the human recombinant S100A14 and Myo117 protein in Escherichia coli was performed as described (14). After dialysis against PBS (pH 7.4) containing 0.1% Triton X-100 overnight at 4 °C using dialysis membrane (Spectrum), the protein was concentrated using PEG 20000 and was equalized for protein concentration using the BCA protein assay (23227, Thermo Scientific).
Immunofluorescence
Cells were grown at low density on sterilized coverslips for 24 h. Cell sections were washed with PBS buffer three times and fixed in 4% paraformaldehyde for 15 min and processed for the immunofluorescence procedure as described previously (15). Primary antibodies used were rabbit anti-S100A14 (1:50) (gift from Dr. Iver Petersen) and mouse anti-HER2 (1:100) (sc-08, Santa Cruz Biotechnology, Inc., Dallas, TX).
Co-Immunoprecipitation and Pull-down Assays
The plasmids were cotransfected into HEK 293T/17 cells, and 48 h after transfection, co-immunoprecipitation was performed essentially as described previously (16), the empty vector was used as a control plasmid. In brief, equal amounts of precleared cellular lysates were incubated with anti-FLAG® M2 affinity gel (A2220, Sigma-Aldrich) or glutathione-Sepharose (GSH) beads (17-5279-01, GE Healthcare) overnight at 4 °C. The immunocomplexes were then washed three times with lysis buffer and analyzed by Western blotting. Nickel-nitrilotriacetic acid pull-down assays were performed as described previously (17). Purified S100A14 protein (10 μg) was incubated with His-resin beads (P6611, Sigma-Aldrich) at 4 °C for 1 h and then incubated with MDA-MB-453 cell lysates overnight at 4 °C. Bound proteins were eluted with 2× SDS sample buffer and analyzed by Western blotting. The protein Myo117, with a size similar to that of S100A14, was used as a control. In the direct interaction assay, the His-tagged HER2-ICD-1 to -4 were expressed in BL21 cells and washed twice with the wash buffer (1 m NaCl, 50 mm Na2HPO4·12H2O, 50 mm imidazole) and then incubated with purified GST-tagged S100A14 for 24 h. After washing twice with the wash buffer, the samples were detected by Western blotting.
Measurement of HER2 Phosphorylation and Signaling
MCF-7, BT474, and SK-BR3 cells were transiently transfected with ON-TAGETplus S100A14 pool siRNAs or ON-TAGETplus non-targeting siRNAs; 24 h after transfection, the plasmid expressing FLAG or FLAG-HER2 was transfected into the cells; and 48 h after transfection, cell monolayers were placed on ice, washed twice with ice-cold PBS, and lysed in 100 μl/well lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 50 mm NaF, 0.01 mm Na3VO4) supplemented with EDTA-free protease inhibitor mixture (11873580001, Roche Applied Science). Clarified cell lysates were equalized for protein concentration using the BCA protein assay, resolved by SDS-PAGE, and processed by Western blotting. HER2, AKT, and ERK phosphorylation were measured by probing blots with phospho-specific HER2 (Tyr-1248, 06-229, Millipore), AKT (Ser-473, sc-33427, Santa Cruz Biotechnology, Inc.), and ERK (Tyr-204, sc-7383, Santa Cruz Biotechnology, Inc.) antibodies, respectively. The blots were then reprobed with antibodies against HER2 (06-562, Millipore), AKT (9272, Cell Signaling), and ERK (sc-94, Santa Cruz Biotechnology) after stripping. All blots were also probed with anti-S100A14 antibodies and anti-β-actin antibody (A5316, Sigma-Aldrich).
Measurement of HER2-stimulated Cell Growth
MCF-7, BT474, and SK-BR3 cells were plated in 96-well plates at a density of 4 × 103 cells/well and then transfected as previously described. After transfection, MTS assays (G3580, Promega) were performed as described previously (18) for 4 days.
Statistical Analysis
Data were presented as mean ± S.D., and Student's t test analysis was performed using SPSS version 17.0 software. The correlation between S100A14 and HER2 was assessed using Spearman correlation analysis. Statistical significance was set at p < 0.05.
RESULTS
Expression of S100A14 and HER2 in Breast Cancer Tissues
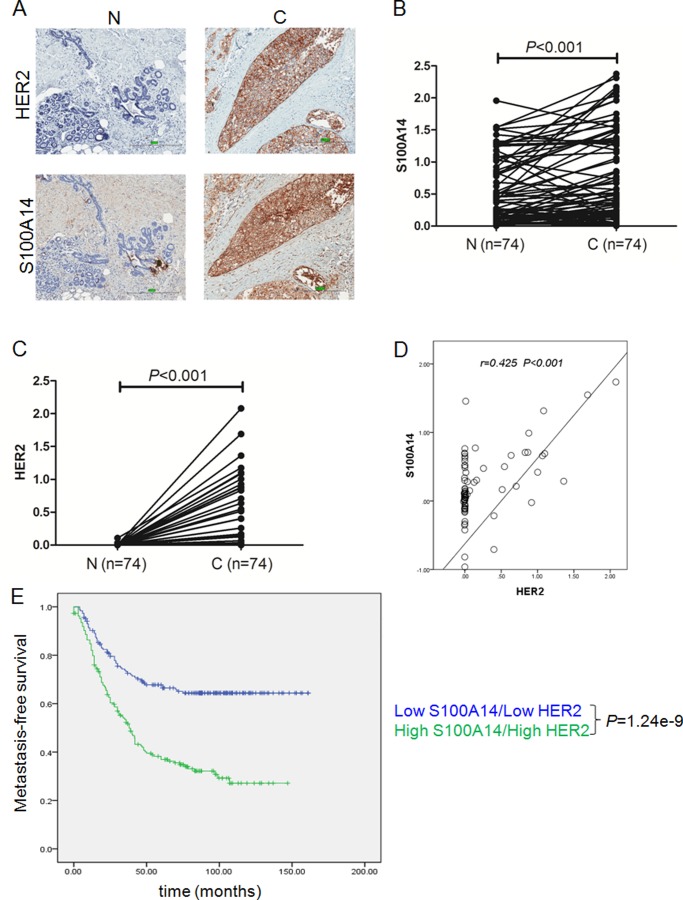
The expression of HER2 (Fig. 1A, top panels) and S100A14 (Fig. 1A, bottom panels) were analyzed in 74 formalin-fixed, paraffin-embedded breast cancer specimens and their adjacent normal tissues by immunohistochemistry staining. Quantitative analysis of the immunostaining sections showed that the immunostaining for S100A14 (Fig. 1B) and HER2 (Fig. 1C) in tumor tissues was significantly higher than that in the adjacent normal tissues. No statistical significance was found between S100A14/HER2 expression level and estrogen receptor, progesterone receptor, or lymph node metastasis status (Tables 2 and 3). However, there is a significant association (r = 0.425, p = 1.67e−4) between the expression levels of HER2 and S100A14 in the tissues (Fig. 1D). Interestingly, in most breast carcinoma specimens, S100A14 displayed prominent plasma membrane, which is similar to cellular distribution of HER2 (13). To further analyze the correlation between S100A14/HER2 expression levels and clinicopathological features, we combined four microarray data sets (GSE2034, GSE2603, GSE5327, and GSE12776) downloaded from the GEO database according to the literature (19, 20). Ultimately 630 breast tumors from genome-wide gene expression data were available. As expected, we found that there was a positive correlation between S100A14 and HER2 mRNA expression (p = 5.73e−7). Then patients were divided into two groups (high or low), depending upon the median S100A14 or HER2 expression level. Patients were further stratified into the S100A14 low/HER2 low group and S100A14 high/HER2 high group. Next, we investigated the correlation between the expression of S100A14/HER2 and clinicopathological features using these data. Notably, we found a significant correlation between S100A14/HER2 gene expression signature with tumor stage (p = 0.008), relapse (p = 1.63e−9), and metastasis-free survival time (p = 1.24e−9, log rank test). Kaplan-Meier survival analyses demonstrated that S100A14/HER2 was inversely correlated with metastasis-free survival time (Fig. 1E). These results strongly support a potential role for S100A14/HER2 in breast cancer metastasis and its predictive capability for prognosis.
FIGURE 1.
Overexpression of S100A14/HER2 and their correlation with metastasis-free survival in breast cancer. Shown is immunohistochemistry analysis of a primary invasive ductal carcinoma specimen stained with anti-HER2 antibody (A, top panels) or anti-S100A14 antibody (A, bottom panels) (bar, 300 μm). N, normal; C, cancer. B and C, determination of the percentage of positive cell staining was performed by ImageScope software (Aperio). Both HER2 and S100A14 staining were significantly increased in breast cancer. D, Spearman correlation analysis showed that there was a significant positive correlation between the expression of S100A14 and HER2 (n = 74, r = 0.425, p = 1.67e−4). E, Kaplan-Meier analysis for metastasis-free survival of 598 patients with breast cancer in the GEO database (GSE12276, GSE2034, GSE2603, and GSE5327). Patients were divided into two groups based on the median value for S100A14/HER2. The log-rank test p values are shown.
TABLE 2.
Comparison of expression levels of S100A14 with clinicopathological features in patients with invasive ductal carcinoma
For S100A14 expression levels, a matched cancer/normal ratio of ≤1 was taken as the non-overexpressed group, and a ratio of >1 was taken as the overexpressed group. +, positive; −, negative.
| Characteristics | Cases | S100A14 |
p | |
|---|---|---|---|---|
| Non-overexpression | Overexpression | |||
| % | ||||
| Age (years) | ||||
| >50 | 29 (39.2) | 9 (31.0) | 20 (69.0) | 0.533 |
| ≤50 | 45 (60.8) | 11 (24.4) | 34 (75.6) | |
| Estrogen receptor | ||||
| + | 54 (73.0) | 14 (25.9) | 40 (74.1) | 0.442 |
| − | 20 (27.0) | 7 (35.0) | 13 (65.0) | |
| Progesterone receptors | ||||
| + | 48 (64.9) | 12 (25.0) | 36 (75.0) | 0.381 |
| − | 26 (35.1) | 9 (34.6) | 17 (65.4) | |
| HER2 | ||||
| + | 50 (67.6) | 11 (22.0) | 39 (78.0) | 0.036a |
| − | 24 (32.4) | 11 (45.8) | 13 (54.2) | |
| Lymph node metastasis | ||||
| + | 34 (45.9) | 9 (26.5) | 25 (73.5) | 0.921 |
| − | 40 (54.1) | 11 (27.5) | 29 (72.5) | |
a Statistical significance (p < 0.05) by the Pearson χ2 test.
TABLE 3.
Comparison of expression levels of HER2 with clinicopathological features in patients with invasive ductal carcinoma
For HER2 expression levels, a matched cancer/normal ratio of ≤1 was taken as the non-overexpressed group, and a ratio of >1 was taken as the overexpressed group. +, positive; −, negative.
| Characteristics | Cases | HER2 |
p | |
|---|---|---|---|---|
| Non-overexpression | Overexpression | |||
| % | ||||
| Age (years) | ||||
| >50 | 29 (39.2) | 12 (41.4) | 17 (58.6) | 0.527 |
| ≤50 | 45 (60.8) | 22 (48.9) | 23 (51.1) | |
| Estrogen receptor | ||||
| + | 54 (73.0) | 24 (44.4) | 30 (55.6) | 0.670 |
| − | 20 (27.0) | 10 (50.0) | 10 (50.0) | |
| Progesterone receptors | ||||
| + | 48 (64.9) | 21 (43.8) | 27 (56.2) | 0.843 |
| − | 26 (35.1) | 12 (46.2) | 14 (53.8) | |
| Lymph node metastasis | ||||
| + | 34 (45.9) | 15 (44.1) | 19 (55.9) | 0.771 |
| − | 40 (54.1) | 19 (47.5) | 21 (52.5) | |
Colocalization of S100A14 and HER2
The function of protein is usually related to its subcellular localization (21). A previous study (22) showed that transient transfected autofluorescent protein-tagged S100A14 was localized in the plasma membrane in both MDA-MB-468 and T-47D cancer cells. This, together with a similar expression pattern of S100A14/HER2 in breast cancer tissues, prompted us to further investigate whether S100A14 expression is colocalized with HER2 in breast cancer cells. We examined the expression and subcellular localization of human S100A14 and HER2 in two S100A14-expressing breast cancer cell lines (BT474 and SK-BR3) using confocal microscopy. Both S100A14 and HER2 were predominantly localized at the membrane, although S100A14 was partially localized in the cytoplasmic compartment. Digital merging of confocal microscopic images of S100A14 (red) and HER2 (green) depicted a considerable colocalization (yellow) of these proteins in BT474 (Fig. 2A) and SK-BR3 (Fig. 2B) cells.
FIGURE 2.
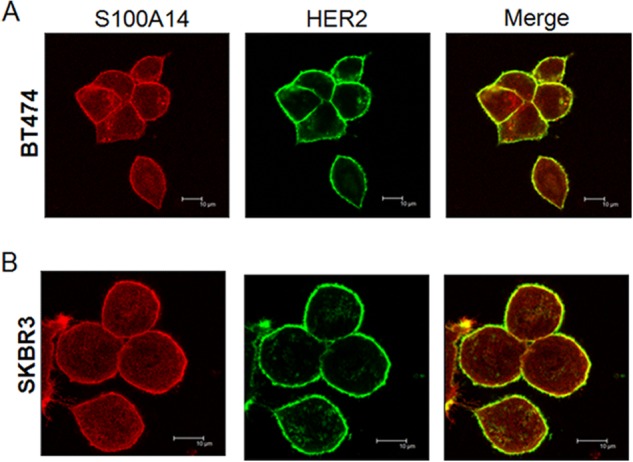
Confocal analysis showing the colocalization of HER2 and S100A14 in BT474 (A) and SK-BR3 (B) cells. Cells were stained with HER2 antibody (green) or S100A14 (red). Merged images are shown on the right. Representative cells are shown in the confocal micrographs. Bars, 10 μm.
S100A14 Binds HER2
To determine whether the colocalization of S100A14 and HER2 can lead to the interaction in BT474 and SK-BR3 cells, we lysed BT474 cells or SK-BR3 cells and incubated the lysate with anti-S100A14 antibody or IgG in the presence of either 0.5 mm Ca2+ or 1 mm EGTA. The results showed that the interaction between endogenous S100A14 and HER2 is weak. However, the interaction is dramatically enhanced by calcium treatment and completely inhibited by EGTA treatment (Fig. 3, A and B). Based on these results, the interaction of S100A14 with HER2 is specific and apparently Ca2+-dependent.
FIGURE 3.
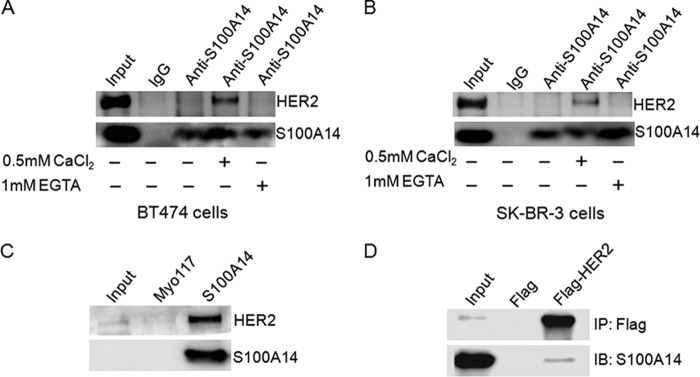
S100A14 associates with HER2. A and B, S100A14 binds HER2 in a Ca2+-dependent manner. Cell lysates from BT474 (A) or SK-BR3 (B) were incubated with anti-S100A14 antibody or IgG in the presence of either Ca2+ or EGTA. The samples were examined by Western blotting using anti-S100A14 antibody or anti-HER2 antibody. C, nickel-nitrilotriacetic acid pull-down assays were performed by incubation of immobilized His-tagged recombinant Myo117 or immobilized His-tagged S100A14 with MDA-MB-453 cell lysates. Bound S100A14 and HER2 were detected by Western blotting using specific antibodies. D, co-IP experiments in HEK 293T/17 cells. FLAG-HER2 and GST-S100A14 were cotransfected into HEK 293T/17 cells as indicated, and cell lysates were then immunoprecipitated using anti-FLAG® M2 affinity gel. The immunoprecipitates were examined by Western blotting (IB) using specific antibodies. Input represented 5% of cell lysates used in the co-IP experiment.
Next, we further confirmed whether S100A14 and HER2 were capable of interacting with each other. To do this, we lysed MDA-MB-453, a malignant human breast epithelial cell line that overexpresses HER2, and incubated the lysate with affinity-purified His6-tagged S100A14 (His-S100A14) and subsequently performed Western blotting analysis with anti-HER2. HER2 can be detected in precipitated complex, suggesting that S100A14 can interact with HER2 (Fig. 3C). The specificity of the binding is validated by the absence of His-S100A14 from samples that were incubated with control Myo117 protein (recombinant 117-amino acid fragment of heavy chain of non-muscle myosin). To further confirm the above results, we cotransfected FLAG-HER2 plasmid or control vector with GST-S100A14 plasmid into HEK 293T/17 cells and performed a co-immunoprecipitation assay with anti-FLAG® M2 affinity gel. S100A14 could be detected in FLAG-HER2 and not FLAG-precipitated complex by Western blotting (Fig. 3D). Collectively, these data reveal an interaction between S100A14 and HER2.
Identification of the Residues in HER2 or S100A14 Important for S100A14/HER2 Interaction
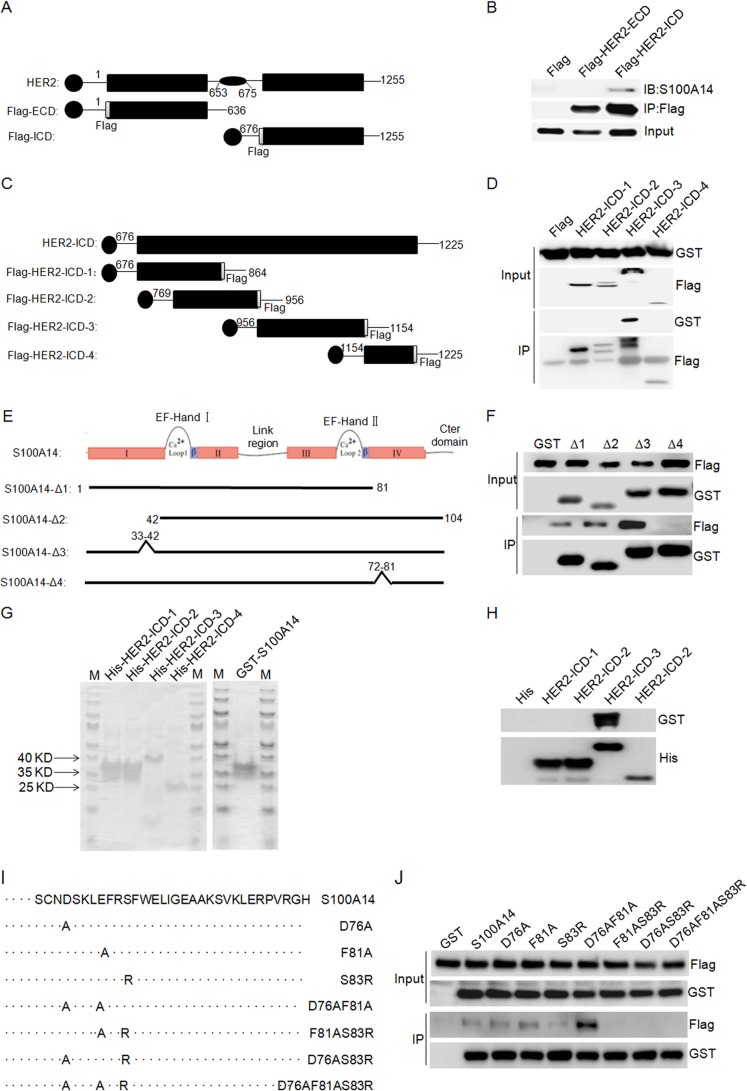
To identify the region of HER2 to which S100A14 binds, we performed transient transfection experiments in HEK 293T/17 cells with FLAG-tagged ECD (extracellular domain, residues 1–636) and ICD (intracellular domain, residues 676–1255) of HER2 and GST-tagged S100A14 and then examined the interaction by co-immunoprecipitation assays. Lysates of these cells were immunoprecipitated with anti-FLAG® M2 affinity gel, and the precipitates were analyzed by immunoblotting with anti-S100A14 antibody. An immunoreactive band recognized by the anti-S100A14 antibody was observed in the anti-FLAG immunoprecipitates from cells coexpressing FLAG-HER2-ICD, but no interaction between S100A14 and FLAG-HER2-ECD (residues 1–636) was detected (Fig. 4, A and B). The amount of HER2 in each sample was essentially identical. These data suggest that the region of HER2 ICD containing residues 676–1255 is necessary for S100A14 binding. To narrow the S100A14 binding site, we constructed four HER2-ICD truncation mutants (HER2-ICD-1 (residues 676–864), HER2-ICD-2 (residues 769–956), HER2-ICD-3 (residues 956–1154), and HER2-ICD-4 (residues 1154–1255) (Fig. 4C)) and examined their interaction with S100A14. As shown in Fig. 4D, only the region of HER2 containing residues 956–1154 (HER2-ICD-3) is necessary for binding with S100A14 (Fig. 4D); no interaction between S100A14 and other HER2-ICD truncations was detected.
FIGURE 4.
Mapping of the S100A14 binding site in HER2 and the HER2 binding site in S100A14. A, schematic overview of HER2 ECD and ICD that were tested for S100A14 binding. The specific residues present in each fragment are indicated. HER2-ECD, HER2 extracellular domain (residues 1–636); HER2-ICD, HER2 intracellular domain (residues 676–1255). B, HER2 truncation mutants bind to immobilized S100A14. The indicated FLAG-HER2 truncations and GST-S100A14 were cotransfected into HEK 293T/17 cells as indicated, and cell lysates were then immunoprecipitated using anti-FLAG® M2 affinity gel. The immunoprecipitates were examined by Western blotting (IB) using anti-S100A14 antibody. Input represented 5% of cell lysates used in the co-IP experiment. C, schematic representation of HER2 constructs depicting the HER2 intracellular domain and HER2 intracellular domain fragments. The specific residues present in each fragment are indicated: HER2-ICD-1 (residues 676–864), HER2-ICD-2 (residues 769–956), and HER2-ICD-3 (residues 956–1154), HER2-ICD-4 (residues 1154–1255). D, HER2-ICD truncation mutants bind to immobilized S100A14. The indicated FLAG-HER2-ICD truncations and GST-S100A14 were cotransfected into HEK 293T/17 cells as indicated, and cell lysates were then immunoprecipitated using anti-FLAG® M2 affinity gel. The immunoprecipitates were examined by Western blotting using anti-S100A14 antibody. Input represented 5% of cell lysates used in the co-IP experiment. E, schematic overview of S100A14 truncation mutants that were tested for HER2 binding. S100A14-Δ1, -Δ2, -Δ3, and -Δ4, S100A14 with residues 82–104, 1–41, 33–42, and 72–81 deleted, respectively. F, S100A14 truncation mutants bind to immobilized HER2. The indicated GST-S100A14 truncations and FLAG-HER2 were cotransfected into HEK 293T/17 cells as indicated, and cell lysates were then immunoprecipitated using glutathione-Sepharose (GSH) beads. The immunoprecipitates were examined by Western blotting using anti-FLAG antibody. Input represented 5% of cell lysates used in the co-IP experiment. G, purified recombinant proteins used in the direct interaction assay. Approximately 0.2 μg of each protein was separated by SDS-PAGE and stained with Coomassie Brilliant Blue. M, molecular weight markers. H, direct interaction between GST-tagged S100A14 and His-tagged HER2 intracellular domain fragments. I, overview of the different S100A14 mutants that were tested for HER2 binding. S100A14 mutants containing point mutations in helix 4 (D76A, F81A, S83R, D76A/F81A, F81A/S83R, D76A/S83R, and D76A/F81A/S83R) are shown. J, amino acid residue Ser-83 of S100A14 is essential for HER2 binding. GST-S100A14 and corresponding mutants and FLAG-HER2 were cotransfected into HEK 293T/17 cells as indicated, and cell lysates were immunoprecipitated using glutathione-Sepharose (GSH) beads. The immunoprecipitates were examined by Western blotting using anti-FLAG antibody. Input represented 5% of cell lysates used in the co-IP experiment.
S100 proteins usually contain two EF-hand motifs. The N-terminal EF-hand comprises helix I, pseudo-calcium-binding site I, and helix II, separated by a flexible linker from the C-terminal EF-hand that includes helix III, calcium-binding site II, and helix IV. Also, the EF-hand domain is critical for the interaction with target proteins (5). In order to identify the region of S100A14 to which HER2 binds, we individually deleted each region from GST-S100A14 (Fig. 4E) and found that only deletion of the C-terminal EF-hand markedly attenuates the interaction of HER2 with S100A14 (Fig. 4F). These data confirmed that the C-terminal EF-hand of S100A14 is necessary for binding HER2. To further verify that S100A14 can directly interact with HER2, we purified S100A14 and four HER2-ICD truncation mutant proteins (HER2-ICD-1 (residues 676–864), HER2-ICD-2 (residues 769–956), HER2-ICD-3 (residues 956–1154), and HER2-ICD-4 (residues 1154–1255)) in a prokaryotic expression system (Fig. 4G). Next, we confirmed the direct interaction between HER2 and S100A14. Consistent with the results from in vivo co-IP, only the interaction between HER2-ICD-3 and S100A14 was detected. These results further confirmed that S100A14 can directly interact with HER2, and residues 956–1154 of HER2 are necessary for the interaction with S100A14 (Fig. 4H). To further define residues critical for HER2 binding, site-directed mutagenesis was performed on the relatively conserved residues of S100A14, and seven mutants (D76A, F81A, S83R, D76A/F81A, F81A/S83R, D76A/S83R, and D76A/F81A/S83R) of S100A14 were obtained (Fig. 4I). Among these mutants, the S83R mutant of S100A14 exhibited a drastically reduced interaction with HER2 (Fig. 4J), suggesting that amino acid residue Ser-83 of S100A14 is required for the interaction with HER2.
S100A14 Regulates HER2 Phosphorylation and Signaling and HER2-stimulated Cell Growth
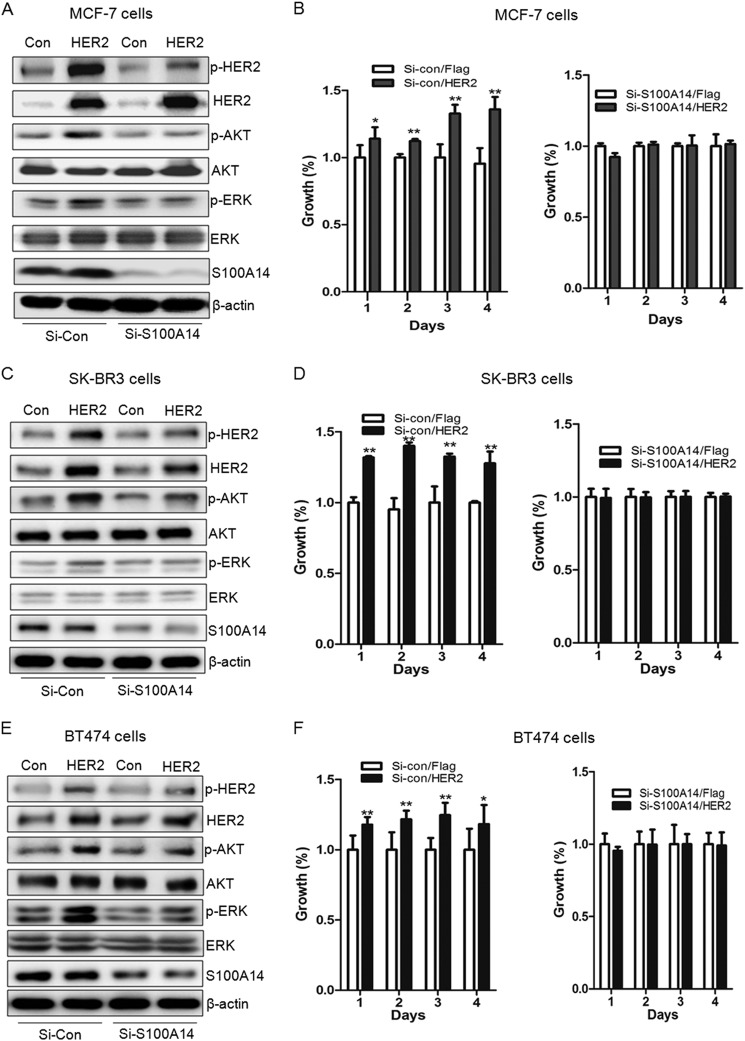
HER2 overexpression can lead to the formation of HER2 homodimer, which may transduce oncogenic signal, such as the PI3K/AKT and Ras/Raf/MEK/MAPK pathway, in the absence of specific ligands (18, 21). To test whether S100A14 is involved in the HER2 signaling pathway, siRNAs against S100A14 were transfected into MCF-7, SK-BR3, and BT474 cells, and 24 h after transfection, the plasmid expressing FLAG or FLAG-HER2 was transfected into these cells. HER2 expression, phosphorylation, and signaling were evaluated by Western blotting. As shown in Fig. 5A, knockdown of S100A14 significantly attenuates pHER2 levels, and there are no accompanying significant changes in total HER2. To evaluate a possible role for S100A14 in HER2 signaling, the activation status of the PI3K/AKT and MAPK pathways was determined. Knockdown of S100A14 significantly decreases pAKT and pERK levels (Fig. 5A). Similar results are also observed in SK-BR3 and BT474 cells (Fig. 5, C and E). Overall, inhibition of S100A14 induces a sustained blockade of the HER2 signaling pathway.
FIGURE 5.
Knockdown of S100A14 reduces HER2 phosphorylation and downstream signaling and HER2-stimulated cell proliferation. A, MCF-7 cells were transiently transfected with the ON-TARGETplus S100A14 pool siRNA or ON-TARGETplus non-targeting siRNA, and 24 h after transfection, the plasmid expressing FLAG or FLAG-HER2 was transfected into the cells. Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and probed with anti-S100A14, anti-pHER2, anti-HER2, anti-pAKT, anti-AKT, anti-pERK, anti-ERK, and anti-β-actin antibodies. B, 24 h after ON-TARGETplus S100A14 pool siRNA or ON-TARGETplus non-targeting siRNA transfection, the plasmid expressing FLAG or FLAG-HER2 was transfected into the cells, and the viability of MCF-7 cells was measured using an MTS assay. *, p < 0.05; **, p < 0.01. C, siRNAs and plasmids were transfected into SK-BR3 cells according to the above method. The levels of pHER2, HER2, pAKT, AKT, pERK, ERK, and β-actin were detected. D, cell proliferation assay was performed in SK-BR3 cells. **, p < 0.01. E, siRNAs and plasmids were transfected into BT474 cells according to the above method. The levels of pHER2, HER2, pAKT, AKT, pERK, ERK, and β-actin were detected. F, cell proliferation assay was performed in BT474 cells. *, p < 0.05; **, p < 0.01. Error bars, S.D.
In HER2-positive breast tumors, activation of pHER2 stimulates cell cycle progression and uncontrolled cell proliferation (23). To evaluate whether S100A14 plays a functional role in HER2-induced cell growth, we compared HER2-stimulated cell growth in siS100A14- and siControl-transfected MCF-7, BT474, and SK-BR3 cells. The results demonstrated that overexpression of HER2 increases siControl-transfected MCF-7 cell growth (up to 36% at day 4) (Fig. 5B). In contrast, HER2 overexpression has no significant effect on siS100A14-transfected MCF-7 cell growth. These results strongly support a critical role for S100A14 in HER2-induced cell proliferation. We also found a similar function of S100A14 in HER2-stimulated cell growth in SK-BR3 cells (Fig. 5D) and BT474 cells (Fig. 5F).
DISCUSSION
HER2 amplification and overexpression have been reported in a number of human tumors, including breast cancer, ovarian cancer, gastric carcinoma, and salivary gland tumor (24). Overexpression and amplification of HER2 is commonly associated with increased tumor aggressiveness, high recurrence rates, and reduced survival (25). The S100 protein family is one of the largest subgroups of EF-hand calcium-binding proteins, and several members have been reported to be overexpressed and associated with tumor progression, prognosis, and survival (26–28). Our previous study showed that S100A14 is overexpressed in breast cancer and contributes to cell invasion by regulating MMP2 (matrix metalloproteinase 2) (12). Therefore, we first analyzed the correlation between S100A14/HER2 expression and clinicopathological features of breast cancer. However, no significant difference was found between the expression of S100A14/HER2 and clinicopathological features, such as lymph node metastasis, prognosis, etc., in the present study. This result may be explained by several reasons, such as the following. 1) The sample size used in this study are not large enough to estimate the results. 2) The value of HER2 overexpression in predicting lymph node metastasis is controversial. An association between HER2 overexpression and lymph node metastasis was found in some studies (29, 30), whereas this association was not found in other studies (31–35). 3) We cannot determine the correlation between S100A14/HER2 and prognosis because the specimens were collected recently, and the follow-up study is too short to draw conclusions on patient outcomes. To further verify the correlation between S100A14/HER2 expression levels and clinicopathological parameters in breast cancer, we used four microarray data sets to perform statistical analysis. Indeed, we found a significant correlation between S100A14/HER2 gene expression signature and tumor stage, relapse, and metastasis-free survival time. Kaplan-Meier survival analyses demonstrated that S100A14/HER2 was inversely correlated with metastasis-free survival time. These results strongly support a potential role of S100A14/HER2 in breast cancer metastasis and prognosis. Moreover, we found that there was a tight correlation between S100A14 and HER2. Consistently, our data also exhibited a strong correlation between S100A14 and HER2 in breast cancer specimens and a similar expression pattern of S100A14/HER2. This observation prompted us to further investigate the association between S100A14 and HER2. We demonstrated that S100A14 can bind HER2 by pull-down and co-immunoprecipitation assays and showed that residues 956–1154 located in the ICD of HER2 are essential for S100A14 binding, and residue 83 in the C-terminal EF-hand of S100A14 is critical for HER2 binding. Although no ligand has been identified for the HER2 receptor, it can interact reversibly with the other family members and form active heterodimers that govern cell proliferation and migration (24). Given that S100A14 binds to the intracellular domain of HER2 and HER2 is the preferred and most mitogenic dimerization partner for EGFR, HER3, and HER4 (1, 3, 4), we speculate that S100A14 may be involved in the homodimerization and/or heterodimerization of HER2. Further study will be carried out to verify this speculation.
Phosphorylation of HER2 plays an essential role in HER2 signaling (23). A number of studies have revealed that reducing HER2 phosphorylation inhibits activation of Ras/Raf/MAPK and PI3K pathways (36–40). In this study, we found that S100A14 knockdown significantly attenuated HER2 phosphorylation, thereby decreasing AKT and ERK phosphorylation that was induced by overexpression of HER2 in MCF-7, BT474, and SK-BR3 cells. Furthermore, S100A14 knockdown also significantly reduced HER2-stimulated cell proliferation. These results indicate that S100A14 acts as a functional partner of HER2. These results are in concordance with previous findings that several S100 family members are involved in the EGF/EGFR signaling pathway. For instance, extracellular S100A4 was found to interact with a variety of EGFR ligands and has the highest affinity for amphiregulin, and it stimulates EGFR/ErbB2 receptor signaling and enhances the amphiregulin-mediated proliferation of mouse embryonic fibroblasts (41). Also, levels of HER2 and S100A4 were tightly correlated in samples of primary medulloblastoma, and HER2 was found to up-regulate the expression of S100A4 in medulloblastoma cells (42). Previous literature has also demonstrated that EGF treatment significantly induces S100A7 expression and that S100A7 plays a functional role in the EGF-induced signaling pathway (43).
In summary, the present study revealed an important role of S100A14 in the HER2 signaling pathway. We have for the first time demonstrated that S100A14 can interact with HER2, and further study showed that residues 956–1154 of HER2 and residue 83 of S100A14 are essential for the two proteins binding. We found that S100A14 knockdown can attenuate the phosphorylation of HER2 and its downstream factors AKT and ERK and further reduce HER2-stimulated cell proliferation. Elucidating the mechanisms of S100 signaling in cancer will increase our understanding of the tumorigenesis of breast cancer.
Acknowledgments
We thank Dr. Iver Petersen and Dr. Youyong Lü for the generous gifts of the S100A14 antibodies, Dr. Mariam Grigorian for the generous gift of pQE30-Myo117 plasmid, and Dr. Espen Stang for the generous gift of pcDNA3-Zeo(+)-ErbB2 plasmid.
This work was supported by National Basic Research Program of China Grant 2011CB504205, National Natural Science Foundation of China Grants 81000954 and 81130043, and Doctoral Fund of Ministry of Education of China Grant 20101106120012.
- EGFR
- EGF receptor
- ECD
- extracellular domain
- ICD
- intracellular domain
- IP
- immunoprecipitation
- pERK
- pAKT, and pHER2, phosphorylated ERK, AKT, and HER2, respectively
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5(3-carboxymethonyphenol)-2-(4-sulfophenyl)-2H-tetrazolium.
REFERENCES
- 1. Yarden Y., Sliwkowski M. X. (2001) Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 2. Nahta R., Esteva F. J. (2006) Herceptin. Mechanisms of action and resistance. Cancer Lett. 232, 123–138 [DOI] [PubMed] [Google Scholar]
- 3. Olayioye M. A., Neve R. M., Lane H. A., Hynes N. E. (2000) The ErbB signaling network. Receptor heterodimerization in development and cancer. EMBO J. 19, 3159–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Graus-Porta D., Beerli R. R., Daly J. M., Hynes N. E. (1997) ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 16, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santamaria-Kisiel L., Rintala-Dempsey A. C., Shaw G. S. (2006) Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 396, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salama I., Malone P. S., Mihaimeed F., Jones J. L. (2008) A review of the S100 proteins in cancer. Eur. J. Surg. Oncol. 34, 357–364 [DOI] [PubMed] [Google Scholar]
- 7. Donato R. (2001) S100. A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 33, 637–668 [DOI] [PubMed] [Google Scholar]
- 8. Pietas A., Schlüns K., Marenholz I., Schäfer B. W., Heizmann C. W., Petersen I. (2002) Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics 79, 513–522 [DOI] [PubMed] [Google Scholar]
- 9. Wang H. Y., Zhang J. Y., Cui J. T., Tan X. H., Li W. M., Gu J., Lu Y. Y. (2010) Expression status of S100A14 and S100A4 correlates with metastatic potential and clinical outcome in colorectal cancer after surgery. Oncol. Rep. 23, 45–52 [PubMed] [Google Scholar]
- 10. Leth-Larsen R., Terp M. G., Christensen A. G., Elias D., Kühlwein T., Jensen O. N., Petersen O. W., Ditzel H. J. (2012) Functional heterogeneity within the CD44 high human breast cancer stem cell-like compartment reveals a gene signature predictive of distant metastasis. Mol. Med. 18, 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smirnov D. A., Zweitzig D. R., Foulk B. W., Miller M. C., Doyle G. V., Pienta K. J., Meropol N. J., Weiner L. M., Cohen S. J., Moreno J. G., Connelly M. C., Terstappen L. W., O'Hara S. M. (2005) Global gene expression profiling of circulating tumor cells. Cancer Res. 65, 4993–4997 [DOI] [PubMed] [Google Scholar]
- 12. Chen H., Yuan Y., Zhang C., Luo A., Ding F., Ma J., Yang S., Tian Y., Tong T., Zhan Q., Liu Z. (2012) Involvement of S100A14 protein in cell invasion by affecting expression and function of matrix metalloproteinase (MMP)-2 via p53-dependent transcriptional regulation. J. Biol. Chem. 287, 17109–17119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jin Q., Chen H., Luo A., Ding F., Liu Z. (2011) S100A14 stimulates cell proliferation and induces cell apoptosis at different concentrations via receptor for advanced glycation end products (RAGE). PLoS One 6, e19375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arumugam T., Simeone D. M., Schmidt A. M., Logsdon C. D. (2004) S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE). J. Biol. Chem. 279, 5059–5065 [DOI] [PubMed] [Google Scholar]
- 15. Tulchin N., Chambon M., Juan G., Dikman S., Strauchen J., Ornstein L., Billack B., Woods N. T., Monteiro A. N. (2010) BRCA1 protein and nucleolin colocalize in breast carcinoma tissue and cancer cell lines. Am. J. Pathol. 176, 1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duverger O., Lee D., Hassan M. Q., Chen S. X., Jaisser F., Lian J. B., Morasso M. I. (2008) Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. J. Biol. Chem. 283, 20198–20208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J. W., Elledge S. J., Kirschner M. W., Rape M. (2010) The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24, 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo W., Liu J., Li J., Zhang D., Liu M., Addo J. K., Patil S., Zhang L., Yu J., Buolamwini J. K., Chen J., Huang C. (2008) Anti-cancer effects of JKA97 are associated with its induction of cell apoptosis via a Bax-dependent and p53-independent pathway. J. Biol. Chem. 283, 8624–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang X. H., Wang Q., Gerald W., Hudis C. A., Norton L., Smid M., Foekens J. A., Massagué J. (2009) Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell 16, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bos P. D., Zhang X. H., Nadal C., Shu W., Gomis R. R., Nguyen D. X., Minn A. J., van de Vijver M. J., Gerald W. L., Foekens J. A., Massagué J. (2009) Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu C. S., Chen Y. C., Lu C. H., Hwang J. K. (2006) Prediction of protein subcellular localization. Proteins 64, 643–651 [DOI] [PubMed] [Google Scholar]
- 22. Adam P. J., Boyd R., Tyson K. L., Fletcher G. C., Stamps A., Hudson L., Poyser H. R., Redpath N., Griffiths M., Steers G., Harris A. L., Patel S., Berry J., Loader J. A., Townsend R. R., Daviet L., Legrain P., Parekh R., Terrett J. A. (2003) Comprehensive proteomic analysis of breast cancer cell membranes reveals unique proteins with potential roles in clinical cancer. J. Biol. Chem. 278, 6482–6489 [DOI] [PubMed] [Google Scholar]
- 23. Moasser M. M. (2007) The oncogene HER2. Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 26, 6469–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baselga J., Swain S. M. (2009) Novel anticancer targets. Revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 9, 463–475 [DOI] [PubMed] [Google Scholar]
- 25. Cianfrocca M., Goldstein L. J. (2004) Prognostic and predictive factors in early-stage breast cancer. Oncologist 9, 606–616 [DOI] [PubMed] [Google Scholar]
- 26. Wang Y. Y., Ye Z. Y., Zhao Z. S., Tao H. Q., Chu Y. Q. (2010) High-level expression of S100A4 correlates with lymph node metastasis and poor prognosis in patients with gastric cancer. Ann. Surg. Oncol. 17, 89–97 [DOI] [PubMed] [Google Scholar]
- 27. Zhang H. Y., Zheng X. Z., Wang X. H., Xuan X. Y., Wang F., Li S. S. (2012) S100A4 mediated cell invasion and metastasis of esophageal squamous cell carcinoma via the regulation of MMP-2 and E-cadherin activity. Mol. Biol. Rep. 39, 199–208 [DOI] [PubMed] [Google Scholar]
- 28. Vimalachandran D., Greenhalf W., Thompson C., Lüttges J., Prime W., Campbell F., Dodson A., Watson R., Crnogorac-Jurcevic T., Lemoine N., Neoptolemos J., Costello E. (2005) High nuclear S100A6 (Calcyclin) is significantly associated with poor survival in pancreatic cancer patients. Cancer Res. 65, 3218–3225 [DOI] [PubMed] [Google Scholar]
- 29. Anan K., Morisaki T., Katano M., Ikubo A., Tsukahara Y., Kojima M., Uchiyama A., Kuroki S., Torisu M., Tanaka M. (1998) Assessment of c-erbB2 and vascular endothelial growth factor mRNA expression in fine-needle aspirates from early breast carcinomas. Pre-operative determination of malignant potential. Eur. J. Surg. Oncol. 24, 28–33 [DOI] [PubMed] [Google Scholar]
- 30. Mittra I., Redkar A. A., Badwe R. A. (1995) Prognosis of breast cancer. Evidence for interaction between c-erbB-2 overexpression and number of involved axillary lymph nodes. J. Surg. Oncol. 60, 106–111 [DOI] [PubMed] [Google Scholar]
- 31. Tokatli F., Altaner S., Uzal C., Ture M., Kocak Z., Uygun K., Bilgi S. (2005) Association of HER-2/neu overexpression with the number of involved axillary lymph nodes in hormone receptor positive breast cancer patients. Exp. Oncol. 27, 145–149 [PubMed] [Google Scholar]
- 32. Schneider J., Pollán M., Tejerina A., Sánchez J., Lucas A. R. (2003) Accumulation of uPA-PAI-1 complexes inside the tumour cells is associated with axillary nodal invasion in progesterone-receptor-positive early breast cancer. Br. J. Cancer 88, 96–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bader A. A., Tio J., Petru E., Bühner M., Pfahlberg A., Volkholz H., Tulusan A. H. (2002) T1 breast cancer. Identification of patients at low risk of axillary lymph node metastases. Breast Cancer Res. Treat. 76, 11–17 [DOI] [PubMed] [Google Scholar]
- 34. Sütterlin M. W., Haller A., Gassel A. M., Peters K., Caffier H., Dietl J. (2000) The correlation of c-erbB-2 oncoprotein and established prognostic factors in human breast cancer. Anticancer Res. 20, 5083–5088 [PubMed] [Google Scholar]
- 35. Arisio R., Sapino A., Cassoni P., Accinelli G., Cuccorese M. C., Mano M. P., Bussolati G. (2000) What modifies the relation between tumour size and lymph node metastases in T1 breast carcinomas? J. Clin. Pathol. 53, 846–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Erjala K., Sundvall M., Junttila T. T., Zhang N., Savisalo M., Mali P., Kulmala J., Pulkkinen J., Grenman R., Elenius K. (2006) Signaling via ErbB2 and ErbB3 associates with resistance and epidermal growth factor receptor (EGFR) amplification with sensitivity to EGFR inhibitor gefitinib in head and neck squamous cell carcinoma cells. Clin. Cancer Res. 12, 4103–4111 [DOI] [PubMed] [Google Scholar]
- 37. Li H., Sánchez-Torres J., Del Carpio A., Salas V., Villalobo A. (2004) The ErbB2/Neu/HER2 receptor is a new calmodulin-binding protein. Biochem. J. 381, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nahta R., Hung M. C., Esteva F. J. (2004) The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res. 64, 2343–2346 [DOI] [PubMed] [Google Scholar]
- 39. Shattuck D. L., Miller J. K., Carraway K. L., 3rd, Sweeney C. (2008) Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 68, 1471–1477 [DOI] [PubMed] [Google Scholar]
- 40. Wainberg Z. A., Anghel A., Desai A. J., Ayala R., Luo T., Safran B., Fejzo M. S., Hecht J. R., Slamon D. J., Finn R. S. (2010) Lapatinib, a dual EGFR and HER2 kinase inhibitor, selectively inhibits HER2-amplified human gastric cancer cells and is synergistic with trastuzumab in vitro and in vivo. Clin. Cancer Res. 16, 1509–1519 [DOI] [PubMed] [Google Scholar]
- 41. Klingelhöfer J., Møller H. D., Sumer E. U., Berg C. H., Poulsen M., Kiryushko D., Soroka V., Ambartsumian N., Grigorian M., Lukanidin E. M. (2009) Epidermal growth factor receptor ligands as new extracellular targets for the metastasis-promoting S100A4 protein. FEBS J. 276, 5936–5948 [DOI] [PubMed] [Google Scholar]
- 42. Hernan R., Fasheh R., Calabrese C., Frank A. J., Maclean K. H., Allard D., Barraclough R., Gilbertson R. J. (2003) ERBB2 up-regulates S100A4 and several other prometastatic genes in medulloblastoma. Cancer Res. 63, 140–148 [PubMed] [Google Scholar]
- 43. Paruchuri V., Prasad A., McHugh K., Bhat H. K., Polyak K., Ganju R. K. (2008) S100A7-downregulation inhibits epidermal growth factor-induced signaling in breast cancer cells and blocks osteoclast formation. PLoS One 3, e1741. [DOI] [PMC free article] [PubMed] [Google Scholar]