Background: Snail1 and Snail2 are highly homologous zinc finger transcriptional repressors exhibiting divergent functions.
Results: Snail1 and Snail2 use a unique set of zinc fingers to execute their biological activity.
Conclusion: The use of different zinc fingers could explain the functional divergence between Snail transcription factors.
Significance: This is the first study dissecting the structural/functional differences between zinc fingers of Snail1 and Snail2 factors.
Keywords: DNA-binding Protein, E-cadherin, EMT, Repressor Protein, Zinc Finger
Abstract
Snail1 (Snail) and Snail2 (Slug) are transcription factors that share a similar DNA binding structure of four and five C2H2 zinc finger motifs (ZF), respectively. Both factors bind specifically to a subset of E-box motifs (E2-box: CAGGTG/CACCTG) in target promoters like the E-cadherin promoter and are key mediators of epithelial-to-mesenchymal transition (EMT). However, there are differences in the biological actions, in binding affinities to E-cadherin promoter, and in the target genes of Snail1 and Snail2, although the molecular bases are presently unknown. In particular, the role of each Snail1 and Snail2 ZF in the binding to E-boxes and in EMT induction has not been previously explored. We have approached this question by modeling Snail1 and Snail2 protein-DNA interactions and through mutational and functional assays of different ZFs. Results show that Snail1 efficient repression and binding to human and mouse E-cadherin promoter as well as EMT-inducing ability require intact ZF1 and ZF2, while for Snail2, either ZF3 or ZF4 is essential for those functions. Furthermore, the differential distribution of E2-boxes in mouse and human E-cadherin promoters also contributes to the differential Snail factor activity. These data indicate a non-equivalent role of Snail1 and Snail2 ZFs in gene repression, contributing to the elucidation of the molecular differences between these important EMT regulators.
Introduction
The Snail superfamily is divided into the Snail and Scratch families, and at present three members of the Snail family have been described in vertebrates: SNAI1, SNAI2, and SNAI3 (1, 2). Snail family members share a common structure with a highly conserved C-terminal domain and a divergent N-terminal region (2). All Snail members, except for Dmsnail, contain a conserved SNAG domain: 9 N-terminal amino acids (3) essential for transcriptional repressor activity (4–7). The central region of the Snail proteins has a serine-proline-rich region that is highly divergent among Snail members (8): Snail2 contains the so-called SLUG domain, required for efficient Snail2-mediated repression (7), while Snail1 presents a regulatory domain containing a nuclear export signal (NES)4 (9) and a destruction box domain (10). Phosphorylation of serine residues in the regulatory regions and potential modification of adjacent lysine residues have been implicated in the subcellular localization, protein stability, and repressor activity of Snail1 and Snail2 (7, 9–12). The C-terminal domain of Snail factors comprises the DNA-binding domain (DBD), formed by four to six C2H2 zinc fingers (ZFs), which recognizes consensus E2-box type elements (C/ACAGGTG) (2–4) and also includes nuclear translocation signals (13, 14). The classical C2H2 zinc finger motif, initially discovered in the transcription factor TFIIIA of Xenopus laevis (15), contains a repeated 28–30 amino acid sequence conforming a secondary structure of a β-hairpin followed by an α-helix that arrange in a left-handed ββα unit, and typically occurs as tandem repeats (16, 17). The two cysteines are near a turn in the antiparallel β-sheet, and the two histidines are in the C-terminal portion of the α-helix and together coordinate a single zinc ion. The C2H2 fingers predominantly participate in protein-DNA recognition via binding to the major groove of the DNA through the N terminus of the α-helix and occupy a subsite of 3–4 base pairs (16). The zinc fingers of Snail proteins have shorter tandem repeats of 22–25 amino acids, but maintain the structural ββα unit organization of the classical zinc finger motif (2, 8, 13). The C-terminal DBD region of Snail1 and Snail2 differs in the number of zinc fingers: four (ZF1 to ZF4) in Snail1, and five (ZF1 to ZF5) in Snail2 (1, 2) that might provide differential interactions and/or binding affinities to target genes. The ZF domain of Snail factors has been further proposed to classify the Snail superfamily: ZF3 and ZF4 have a consensus sequence in all family members, ZF2 and ZF5 discriminate the Snail and Scratch families (1, 2, 8), and the ZF1 of Snail2 and Dmsnail has been suggested to be not functional (18).
At the cellular level, Snail factors regulate cell movements and trigger the Epithelial-to-Mesenchymal Transition (EMT) process, converting almost static epithelial cells into motile and invasive mesenchymal cells with stem cell properties (1, 2, 19, 20). EMT is an essential process during embryonic development and has proved to be a key event in tumor invasion and metastasis (21–23). One of the hallmarks of EMT is the loss of E-cadherin function, and in fact it is generally accepted that EMT-inducing factors initiate epithelial disorganization by impairing the expression or function of E-cadherin (21, 24). Indeed, E-cadherin was the first target described for Snail1 and Snail2 (5, 25–27), both factors bind to the E2-boxes of the proximal E-cadherin (CDH1) promoter and recruit different co-repressor complexes (5–7, 27). The mouse and human CDH1 promoters have three proximal E2-boxes with a differential distribution: the mouse promoter contains two adjacent E1- and E2-boxes in a palindromic element, called E-pal (−70 to −90) and E3-box (−30), whereas the human promoter lacks the E2-box and has an additional E4-box after the transcription start point (28). Furthermore, distinct in vitro affinities of Snail1 and Snail2 to the E-pal element have been described (27). Other target genes repressed by both Snail1 and Snail2 have been reported, including claudins and other epithelial genes (reviewed in Refs. 20, 29). Snail1 and Snail2 not only directly repress epithelial gene promoters, but also activate the expression of mesenchymal genes, like vimentin, fibronectin, and N-cadherin through indirect mechanisms not yet well understood (30). Nevertheless, and despite the high homology in their DNA binding and SNAG domains, Snail1 and Snail2 induce common and differential gene expression patterns when overexpressed in epithelial cells (31), pointing to distinct structural and/or functional characteristics between both factors. Snail1 and Snail2 are equivalent as EMT inducers when ectopically expressed in proper species and developmental contexts (32, 33) and are involved in morphogenetic processes (1, 34), but they also play divergent functions in development. Thus, in mouse embryos Snail1 is essential for gastrulation (35) while Snail2 is dispensable for embryonic development (36), although both factors are required for left-right hand asymmetry (33). In addition, an increasing number of studies have shown Snail1 and/or Snail2 expression in a variety of tumors (reviewed in Refs. 20, 21, 29), but with differential roles in tumor progression and metastasis (37, 38), indicating specific functions for either factor in those processes.
The molecular bases for the distinct regulation and binding affinity of downstream target genes by Snail1 and Snail2 are still largely unknown. To get insights into this relevant issue, we have investigated in detail the participation of the Snail1 and Snail2 DBD domain to determine which zinc fingers are essential for their repression activity in human and mouse E-cadherin and human claudin-1 promoters. To this end, we developed a computational model that predicts the consequences of mutation in specific Snail1 and Snail2 ZFs in their interaction with the DNA. The specific Snail1 and Snail2 mutants, in which the structure of individual zinc fingers was modified by point mutation, were analyzed for repression activity, DNA binding, and EMT induction. We show that Snail1 and Snail2 ZFs are not functionally equivalent, and identify for the first time the specific ZFs required for repression, DNA binding, and EMT induction by each Snail factor. Our results thus contribute to the knowledge of the molecular bases behind the differential action of Snail1 and Snail2, opening new avenues into the regulation of EMT and other essential processes by both factors.
EXPERIMENTAL PROCEDURES
Computational Method
To analyze the zinc finger-DNA sequence recognition, a three-dimensional model of Snail1 and Snail2 was developed using homology modeling. The amino acid sequence of the proteins was retrieved from the NCBI database (Gene ID: 20613 and 20583). Afterward, the Basic Local Alignment Search Tool (BLAST) was used to perform a search for the closest homologue with an available three-dimensional structure in the Protein Data Bank (PDB). Results indicate that a six zinc finger protein (PDB code 2I13) was a suitable template for modeling with 41% sequence identity. Homology modeling of the target was carried out by the automated server CPH model v3.0 (www.cbs.dtu.dk/services/CPHmodels/). Then, the model was validated using PROCHECK, to obtain the Ramachandran plot, and ERRAT. Finally, the structure was relaxed using an energy minimization protocol of 2000 steps of steepest descent and 1000 steps of Polak-Ribiere conjugate gradients with the AMBER 99sb force field. These calculations were performed with the GROMACS 4.5 package. The DNA sequence of the template protein was manually mutated to resemble the possible recognition pattern of the E2-box sequence CAGGTG and adjusted to the model. Then, a new minimization protocol, homologous to the first one, was carried out. Finally, protein-DNA interactions were described using the molecular viewer PyMOL.
Generation of Plasmids and Expression Vectors
Mutants of each Snail1 and Snail2 ZF have been generated by point mutation from the pcDNA3-mSnail1, pcDNA3-mSnail1-HA and pcDNA3-mSnail2-HA plasmids, previously generated in our group (25, 27) by site-directed mutagenesis (39). Due to the high degree of identity between human and mouse Snail1 (96% identity) and Snail2 (100% identity) ZFs we restricted this study to the mouse proteins. The mutation consists of the substitution of the first cysteine of each ZF by alanine that avoids the formation of the zinc finger tri-dimensional structure. The specific C to A mutants correspond to Snail1: ZF1 (C156A), ZF2 (C182A), ZF3 (C210A), and ZF4 (C238A); and to Snail2: ZF1 (C131A), ZF2 (C162A), ZF3 (C188A), ZF4 (C216A), and ZF5 (C244A). Combined mutations of Snail1 were obtained by site-directed mutagenesis using pcDNA3-mSnail1 as a template. Oligonucleotide sequences for each mutation will be provided upon request. After mutagenesis and subcloning, the entire Snail2-HA and Snail1 cDNAs were sequenced to verify that only the nucleotide changes introduced by the mutagenic oligonucleotides were obtained. Mouse and human E-cadherin promoter both containing the three proximal E2-boxes (−178 to +92 nucleotides), and the human claudin-1 reporter, containing two E2-boxes in the proximal promoter and first intron (−82 and +236) fused to the luciferase gene reporter have been previously described (12, 25, 40).
Cell Culture and Transfections
Human HEK293T, canine MDCK, and mouse NMuMG cells were obtained from the ATCC (CRL-11268, CRL-2936, CRL-1636, respectively). HEK293T and MDCK cells were maintained in DMEM medium supplemented with 10% fetal calf serum (Invitrogen) and antibiotics (100 μg/ml ampicillin, 32 μg/ml gentamicin, Sigma-Aldrich); NMuMG were grown in DMEM supplemented also with insulin. Stable and transient transfections were performed using Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. For promoter assays, MDCK or HEK293T cells were transiently transfected in 24-well plates. 5 × 104 HEK293T or 2 × 104 MDCK cells per well were seeded, and after 24 h the cells were co-transfected with the indicated human or mouse E-cadherin or human claudin-1 promoter constructs (200 ng/well) and 75–100 ng of the wild type or mutant Snail1 or Snail2 vectors; 10 ng/well of the pCMV-βGal vector (Promega) was co-transfected as a control of transfection efficiency. For immunofluorescence analyses 5 × 104 MDCK or HEK293T cells were spread in 6-well plates over 12-mm diameter coverslips, and then were transiently transfected with 1.5 μg of wild type Snail1, Snail2, or their different mutant constructs. For generation of stable clones, MDCK cells (1 × 106 cells) grown in P60 plates were transfected with pcDNA3-Snail1-HA, pcDNA3-Snail2-HA, the indicated ZF mutants, or control pcDNA3-HA (CMV) and grown in the presence of G418 (500 μg/ml) for 2–3 weeks; individual colonies were then selected and grown individually. At least ten independent clones were selected and characterized from each transfection with the pcDNA3-Snail1 and pcDNA3-Snail2-HA mutants.
Promoter Activity Assays
Promoter activity assays were basically performed as previously reported (6, 27, 41). Luciferase and βGal activities were measured 24 h post-transfection using the Dual-luciferase β-Glo Reporter assay kit (Promega) and normalized to the promoter activity detected in mock-transfected (pcDNA3-HA, pcDNA3) cells as described (41). Reporter assays in the various experimental settings were performed at least five times using triplicate samples. Results represent the mean ± S.D.
Chromatin Immunoprecipitation (ChIP) Assays
Cells were seeded 24 h prior to transfection with the corresponding DNA plasmid; 24 h post-transfection cells were fixed with formaldehyde at room temperature for 15 min, then glycine was added and incubated for 5 min at room temperature, and the cells were washed with PBS (1×). Cells were harvested in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.00, and protease and phosphatase inhibitors (2 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mm PMSF, 1 mm sodium vanadate, 10 mm sodium fluoride)) and sonicated to shear the chromatin (∼500 bp). The soluble fraction was collected by centrifugation and precleared with Sepharose G-beads (Sigma Aldrich), and then incubated with specific antibodies or control IgG at 4 °C overnight. The immune complexes were captured with Sepharose G-beads. After extensive washing, the bound DNA fragments were eluted and purified. The DNA samples were subjected to quantitative PCR analyses using human E-cadherin promoter Fw-CDH1 5′-TCCTTTGTAACTCCATGTCTCCCGT-3′ and Rw-CDH1 5′-CGGGCAGGAGTCTAGCAGAAG-3′, mouse E-cadherin promoter Fw-cdh1 5′-CATGTCTCCGTGGGTCAGA-3′ and Rw-cdh1 5′-AGGTGGCAGCCAAGGAACT-3′; or human claudin-1 promoter Fwd.Cld1 5′-GCCACCTTCGGGAGTCCGGG-3′ and Rv-Cld1 5′-GGCGCCCGCGCTGGCTCAGG-3′ and SYBR green supermix (Bio-Rad) using an iQ5 (Bio-Rad) machine.
Western Blot Assays
Protein samples from whole cell extracts or promoter assays reactions were resolved on SDS-PAGE, transferred to nitrocellulose Inmobilon-P membranes (Millipore), and analyzed by Western blot as described (41). Briefly, membranes were blocked using 5% nonfat dry milk in 0.5% Tween-Tris-glycine buffer, and then incubated at 4 °C overnight with the primary antibodies followed by 1 h incubation at room temperature with the secondary antibodies. After washing, proteins were visualized with ECL detection reagent (Amersham). The primary antibodies were: rat anti-HA (Roche, 1:1000), mouse anti-Snail1 H33 (1:200; provided by I. Virtanen. Institute of Biomedicine/Anatomy, Helsinki, Finland), mouse anti-Snail and anti-Slug (1:1000, Cell Signaling), mouse anti-N-cadherin (Zymed Laboratories Inc., 1:200), mouse anti-fibronectin (1:5000, BD Transduction), mouse anti-vimentin (1:5000, Dako), rat anti-E-cadherin ECCD2 (1:200, produced in our laboratory from the ECCD2 hybridoma, a gift of M.Takeichi, Ricken Center, Japan) and mouse anti-α-tubulin (1:5000, Santa Cruz Biotechnology). The secondary antibodies were HRP-coupled goat anti-rat (1:10000) or sheep anti-mouse (1:5000, Pierce).
Immunofluorescence
Immunofluorescence analyses on cells grown on coverslips were basically performed as described (41). Cells were fixed with cold (−20 °C) methanol for 1 min, or 3.7% formaldehyde for 20 min at room temperature and permeabilized using 0.5% (v/v) Triton X-100. Transiently or stably transfected MDCK or HEK293T cells were processed for indirect immunofluorescence with rat anti-E-cadherin (1:100), mouse anti-vimentin (1:400), mouse anti-Snail1 H33 (1:100), or rat-anti HA (1:500) antibodies. Alexa-488 or Alexa-585 (1:800, Molecular Probes) were used as secondary fluorescent antibodies. Nuclei were detected by DAPI stain. Samples were analyzed using a confocal SP2 Spectral Leica microscope and a ×40 objective.
Statistics
Error bars in the graphical data represent means ± S.D. All in vitro experiments were performed at least in triplicate. p values of p < 0.05 were considered statistically significant by two-tailed Student's t test. Statistical analyses were carried out using Microsoft Excel 2010.
RESULTS
Molecular Analysis of Snail1 and Snail2 ZFs
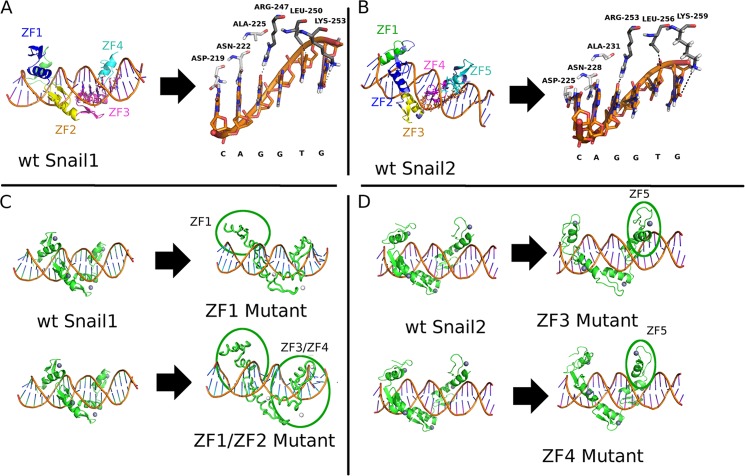
To further ascertain the structure and molecular interactions that Snail1 and Snail2 establish with the DNA consensus sequence, we performed a computational analysis of Snail1 and Snail2 protein-DNA interactions (Fig. 1). The proposed model of specific recognition between the DNA sequence (E2-box) and Snail1/Snail2 proteins includes a classic ZF-DNA interaction pattern, where the residues of the α-helix establish hydrogen bonds and other favorable contacts with the bases of the DNA. The specific interactions are shown in Fig. 1, A and B. The amino acids involved are compatible with other natural recognition sequences (42) and designed ZF domains (43). The four Snail1 ZFs are surrounding the double DNA helix and establish hydrogen bonds through ZF1 and ZF2, whereas ZF3 and ZF4 also establish hydrogen bonds with specific DNA bases of the E2-box sequence (CAGGTG); they involve residues inside the α-helix of ZF3 and ZF4: Asp-219, Asn-222, and Ala-225 within ZF3; and Arg-247, Leu-250, and Lys-253 within ZF4 (Fig. 1A), in agreement with the canonical binding of C2H2 zinc fingers (42). Of note, phosphate and base bonds have the same importance in DNA interaction of zinc fingers, because the phosphates are necessary to reach the most appropriate spatial organization for the base-protein contacts (42). A similar structural organization was observed for Snail2: ZF2 and ZF3 (equivalent to Snail1 ZF1 and ZF2) establish connections with phosphate groups through charge-charge hydrogen bonds, and ZF4 and ZF5 (equivalent to Snail1 ZF3 and ZF4) bind through hydrogen bonds to bases of the E2-box involving equivalent residues inside the α-helix of ZF4 and ZF5 of Snail2: Asp-225, Asn-228, and Ala-231 within ZF4; and Arg-253, Leu-256, and Lys-259 within ZF5 (Fig. 1B). The additional Snail2 ZF1 does not interact with the DNA backbone (Fig. 1B), in agreement with previous proposals (18). Next, we explored the effects of the mutations in several ZFs in Snail1 (ZF1 and double ZF1/ZF2 mutants) and Snail2 (ZF3 and ZF4 mutants) through molecular dynamic simulations. To this end, the proposed complexes for both Snail1 and Snail2 were simulated in their wild type and mutant forms, and a comparison of the final states of simulations was performed. The simulated complexes were found to be stable in the cases of wild type forms (Fig. 1, A and B) while in the case of the ZF mutants, full or partial loss of the modeled specific contacts was found (Fig. 1, C and D). Surprisingly, when the Snail1 ZF1 or both ZF1 and ZF2 are destroyed by point mutation (C156A; C182A) a big displacement from the original position was detected in the whole Snail1 DBD domain, affecting not only to the backbone interactions of mutated ZF1 and ZF2, but also the ZF3 and ZF4 binding that established weaker (ZF1 mutant) or completely lost (ZF1/ZF2 mutant) interactions with the DNA bases (Fig. 1C). In the case of Snail2, mutations in either ZF3 (C188A) or ZF4 (C216A) leads to the complete displacement of interactions with the DNA bases, affecting also ZF5 interactions (Fig. 1D). These structural simulations suggest that integrity of the four ZFs of Snail1 and the equivalent ZFs of Snail2 are required for a proper recognition and binding to E2-box DNA sequences.
FIGURE 1.
Schematic representation of Snail1 and Snail2 structure. A, three-dimensional model of Snail1 DBD; ZF3 and ZF4 bind to DNA bases directly, while the two first ZFs do not establish bonds with bases. B, three-dimensional model of Snail2 DBD; ZF4 and ZF5 bind to DNA bases directly, but ZF1-ZF3 only establish indirect interactions. C, three-dimensional model showing the consequences of ZF1 and ZF1/ZF2 destruction in Snail1, leading to an almost complete absence of interaction between DNA and the protein. D, three-dimensional model showing the consequences of the ZF3 or ZF4 destruction in Snail2, these mutations avoid the proper interaction with the DNA.
Analysis of Snail1 Zinc Fingers Repressor Activity and Binding Affinity
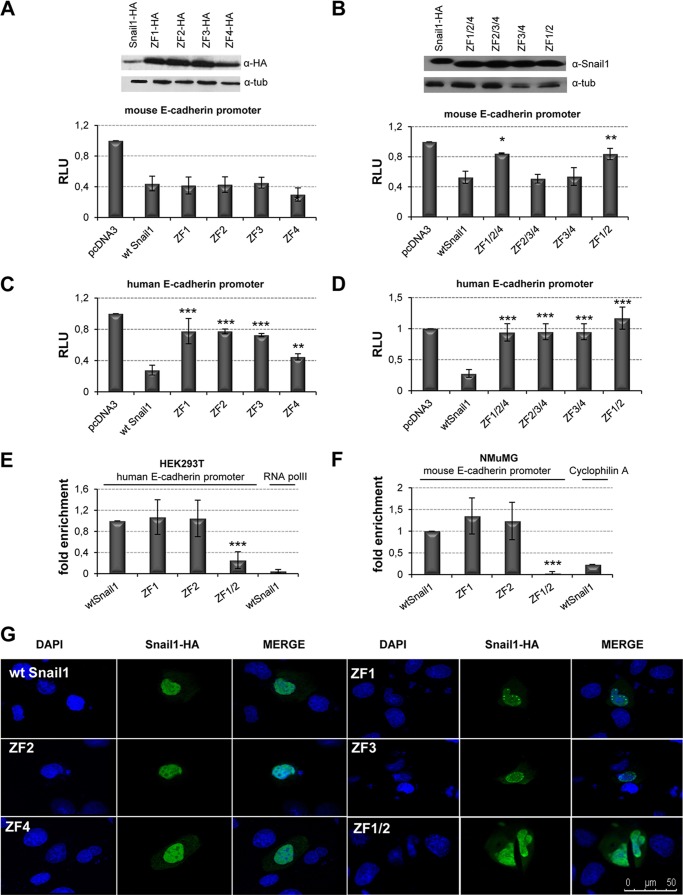
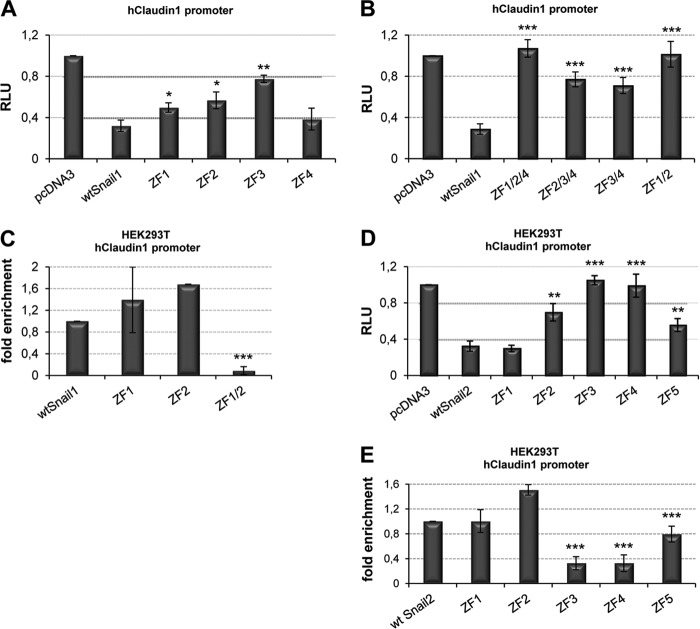
Previous studies have shown the requirement of the full zinc finger domain of Snail1 and Snail2 for the repression of E-cadherin and other promoters (5–7, 27, 40), but the implication of the different zinc fingers and/or the E2-boxes organization of the target promoters have not yet been defined. To get further insights into this subject, we mutated the individual zinc fingers of mouse Snail1 and analyzed their effect in the repression on the mouse and human E-cadherin promoter. Analyses of the Snail1 mutants with the mouse E-cadherin promoter in HEK293T cells indicated that individual mutation of the ZF1, ZF2, or ZF3 did not have a significant effect over the activity of wild type Snail1, while alteration of ZF4 slightly increased repression (Fig. 2A). However, combined destruction of ZF1 and ZF2 greatly decreased Snail1 repressor activity (Fig. 2B), as anticipated in the three-dimensional simulation model, while other mutant combinations had no significant effect. Therefore, both ZF1 and ZF2 are required for Snail1 repression of the mouse E-cadherin promoter. By contrast, similar analyses on the human E-cadherin promoter indicated that mutation of any of the individual ZF significantly decreased Snail1 repressor activity (Fig. 2C), indicating that all zinc fingers contribute to the Snail1-mediated repression of the human E-cadherin promoter. Similar results with the individual and combined ZF's Snail1 mutants were obtained when analyzed in MDCK cells with both E-cadherin promoters (data not shown). Moreover, additional analyses on the human claudin-1 promoter also showed the individual contribution of ZFs to repression mediated by Snail1 (Fig. 3A) and the collaboration of Snail1 ZF1 and ZF2 for complete repression (Fig. 3B). Immunofluorescence analyses showed a diffuse nuclear localization of the wild type and most Snail1 mutants, except for the ZF1 and ZF3 mutants showing a more punctuate pattern (Fig. 2G and data not shown), indicating that the changes in repressor activity are not due to failure of nuclear localization.
FIGURE 2.
Snail1 ZF1 and ZF2 are required for E-cadherin promoter repression and DNA binding activity. A and B, repressor activity of wt Snail1 and individual (A) or combined (B) ZF mutants on the mouse E-cadherin promoter in HEK293T cells. C and D, repressor activity of wt Snail1 and individual (C) or combined (D) ZF mutants on the human E-cadherin promoter in HEK293T cells. Activity (RLU) was normalized to that present in cells transfected with control pcDNA3 vector. Data represent the average ± S.D. of five experiments performed on triplicate samples. *: p < 0.5; **: p < 0.005; ***: p < 0.001 compared with wt Snail1. Western blots shown in the upper panels indicate similar transfection efficiency of wt Snail1 and the different mutants. E and F, ChIP assays in HEK293T (E) and NMuMG (F) cells transfected with wt Snail1 and the indicated ZF mutants. After amplification of the E-cadherin promoter sequences, binding of the various Snail1 forms was quantified as fold induction relative to input. Amplification of PolII (E) and cyclofilin A (F) was used as a nonspecific binding of Snail1 to human and mouse genes, respectively. Data represent the average ± S.D. of three experiments. ***: p < 0.001 compared with wt Snail1. G, subcellular localization of Snail1 mutants. Immunofluorescence images indicate that wt Snail1 and Snail1 ZF mutants mainly localized within the nucleus. Bar, 50 μm.
FIGURE 3.
Repression activity and DNA binding of Snail1 and Snail2 to the human claudin-1 promoter. A and B, promoter assays with wt Snail1 and individual (A) or combined (B) ZF mutants on claudin-1 promoter in HEK293T cells. C, ChIP assay in HEK293T with wt Snail1 and the indicated ZF1 and/or ZF2 mutants. D, promoter assays with wt Snail2 and the individual ZF mutants on claudin-1 promoter in HEK293T. Data represent the average ± S.D. of five experiments performed on triplicate samples. E, ChIP assay in HEK293T cells with wt Snail2 and individual ZF mutants. Data represents the average ± S.D. of three experiments. *: p < 0.5; **: p < 0.005; ***: p < 0.001 compared with wt Snail1 or wt Snail2.
Once we established the relevance of Snail1 ZF1 and ZF2 in promoter repression, we analyzed their requirement for in vivo DNA binding as suggested from the structural analyses. ChIP assays in HEK293T cells indicated that only destruction of both ZF1 and ZF2 leads to an almost complete loss of binding to the endogenous human and mouse E-cadherin promoters, while destruction of individual zinc fingers had no significant effect on DNA binding (Fig. 2, E and F). Similar results were obtained in ChIP analysis for the endogenous claudin-1 promoter (Fig. 3C). Taken together, these data indicate that Snail1 ZF1 and ZF2 are together essential for DNA binding and repression of target human and mouse promoters.
Identification of Snail2 Zinc Fingers Required for Transcriptional Repression
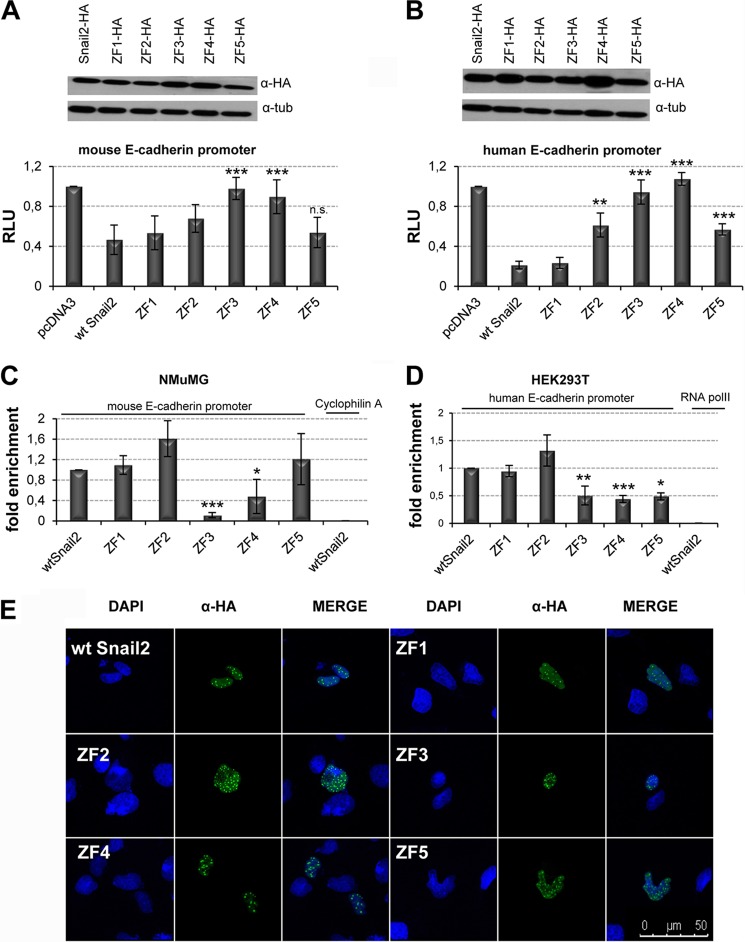
To study the relevance of Snail2 zinc fingers, similar analyses as those described above for Snail1 were performed using Snail2 mutants in each of its five ZFs. Reporter assays were performed with human and mouse E-cadherin promoters in HEK293T and MDCK cells (Fig. 4 and data not shown). The results obtained strongly suggest, first, that ZF1 is not required for the mouse or human E-cadherin promoter repression, confirming previous theoretical proposals (18); second, that individual mutations of the ZF3 or ZF4 lead to the complete loss of Snail2 repressor activity on both human and mouse E-cadherin promoters, thus indicating an essential role of Snail2 ZF3 and ZF4; and third, the partial contribution of Snail2 ZF2 and ZF5 to human E-cadherin repression (Fig. 4, A and B). The predominant role of ZF3 and ZF4 and the partial contribution of Snail2 ZF2 and ZF5 were also observed in the repression of the human claudin-1 promoter (Fig. 3D). All individual Snail2 ZF mutants localized in the nucleus (Fig. 4E), demonstrating that the loss of repression is not due to their mis-localization. We saw a slightly different pattern in the cellular distribution of Snail1 compared with Snail2; nuclear speckles were observed in the case of Snail2 and its mutants compatible with nuclear accumulation and interchromatin granule clusters as previously reported (44). ChIP assays with wild type Snail2 and all its mutants in human (HEK293T cells) and mouse (NMuMG cells) showed strongly decreased binding of Snail2 ZF3 and ZF4 mutants to the endogenous human and mouse E-cadherin promoters (Fig. 4, C and D) in complete agreement with the promoter activity data. A distinct situation was observed for Snail2 ZF5 mutant since it maintains partial repression activity while apparently losing DNA binding to the endogenous human E-cadherin promoter (Fig. 4, B and D) but did not affect the repressor activity or binding to endogenous mouse E-cadherin promoter (Fig. 4, A and C). ChIP assays on the endogenous human claudin-1 promoter confirmed the complete or partial loss of DNA binding of ZF3 and ZF4 or ZF5 mutants, respectively, in agreement with the repressor activity exhibited by these Snail2 mutants (Fig. 3, D and E).
FIGURE 4.
Snail2 ZF3 and ZF4 are independently required for E-cadherin promoter repression and DNA binding activity. A and B, repressor activity of wt Snail2 and individual ZF mutants on the mouse (A) and human (B) E-cadherin promoters in HEK293T cells. Activity (RLU) was normalized to that present in cells transfected with control pcDNA3 vector. Data represent the average ± S.D. of five experiments performed on triplicate samples. **: p < 0.005; ***: p < 0.001; n.s.: nonsignificant compared with wt Snail2. Western blots shown in the upper panels indicate similar transfection efficiency of wt Snail2 and the different mutants. C and D, ChIP assays in NMuMG (C) and HEK293T (D) cells transfected with wt Snail2-HA and the indicated ZF mutants. After amplification of E-cadherin promoter sequences, binding of the various Snail2 forms was quantified as fold induction relative to input. Amplification of cyclofilin A (C) and PolII (D) was used as a nonspecific binding of Snail2 to mouse and human genes, respectively. Data represent the average ± S.D. of three experiments. *: p < 0.5; **: p < 0.005; ***: p < 0.001 compared with wt Snail2. E, subcellular localization of Snail2 mutants. Immunofluorescence images show that wt Snail2 and Snail2 ZF mutants localized within the nucleus in a punctuate pattern. Bar, 50 μm.
Overall, these data indicate a predominant role for Snail2 ZF3 and ZF4 in DNA binding and repressor activity on target gene promoters and the partial contribution of ZF5 in human promoters. Together with the analyses on Snail1, these results indicate a differential contribution of the individual zinc fingers of Snail1 and Snail2 factors.
Specific Zinc Fingers of Snail1 and Snail2 Are Required for in Vivo Induction of EMT
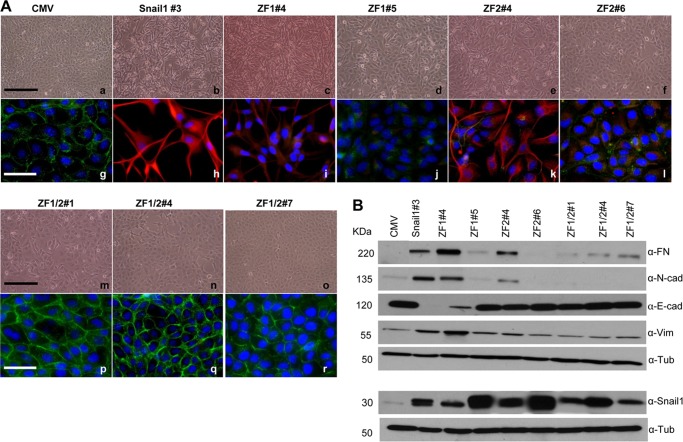
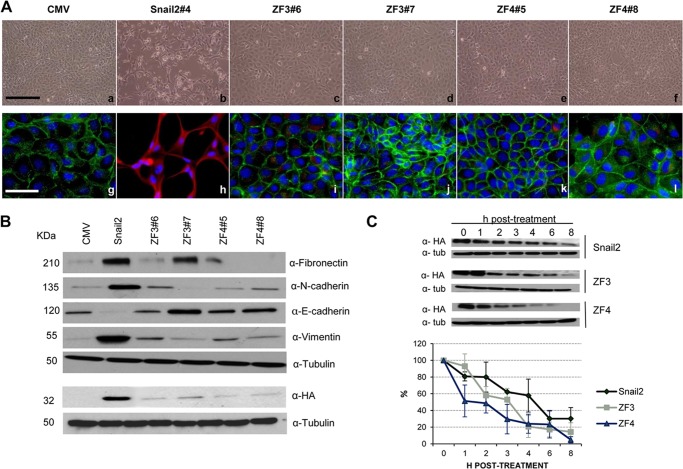
Once the specific zinc fingers of Snail1 and Snail2 required for E-cadherin and claudin-1 promoter repression and DNA binding were established, we wondered whether those zinc fingers were also required for the in vivo biological action of Snail factors, that is, their EMT inducing capacity. For this purpose, stable transfectants expressing Snail1-HA or Snail2-HA wt and the mutant versions in the identified zinc fingers (ZF1 and/or ZF2 for Snail1; ZF3 or ZF4 for Snail2) were generated in epithelial MDCK cells. As previously described (6, 7, 25, 27), expression of Snail1-HA or Snail2-HA induced a phenotypic change compatible with a complete EMT process as observed from the mesenchymal phenotype associated with the loss of E-cadherin and increased expression of mesenchymal markers, such as N-cadherin, fibronectin, and vimentin (Figs. 5 and 6). Stable expression of the Snail1 single ZF1 or ZF2 mutants lead to a partial EMT process as indicated by the variable phenotype and expression of E-cadherin and mesenchymal markers in different clones; interestingly, a much more attenuated EMT phenotype was observed in those clones expressing higher amounts of ZF1 and ZF2 mutants (Fig. 5, A and B; compare clone ZF1#5 to ZF1#4, and ZF2#6 to ZF2#4, respectively) in which partial localization of E-cadherin at cell-cell contacts and the absence of vimentin together with decreased mesenchymal marker expression was detected (Fig. 5, A, panels d, j, and f, l, and B). However, stable expression of Snail1 double ZF1/ZF2 mutant fully blocked the Snail1-EMT induction capacity as determined by the epithelial phenotype; high levels of E-cadherin organized at cell-cell contacts and low or absent expression of mesenchymal markers observed in all analyzed clones (Fig. 5A, panels m–r, and B). Interestingly, similar analyses for Snail2 showed that individual mutation of either Snail2 ZF3 or ZF4 fully abolished the Snail2-EMT-inducing capacity as indicated by the epithelial phenotype, expression, and organization of E-cadherin, and the absence or low levels of mesenchymal markers obtained in all clones derived from either of the two Snail2 mutants (Fig. 6, A and B). Remarkably, the Snail2 ZF3 and ZF4 mutants were expressed at low levels in all the analyzed transfectants compared with Snail2 wt (Fig. 6B), suggesting that those mutations confer increased protein instability. Cycloheximide pulse-chase assays showed that indeed Snail2 mutation in ZF3 or ZF4 decreased by about 50% the half-life of the proteins compared with wild type Snail2 that exhibited an estimated half-life of 5 h in HEK293T cells (Fig. 6C), in agreement with recent observations (7).
FIGURE 5.
Snail1 ZF1 and ZF2 are required for EMT induction. A, phenotypic characterization of one representative wt Snail1-HA clone, two representative Snail1-ZF1 (#4 and #5) and Snail1-ZF2 (#4 and #6) clones, and three representative clones for Snail1-ZF1/2 (#1, #4, and #7) mutants compared with control MDCK-CMV cells. Panels a–f and m–o, phase contrast images; g–l and p–r, immunofluorescence staining for E-cadherin (green) and vimentin (red). Nuclei were stained with DAPI (blue). Bars, panels a–f and m–o: 600 μm; g–l and p–r, 300 μm. B, Western blot showing the levels of the epithelial (E-cadherin) and mesenchymal (fibronectin, N-cadherin, and vimentin) markers (upper panels) and wt Snail1-HA or mutants using anti-Snail1 (lower panels) in MDCK cells stably transfected with wt Snail1 and the indicated ZF-mutants; α-tubulin was used as loading control.
FIGURE 6.
Snail2 ZF3 and ZF4 are independently required for EMT induction. A, phenotypic characterization of one representative Snail2-HA clone, two representative Snail2-ZF3 (#6 and #7), and Snail2-ZF4 clones (#5 and #8) mutants compared with control MDCK-CMV cells. Panels a–f, phase contrast images; g–l, immunofluorescence staining for E-cadherin (green) and vimentin (red). Nuclei were stained with DAPI (blue). Bars a–f: 600 μm; g–l: 300 μm. B, Western blot showing the levels of the epithelial (E-cadherin), and mesenchymal (fibronectin, N-cadherin, and vimentin) markers (upper panels) and wt Snail2-HA or mutants using anti-HA (lower panels) in MDCK cells stably transfected with wt Snail2 and the indicated ZF-mutants; α-tubulin was used as loading control. C, stability of Snail2-HA and the indicated ZF mutants in HEK293T cells determined by incubation with cycloheximide for the indicated time periods. Upper, Western blot analysis of wt Snail2-HA and ZF3 or ZF4 mutants levels of one representative experiment; α-tubulin was used as loading control. Bottom, densitometric quantification of the relative amount of wt Snail2-HA and the ZF mutants at the indicated time points. Results show the mean of three independent experiments ± S.D.
These results confirm the in vivo relevance of the Snail1 and Snail2 zinc fingers identified in the modeling and in vitro studies on E-cadherin and claudin-1 promoters, and strongly support the differential contribution of specific zinc fingers in Snail1 (ZF1 and ZF2) and Snail2 (ZF3 or ZF4) for their biological function.
DISCUSSION
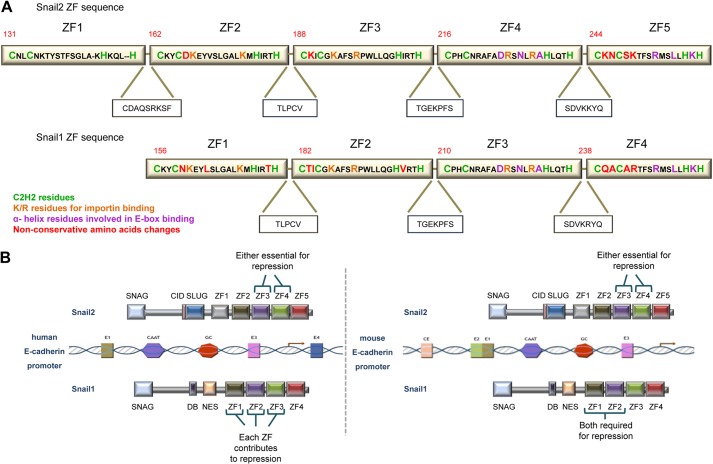
The molecular mechanisms responsible for the different biological actions of Snail1 and Snail2 are still poorly understood. Using E-cadherin as the prototypical gene target of Snail factors and careful dissection of the zinc fingers of Snail1 (ZF1-ZF4) and Snail2 (ZF1-ZF5), we have uncovered non-equivalent roles for the zinc fingers of both factors in epithelial promoter repression, DNA binding, and EMT-inducing ability. The main difference between Snail1 and Snail2 zinc finger region is the unique presence of the first zinc finger in Snail2 (45). The other four zinc fingers (ZF1-ZF4 in Snail1; ZF2-ZF5 in Snail2) are highly conserved between both factors (8); however, a careful inspection of the ZF sequences indicates the presence of several non-conservative changes between some equivalent ZFs in Snail1 and Snail2 (Fig. 7A), not previously observed. In agreement with the conserved global organization, three-dimensional modeling of Snail1 and Snail2 ZF domains bound to an E2-box sequence showed an overall similar structure, with the last two zinc fingers of either factor establishing ionic interactions with specific DNA bases through conserved residues, while the ZF1/ZF2 and ZF2/ZF3 of Snail1 and Snail2, respectively, establishing hydrogen bonds with the DNA backbone. Despite the similar structural organization, analyses of the Snail1 and Snail2 zinc finger mutants indicate a differential participation of structurally equivalent zinc fingers between both factors.
FIGURE 7.
Model of the involvement Snail1 and Snail2 zinc fingers in E-cadherin promoter repression. A, comparison of amino acid sequence of mouse Snail2 and Snail1 ZFs. The C2H2 motives, K/R residues essential for importin binding, α-helix residues that recognize the E2-boxes and the non-conservative amino acid changes between Snail1 and Snail2 ZFs are indicated by color code. The sequences of the linker regions between ZFs and the C-terminal amino acids of Snail1 and Snail2 are also shown. B, schematic diagram showing the participation of specific Snail1 and Snail2 ZFs in the repression of human and mouse E-cadherin promoters. For the human promoter, Snail2 ZF3 and ZF4 and Snail1 ZF1, ZF2, and ZF3 are independently required for efficient promoter repression. In the case of the mouse promoter, Snail2 ZF3 and ZF4 are essential for repression, while the combined action of Snail1 ZF1 and ZF2 is required for efficient repression. The last zinc fingers of Snail2 (ZF5) and Snail1 (ZF4) are dispensable or play a minor role in the repression activity of each factor.
The simulated three-dimensional models of Snail1 mutants support the essential role played by Snail1 ZF1 and ZF2 in promoter repression; thus, mutation in both ZF1 and ZF2 leads to a profound alteration in Snail1 interaction with E2-boxes that results in almost complete lack of interaction of the ZF3/ZF4 domain with the corresponding DNA bases. Indeed, ChIP assays further supported the three-dimensional model, since disruption of both Snail1 ZF1 and ZF2 leads to the loss of interaction with the endogenous human and mouse promoters, while individual ZF1 and ZF2 mutations does not alter interaction with promoters. Interestingly, promoter assays for Snail1 repression revealed important differences between human and mouse promoters; assays with human promoters (E-cadherin and claudin-1) showed that individual mutations in the first two zinc fingers (ZF1, ZF2) decreased the Snail1 repressor activity, although both mutants conserved intact their DNA binding ability (Fig. 2, C and E). By contrast, in the mouse E-cadherin promoter context, the individual Snail1 ZF mutants maintain their repression activity, in agreement with their preserved DNA binding. These results could suggest that binding to the human promoter of the single ZF1 and ZF2 mutants leads to Snail1 to adopt spatial structures unable to recruit the appropriate co-repressor machinery, and, therefore, that differential organization of the proximal E2-boxes in the human and mouse E-cadherin promoters also contributes to the Snail1/Snail2 repressive action.
On the other hand, the results obtained with Snail2 mutants, highlight the relevance of individual zinc fingers ZF3 and ZF4, since mutation in only one of them fully abolished Snail2 DNA binding and repression in all analyzed promoters in the two cell systems model. In contrast, Snail2 ZF2 and ZF5 have a minor or nonsignificant contribution to repression of human and/or mouse promoters, respectively. Indeed, ChIP assays corroborate the prominent role of Snail2 ZF3 and ZF4 in binding to endogenous human and mouse promoters and confirm the dispensable or mild participation of Snail2 ZF2 and ZF5 for DNA binding. The simulated three-dimensional models fully agree with the experimental data, since mutation of Snail2 in either ZF3 or ZF4 provokes a strong displacement on the Snail2 interaction with E2-box. Therefore, these data indicate that Snail2 ZF3 or ZF4 is a key domain in the action of Snail2.
The data obtained in analyses of the single and double mutants of Snail1 and Snail2 on different promoters indicate for the first time meaningful functional differences in the ZF region of Snail1 and Snail2 factors, with a predominant role of ZF1 and ZF2 for Snail1 transcriptional activity and a key function of ZF3 or ZF4 for Snail2 activity and binding to endogenous promoters (Fig. 7B). In addition, our data provide experimental evidence for the lack of function of Snail2 ZF1, confirming the theoretical proposal that the first zinc finger is not functional when the protein has more than four ZF (18). It could be speculated that non-conservative changes in adjacent cysteine residues present in Snail1 ZF1 and ZF2 regarding the equivalent Snail2 ZF2 and ZF3 (N160 to D166, and T183 to K189, respectively) (Fig. 7A) confer distinct spatial organization to the corresponding α-helix affecting the overall three-dimensional organization of the zinc fingers and DNA binding capacity. The three-dimensional simulation models indicate that indeed Snail1 ZF1 and ZF2 adopt a distinct spatial organization with almost a perpendicular arrangement over the DNA double helix while the equivalent Snail2 ZF2 and ZF3 exhibit a linear arrangement on the DNA (Fig. 1, A and B). These differences could explain the differential effects of individual mutations in those fingers for promoter repression and binding capacities. Although further experimental work and refined modeling is required, the present data indicate a distinctive structural organization and functional properties of, as yet, considered equivalent ZF fingers in Snail1 (ZF1, ZF2) and Snail2 (ZF2, ZF3) that influence DNA binding and likely co-repressor recruitment (Fig. 7B). Furthermore, structural analyses support an equivalent spatial three-dimensional organization of Snail1 ZF3 and Snail2 ZF4, in agreement with their strictly conserved amino acid sequence (only one conservative change between both factors, Fig. 7A) and with the deleterious effect of their individual mutation on repression of human promoters. The present data also indicate, unexpectedly, the lack or minor contribution of the last ZF in either Snail1 (ZF4) or Snail2 (ZF5) for repression activity or DNA binding, pointing to a milder, or even dispensable, function of these fingers than previously suggested based on sequence conservation (8, 45).
Taken together, the present findings indicate that the overall zinc finger domains of Snail1 and Snail2 are not equivalent, with specific zinc finger combinations (ZF1/ZF2 in Snail1, and ZF3/ZF4 in Snail2) being required for a full repression activity depending on the specific context of the E2-boxes on the human or mouse E-cadherin promoters (Fig. 7B). They further suggest that those structural differences could be behind the differential regulation of a specific set of genes and the distinct biological behavior of Snail1 and Snail2 factors (31, 38, 45). The biological relevance of Snail1 ZF1 and ZF2 and Snail2 ZF3 or ZF4 was confirmed by the loss of EMT-inducing capacity of the corresponding mutants when expressed in MDCK cells, with a proximal E-cadherin promoter organization of E2-boxes similar to the human promoter (28). In the case of Snail2, the loss of biological activity should be attributed to both the loss of binding/repressor activity and the decreased Snail2 half-life caused by the mutations. All together, these observations suggest that potential mutations in Snail1 ZF1 and ZF2 and Snail2 ZF3 or ZF4 or in other regions affecting the overall three-dimensional organization of these specific ZF domains could convey the loss of function of Snail factors. Interestingly, and in agreement with the lack of EMT induction, loss of function in Snail1 or Snail2 ZFs mutants also alter the expression of mesenchymal markers, suggesting direct or indirect effects on additional genes beyond E-cadherin and claudin-1 expression. Such mutations in specific ZFs could also explain previous observations on Snail1 or Snail2 overexpression in several tumors and cell lines without correlation with an overt EMT or pathological behavior (9, 46–49). In addition, the distinct behavior of specific Zn fingers in Snail1 and Snail2 could also explain the remarkable tumorigenic and metastatic differences detected when silencing Snail1 or Snail2 in skin carcinoma cells (38). Although further pre-clinical studies will be necessary to address the involvement of the herein identified Zn fingers, the present findings provide new avenues for therapeutic interventions addressed to target specific ZFs of Snail1 and Snail2.
Acknowledgments
We thank Amalia Montes for technical support and members of the A. Cano laboratory for helpful discussions.
This work was supported by the Spanish Ministry of Economy and Competitiveness (formerly Innovation and Sciences) [SAF2010-21143; Consolider-Ingenio CSD2007-00017]; Comunidad de Madrid [S2010/BMD-2303]; and the Instituto de Salud Carlos III [RETIC-RD12/0036/0007].
- NES
- nuclear export signal
- ZF
- zinc finger
- DBD
- DNA binding domain
- EMT
- epithelial to mesenchymal transition
- RLU
- relative luciferase units.
REFERENCES
- 1. Nieto M. A. (2002) The Snail superfamily of zinc finger transcription factors. Nat. Rev. Mol. Cell Biol. 3, 155–166 [DOI] [PubMed] [Google Scholar]
- 2. Barrallo-Gimeno A., Nieto M. A. (2009) Evolutionary history of the Snail/Scratch superfamily. Trends Genet. 25, 248–252 [DOI] [PubMed] [Google Scholar]
- 3. Grimes H. L., Chan T. O., Zweidler-McKay P. A., Tong B., Tsichlis P. N. (1996) The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell Biol. 16, 6263–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hemavathy K., Ashraf S. I., Ip Y. T. (2000) Snail/slug family of repressors: slowly going into the first lane of development and cancer. Gene. 257, 1–12 [DOI] [PubMed] [Google Scholar]
- 5. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. (2000) The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 6. Peinado H., Ballestar E., Esteller M., Cano A. (2004) Snail mediates E-cadherin repression by the recruitment of the Sin3A/ Histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell Biol. 24, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Molina-Ortiz P., Villarejo A., MacPherson M., Santos V., Montes A., Souchelnytskyi S., Portillo F., Cano A. (2012) Characterization of the SNAG and SLUG domains of Snail2 in the repression of E-cadherin and EMT induction. Modulation by serine 4 phosphorylation. PLoS ONE 7, e36132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sefton M., Sánchez S., Nieto M. A. (1998) Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 125, 3111–3121 [DOI] [PubMed] [Google Scholar]
- 9. Domínguez D., Montserrat-Sentís B., Virgós-Soler A., Guaita S., Grueso J., Porta M., Puig I., Baulida J., Francí C., García de Herreros A. (2003) Phosphorylation regulates the subcellular localization and activity of the snail transcriptional repressor. Mol. Cell Biol. 23, 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., Hung M. C. (2004) Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial mesenchymal transition. Nat. Cell Biol. 6, 931–940 [DOI] [PubMed] [Google Scholar]
- 11. Peinado H., Portillo F., Cano A. (2005) Switching on-off Snail: LOXL2 versus GSK3β. Cell Cycle 12, 1749–1752 [DOI] [PubMed] [Google Scholar]
- 12. MacPherson M. R., Molina P., Souchelnytskyi S., Wernstedt C., Martin-Pérez J., Portillo F., Cano A. (2010) Phosphorylation of serine 11 and 92 as new positive regulators of human Snail1 function: potential involvement of csaein kinase-2 amd the c-AMP- activated kinase protein kinase A. Mol. Biol. Cell 21, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mingot J. M., Vega S., Maestro B., Sanz J. M., Nieto M. A. (2009) Characterization of Snail nuclear import pathways as representatives of C2H2 zinc finger transcription factors. J. Cell Sci. 122, 1452–1460 [DOI] [PubMed] [Google Scholar]
- 14. Yamasaki H., Sekimoto T., Ohkubo T., Douchi T., Nagata Y., Ozawa M., Yoneda Y. (2005) Zinc finger domain of Snail functions as a nuclear localization signal for importin beta-mediated nuclear import pathway. Genes Cells. 10, 455–464 [DOI] [PubMed] [Google Scholar]
- 15. Miller J., McLachlan A. D., Klug A. (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pabo C., Peisach E., Grant R. A. (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 70, 313–340 [DOI] [PubMed] [Google Scholar]
- 17. Krishna S., Majumdar I., Grishin N. (2003) Structural classification of zinc fingers. Nucleic Acids Res. 31, 532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pavletich N. P., Pabo C. O. (1993) Crystal structure of a five finger GLI-DNA complex: new perspectives on zinc fingers. Science. 261, 1701–1707 [DOI] [PubMed] [Google Scholar]
- 19. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008) The epithelial mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nieto M. A. (2011) The Ins and Outs of the Epithelial to Mesenchymal Transition in health and disease. Annu. Rev. Gen. Dev. Biol. 27, 347–376 [DOI] [PubMed] [Google Scholar]
- 21. Thiery J. P., Acloque H., Huang R., Nieto M. A. (2009) Epitelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 22. Moreno-Bueno G., Portillo F., Cano A. (2008) Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 27, 6958–6969 [DOI] [PubMed] [Google Scholar]
- 23. Nieto M. A., Cano A. (2012) The epitelial-mesenchymal transition under control: global programs to regulate epithelial plasticity. Sem. Cancer Biol. 22, 361–368 [DOI] [PubMed] [Google Scholar]
- 24. Kalluri R., Weinberg R. A. (2009) The basics of epitelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 26. Hajra K. M., Chen D. Y., Fearon E. R. (2002) The SLUG zinc finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
- 27. Bolós V., Peinado H., Pérez-Moreno M. A., Fraga M. F., Esteller M., Cano A. (2003) The transcription factor Slug represses E-cadherin and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116, 499–511 [DOI] [PubMed] [Google Scholar]
- 28. Peinado H., Portillo F., Cano A. (2004) Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 48, 365–375 [DOI] [PubMed] [Google Scholar]
- 29. Peinado H., Olmeda D., Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 30. Stanisavljevic J., Porta-de-la-Riva M., Batlle R., de Herreros A. G., Baulida J. (2011) The p65 subunit of NFκB and PARP1 assist Snail1 in activating fibronectin transcription. J. Cell Sci. 124, 4161–4171 [DOI] [PubMed] [Google Scholar]
- 31. Moreno-Bueno G., Cubillo E., Sarrió D., Peinado H., Rodríguez-Pinilla S. M., Villa S., Bolós V., Jordá M., Fabra A., Portillo F., Palacios J., Cano A. (2006) Genetic profiling of epitelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epitelial mesenchymal transition. Cancer Res. 66, 9543–9556 [DOI] [PubMed] [Google Scholar]
- 32. del Barrio M. G., Nieto M. A. (2002) Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 129, 1583–1593 [DOI] [PubMed] [Google Scholar]
- 33. Murray S. A., Gridley T. (2006) Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 10300–10304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martínez Álvarez C., Blanco M., Pérez R., Rabadan M., Aparicio M., Resel E., Martínez T., Nieto M. A. (2004) Snail family members and cell survival in physiological and pathological cleft palates. Dev. Biol. 262, 207–218 [DOI] [PubMed] [Google Scholar]
- 35. Carver E. A., Jiang R., Lan Y., Oram K. F., Gridley T. (2001) The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell Biol. 21, 8184–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang R., Lan Y., Norton C. R., Sundberg J. P., Gridley T. (1998) The Slug gene is not essential for mesoderm or neural crest development in mice. Dev. Biol. 198, 277–285 [PubMed] [Google Scholar]
- 37. Olmeda D., Moreno-Bueno G., Flores J. M., Fabra A., Portillo F., Cano A. (2007) Snail is required for tumor growth and lymph node metastasis of human breast carcinoma. Cancer Res. 67, 11721–11731 [DOI] [PubMed] [Google Scholar]
- 38. Olmeda D., Montes A., Moreno-Bueno G., Flores J. M., Portillo F., Cano A. (2008) Snai1 and Snai2 collaborate on tumor growth and metastasis properties of mouse skin carcinoma cell lines. Oncogene. 27, 4690–4701 [DOI] [PubMed] [Google Scholar]
- 39. Zheng L., Baumann U., Reymond J. L. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martínez-Estrada O. M., Cullerés A., Soriano F. X., Peinado H., Bolós V., Martínez F. O., Reina M., Cano A., Fabre M., Vilaró S. (2006) The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem. J. 394, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moreno-Bueno G., Peinado H., Molina P., Olmeda D., Cubillo E., Santos V., Palacios J., Portillo F., Cano A. (2009) The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat. Protoc. 4, 1591–1613 [DOI] [PubMed] [Google Scholar]
- 42. Wolfe S., Nekludova L., Pabo C. O. (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 3, 183–212 [DOI] [PubMed] [Google Scholar]
- 43. Segal D. J., Crotty J. W., Bhakta M. S., Barbas C. F., 3rd, Horton N. C. (2006) Structure of Aart, a Designed Six-finger Zinc Finger Peptide Bound to DNA. J. Mol. Biol. 363, 405–421 [DOI] [PubMed] [Google Scholar]
- 44. Hemavathy K., Guru S. C., Harris J., Chen J. D., Ip Y. T. (2000) Human slug is a repressor that localizes to sites of active transcription. Mol. Cell Biol. 20, 5087–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manzanares M., Locascio A., Nieto M. A. (2001) The increasing complexity of the Snail gene superfamily in metazoan evolution. Trends Genet. 17, 178–181 [DOI] [PubMed] [Google Scholar]
- 46. Viñas-Castells R., Beltran M., Valls G., Gómez I., García J. M., Montserrat-Sentís B., Baulida J., Bonilla F., de Herreros A. G., Díaz V. M. (2010) The hypoxia controlled FBX114 ubiquitin ligase targets SNAI1 for proteasome degradation. J. Biol. Chem. 285, 3794–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elloul S., Silins I., Tropé C. G., Benshushan A., Davidson B., Reich R. (2006) Expression of E-cadherin transcriptional regulators in ovarian carcinoma. Virchows Arch. 449, 520–528 [DOI] [PubMed] [Google Scholar]
- 48. Côme C., Magnino F., Bibeau F., De Santa Barbara P., Becker K. F., Theillet C., Savagner P. (2006) Snail and Slug play distinct roles during breast carcinoma progression. Clin. Cancer Res. 12, 5395–5402 [DOI] [PubMed] [Google Scholar]
- 49. Castro Alves C., Rosivatz E., Schott C., Hollweck R., Becker I., Sarbia M., Carneiro F., Becker K. F. (2007) Slug is overexpressed in gastric carcinomas and may act synergistically with SIP1 and Snail in the down-regulation of E-cadherin. J. Pathol. 211, 507–515 [DOI] [PubMed] [Google Scholar]