Background: The transcription factor HypT is a dynamic oligomer and is activated by methionine oxidation.
Results: Dissociation of dodecamers into tetramers enhances and accelerates HypT activation by HOCl, proving tetramers to be the activation-competent species.
Conclusion: HypT activity is controlled by its oligomeric state and exposure to HOCl.
Significance: Regulation of a transcription factor by two levels of control enables fine-tuned activation under stress.
Keywords: Bacteria, DNA-binding Protein, DNA-Protein Interaction, Methionine, Molecular Biology, Oxidative Stress, Protein Assembly, Protein Structure, Reactive Oxygen Species (ROS), Transcription Factors
Abstract
Hypochlorous acid (HOCl) is an important component of the immune system and is produced by neutrophils to kill invading microorganisms. The transcription factor HypT is specifically activated by HOCl by methionine oxidation and protects Escherichia coli cells from the detrimental effects of HOCl. HypT forms dodecameric ring-like oligomers. Binding of HypT to DNA induces dissociation of the dodecamers into dimers and tetramers, thus forming the DNA-binding species. To dissect HypT dissociation, binding to DNA, and activation, we aimed to dissociate the dodecamers independently of DNA and to analyze HOCl-dependent activation in vitro. We found that HypT dodecamers dissociated into tetramers in the presence of l-arginine and NaCl, which was reversible upon dilution of the additive. Making use of the reversible dissociation, we generated mixed assemblies consisting of wild-type and mutant HypT subunits and determined that mutant subunits with reduced thermal stability were stabilized by wild-type HypT in the mixed assembly. HypT tetramers, as present at high NaCl concentrations, were stabilized against thermal unfolding and aggregation triggered by high HOCl concentrations. Importantly, in vitro activation by HOCl of HypT tetramers was completed within 1 min, whereas activation of dodecamers required 1 h for completion. Furthermore, activation of HypT tetramers required stoichiometric amounts of HOCl instead of an excess of HOCl, as observed for dodecamers. This supports the idea that small HypT oligomers are the activation-competent species, whereas the dodecamers are a storage form. Our study reveals the importance of the dynamic oligomeric state for HypT activation by HOCl.
Introduction
Reactive oxygen species are formed as a byproduct of aerobic respiration and are further generated by the immune system and mucosal barrier epithelia to kill invading bacteria (1, 2). Hypochlorous acid (HOCl) is a strongly antimicrobial reactive oxygen species with high oxidizing potential. Its bactericidal activity is due to oxidative damage to DNA, lipids, and proteins, whose accumulation leads to oxidative stress and damage to cells (3, 4). HOCl damages proteins on a proteome-wide scale; methionine oxidation and oxidative unfolding and concomitant protein aggregation have been described to lead to killing of bacterial cells (5, 6). Besides the damage inflicted upon proteins by HOCl, some proteins are activated by HOCl. These include the chaperone Hsp33, which in its active state prevents irreversible aggregation of proteins, and the regulators NemR, HypR, and HypT, which in turn regulate genes that support survival of the stress (6–10). Whereas Hsp33, NemR, and HypR are activated by oxidation of cysteine thiols (7, 8, 11), HypT is activated by HOCl by simultaneous oxidation of three methionines (Met-123, Met-206, and Met-230) to methionine sulfoxide, thus forming the active HypT species (10). This active state can be mimicked by methionine-to-glutamine substitution, leading to the constitutively active mutant HypTM123,206,230Q (10). Both active HypT and the constitutively active mutant show strong DNA binding.
HypT is a LysR-type transcriptional regulator that specifically protects Escherichia coli cells from HOCl damage (9, 12). LysR-type transcriptional regulators are ubiquitous regulators that typically form dimers and tetramers (e.g. OxyR (13), CysB (14), and NAC (15)) but can also form large oligomers such as octamers (CrgA (16)) and dodecamers (HypT (9)). The reduced HypT dodecamer is inactive. However, it dissociates into dimers and tetramers upon incubation with DNA in vitro, thus forming the DNA-binding species (9). Of note, given the reduced state of the protein, DNA binding is only weak (9, 10). This DNA-induced dissociation is independent of the DNA used and occurs with nonspecific DNA and target DNA alike (9).
Changes in the oligomeric state of proteins can also be enforced by using chemical additives that are known to alter the oligomerization and increase the stability and/or solubility of proteins such as urea, l-arginine, and salt (17). Urea is used to unfold proteins for stability experiments; it denatures protein either by directly interacting with the peptide backbone and polar residues or by altering the structure and dynamics of water molecules (18). l-Arginine is widely used as an additive in refolding buffers, to increase the solubility of refolding proteins, and to stabilize proteins against aggregation (19–22). l-Arginine suppresses intermolecular interactions between aggregation-prone polypeptides (22) and between subunits of oligomeric proteins such as α-crystallin, thus causing dissociation of the oligomer and simultaneously enhancing the chaperone-like activity (23). The effect of salt on protein stability and oligomerization depends largely on the kind of salt, the ionic strength, and the model substrate used (24–28). If salt acts as a stabilizer or destabilizer is determined by the Hofmeister series. Hofmeister ions interact oppositionally with different functional groups by salting out nonpolar groups and salting in peptide groups (29, 30).
In this study, we aimed to analyze the requirements for HypT activation by HOCl. Considering that the DNA-binding species, the in vivo HOCl-activated species, and the constitutively active HypT mutant are small oligomers (9, 10), we wanted to dissociate HypT dodecamers into smaller oligomers prior to activation by HOCl. Although DNA causes dissociation of HypT into smaller oligomers, the presence of DNA complicates the study of HypT activation by HOCl because DNA reacts with HOCl. Therefore, we used additives known to destabilize oligomers as a tool to dissociate the HypT dodecamer (i.e. inactive HypT) into smaller oligomers independently of DNA. We show that l-arginine and NaCl caused reversible dissociation of HypT. Reassociation did not exclusively require identical HypT species but could also be achieved using different subunits, thus allowing for generation of mixed assemblies. l-Arginine and NaCl did not cause significant conformational changes but increased the thermal stability of HypT, and NaCl also increased the stability of HypT against HOCl-induced aggregation. NaCl-induced dissociation into small oligomers strongly accelerated HypT activation by HOCl. This supports the idea that the small oligomers (i.e. the previously identified DNA-binding species (9)) are the activation-competent species and that oxidized small oligomers constitute the active species.
EXPERIMENTAL PROCEDURES
Expression and Purification of HypT and Mutants
HypT and mutants were produced in BL21(DE3) cells (strains KMG89 (wild-type HypT (9)), AD3 (FlAsH-labeled HypT* (9)), AD27 (HypTM123,206,230Q (10)), and YL20 (HypT5C→S (12))). Purification was performed as described (9, 10), and purified proteins were stored in storage buffer (10 mm NaH2PO4 (pH 7.5), 0.4 m NaCl, and 5% (v/v) glycerol) and reduced (1 mm tris(2-carboxyethyl)phosphine or DTT, 37 °C, 1 h) prior to use unless indicated otherwise. FlAsH labeling to generate HypT* was performed as described (9); HypT* does not contain a His tag.
Analytical Ultracentrifugation
Analytical ultracentrifugation analysis was performed as described (9) using 4–10 μm protein, detecting absorbance at 280 nm. Values were corrected for buffer depending on salt and arginine concentrations. The oligomerization state was analyzed by the c(s) method provided with the SedFit software or dc/dt method analyzed with the dc/dt+ software.
Analysis of HypT Dissociation and Reassociation
HypT (0.3–1.2 mg/ml) in storage buffer was incubated with various concentrations of l-arginine or dl-arginine (Sigma-Aldrich), NaCl, or urea (37 °C, 0.5 h). These additives have been described to alter the oligomerization state of certain proteins and increase the stability and/or solubility of proteins (17). Here, these additives were used to test whether HypT dodecamers can be forced to dissociate into smaller oligomers. A 2 m l-arginine or 0.7 m dl-arginine stock solution was prepared, and the pH was adjusted to 7.5 using HCl. dl-Arginine was used for CD analysis and intrinsic fluorescence, whereas l-arginine was used for all other experiments. Reassociation after arginine or NaCl treatment was induced by diluting samples with gel filtration buffer (10 mm NaH2PO4 (pH 7.5) and 0.4 m NaCl) to obtain a final concentration of 0.05 m arginine or 0.4 m NaCl (37 °C, 1 h, final protein concentration of 0.03–0.1 mg/ml).
Generation of Mixed Assemblies
HypT* (without a His tag), His-tagged HypT (HypT (9)), a His-tagged mutant in which all cysteines were replaced by serine (HypT5C→S (12)), and the His-tagged constitutively active HypT mutant (HypTM123,206,230Q (10)) in storage buffer were incubated separately with l-arginine (l-arginine final concentration of 0.3 m, 37 °C, 0.5 h). HypT* was then mixed with one of the His-tagged species. Subsequently, additives were either diluted to a final concentration of 0.05 m, removed with a PD-10 column (GE Healthcare), and protein-eluted with gel filtration buffer or removed using a Centricon 10K tube and washed with gel filtration buffer. The same effects were observed for both procedures. The final protein concentration was 0.03–0.08 mg/ml. Samples were loaded onto a HisTrap column (1-ml column volume), His-tagged assemblies were eluted with 0.5 m imidazole, and buffer was exchanged to gel filtration buffer using a NAP-5 column (GE Healthcare). Using this procedure, exclusively assemblies containing a His tag were purified.
Detection of Fluorescence (HypT*) and Western Blotting
Samples in nonreducing sample buffer were separated on a neutral pH gradient SDS gel (Serva), fluorescence was analyzed using a Typhoon scanner (9), and gels were subsequently blotted onto PVDF membrane followed by decoration with His tag-specific antibodies.
Analysis of Stability
The thermal stability of mixed assembled HypT oligomers was followed by thermal stability assays using SYPRO Orange (Invitrogen) (31). SYPRO Orange (2 μl; 1:100 dilution) was added to 18 μl of 1.5 μm HypT (in storage buffer), and fluorescence intensity was recorded from 25 to 80 °C in 55 cycles (heating rate of 1 °C/min).
Intrinsic Fluorescence
The intrinsic fluorescence intensity was analyzed using a HORIBA Yvon FluoroMax-4 fluorometer. HypT (1 μm) was analyzed in storage buffer (excitation at 295 nm, emission at 305–500 nm, 20 °C).
CD Analysis
Far-UV CD spectra were recorded using a Jasco J-715 spectrometer with 3–6 μm HypT in the absence or presence of tris(2-carboxyethyl)phosphine (1 mm, 1 h). CD spectra were recorded from 205 nm (NaCl) or 215 nm (dl-arginine) to 260 nm at 20 or 37 °C (20 nm/min), accumulating 16 spectra. To determine the thermal stability of proteins, samples were incubated from 20 to 80 °C (heating rate of 20 °C/h), and the CD signal was followed at 222 nm. Curves were fitted using a Boltzmann fit (y = A2 + (A1 − A2)/(1+e((x−x0)/dx))).
Analysis of HypT Solubility upon Incubation with HOCl
Dodecameric HypT (0.4 m NaCl) and tetrameric HypT (1.5 m NaCl) were incubated with various concentrations of HOCl (HOCl:HypT molar ratio of 1:1 to 200:1, 37 °C, 1 h). Samples were centrifuged at 17,000 × g for 20 min at 4 °C, the supernatant and insoluble proteins were separated on a neutral pH gradient SDS gel, and Coomassie Blue-stained bands were quantified using the ImageJ software.
Activation of HypT in Vitro
Glutathionylated HypT (HypTGSSG) was used (10). Dodecameric HypTGSSG (0.4 m NaCl) and tetrameric HypTGSSG (1.5 m NaCl) were incubated with various concentrations of HOCl (yielding defined HOCl:HypT molar ratios, 37 °C), and samples were removed at different time points. HOCl was quenched, and HypT was reduced by the addition of tris(2-carboxyethyl)phosphine (1 mm, 37 °C, 2 h).
Fluorescence Anisotropy Measurements
Fluorescence anisotropy with HypT variants (2 μm final concentration) and Alexa Fluor 488-labeled 158-bp metN DNA (10 nm final concentration) was performed exactly as described (9).
RESULTS
Dodecameric HypT Dissociates in the Presence of l-Arginine and NaCl
HypT forms dodecameric ring-like structures that dissociate into dimers and tetramers upon incubation with DNA, thus forming the DNA-binding species (9). To dissect dissociation of dodecamers and binding to DNA, we aimed to dissociate HypT independently of DNA. We therefore incubated purified HypT with substances known to destabilize intermolecular interactions such as salt, urea, and l-arginine (23) and analyzed the oligomerization state by analytical ultracentrifugation. HypT sediments at 10–11 S under standard conditions (10 mm sodium phosphate (pH 7.5) and 0.4 m NaCl (9)). The addition of urea did not significantly alter the oligomeric state of HypT, and the protein remained a large oligomer sedimenting at 10–11 S in the presence of up to 1 m urea (Fig. 1A). In contrast, incubation of HypT with l-arginine and NaCl, respectively, resulted in gradual dissociation of dodecamers into smaller oligomers (Fig. 1, B and C). Already 0.15–0.2 m l-arginine was sufficient to generate species at 8–9 S, likely corresponding to the hexamer (Fig. 1B). At higher concentrations of l-arginine, species sedimenting at ∼7 S (0.25 m) and 4 S (0.3 m) were detectable, which correspond to tetramers and dimers, respectively (9). Above 0.3 m l-arginine, species sedimenting at ∼3 S were detectable (Fig. 1B), which correspond to monomers (9). In the presence of NaCl, HypT remained a dodecamer until 0.6 m NaCl, whereas 0.8–1.5 m NaCl generated species of ∼6 S, corresponding to tetramers, and 2 m NaCl generated 4 S species, corresponding to dimers (Fig. 1C). Thus, we are able to dissociate the HypT dodecamer into smaller oligomers in a DNA-independent manner.
FIGURE 1.
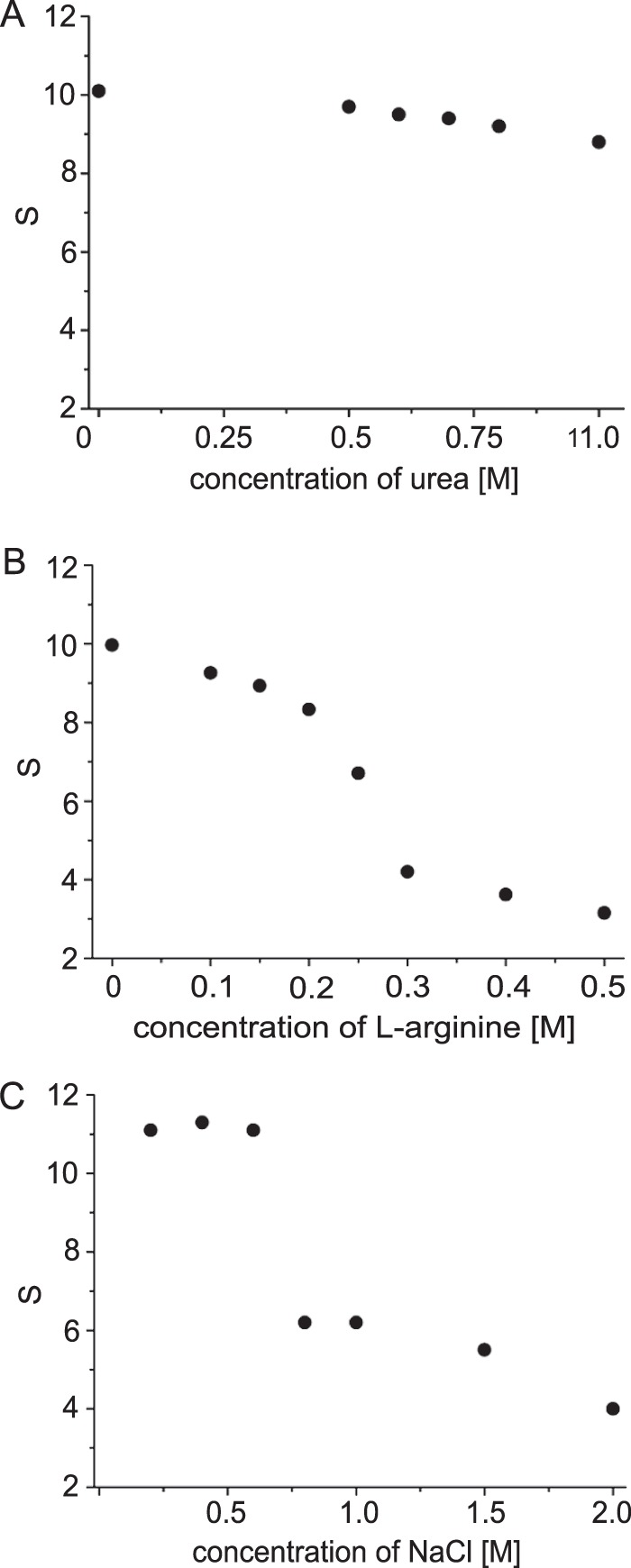
DNA-independent dissociation of HypT. HypT was incubated with the indicated concentrations of urea (A), l-arginine (B), and NaCl (C), and the oligomeric state was determined by analytical ultracentrifugation.
HypT Dissociation Is Reversible
Next, we hypothesized that the observed dissociation into small oligomers should be reversible upon removal of l-arginine or NaCl. Therefore, we used analytical ultracentrifugation to analyze the oligomerization state of HypT before and after dissociation and after removal of additives (Fig. 2). HypT was incubated with l-arginine (0.3 m) or NaCl (1.5 m) to induce dissociation, and samples were then diluted to yield a low final concentration of additives (0.05 m l-arginine or 0.4 m NaCl in gel filtration buffer). HypT dodecamers dissociated into 4.3 S species, corresponding to dimers (0.3 m l-arginine), and 6 S species, corresponding to tetramers (1.5 m NaCl), thus confirming the above observations. Importantly, these small oligomers reformed dodecamers that sedimented at ∼11 S upon dilution of additives (Fig. 2). Noteworthy, dissociation and reassociation of HypT were independent of its prior concentration. Thus, we conclude that dissociation of HypT is reversible and that the different oligomeric species of HypT are in an equilibrium governed by the concentrations of l-arginine and NaCl, respectively.
FIGURE 2.
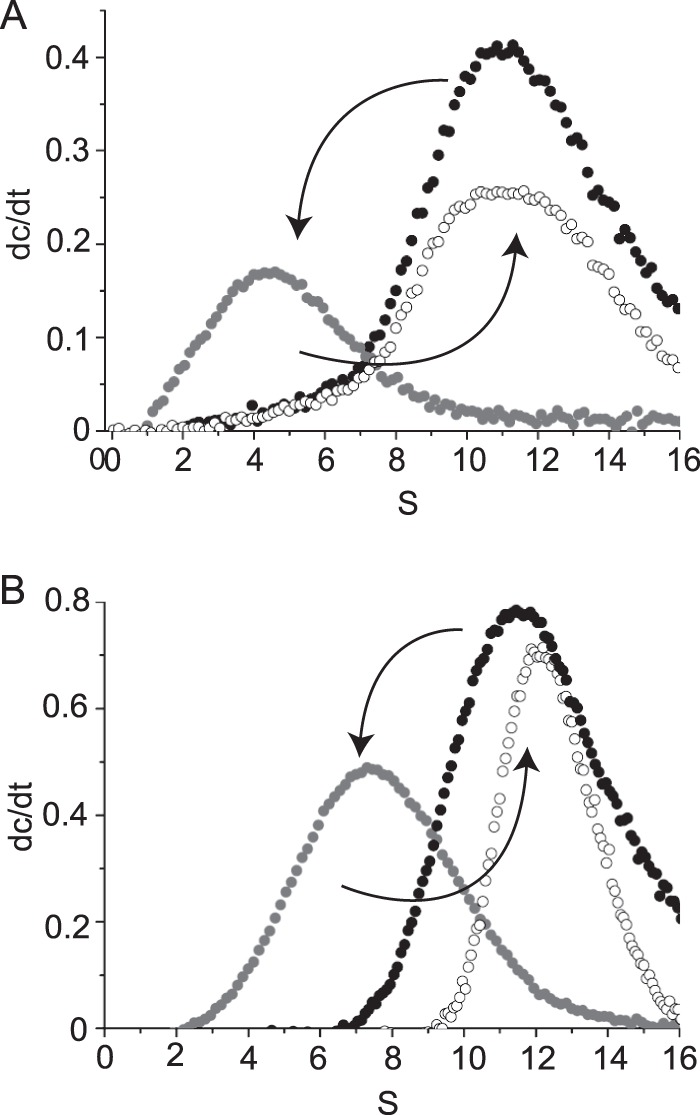
HypT dissociation is reversible. HypT (black circles) was incubated with 0.3 m l-arginine (A) or 1.5 m NaCl (B) to dissociate the dodecamer (gray circles), and additives were diluted to a final concentration of 0.05 m arginine (A) or 0.4 m NaCl (B), thus resulting in reassociation of HypT in its dodecameric state (white circles).
HypT Forms Mixed Oligomers with HypT Mutants
Given that HypT can shift from one oligomeric state to another, we asked whether it is possible to form oligomers consisting of HypT and a HypT variant. To address this question, we tested whether mixed assemblies consisting of fluorescently labeled HypT (HypT*) and His-tagged HypT can be generated (Fig. 3A). This way, we could purify assemblies via the His tag, thus removing any homogenous HypT* assembly that did not contain a His tag. The presence of HypT* in the His-tagged assemblies can be detected via fluorescence. The simultaneous presence of fluorescence and the His tag thus indicates a mixed assembly. For a His-tagged assembly partner, we chose wild-type HypT (HypT), a HypT mutant in which all cysteines were replaced by serine (HypT5C→S), and the constitutively active HypT mutant (HypTM123,206,230Q).
FIGURE 3.
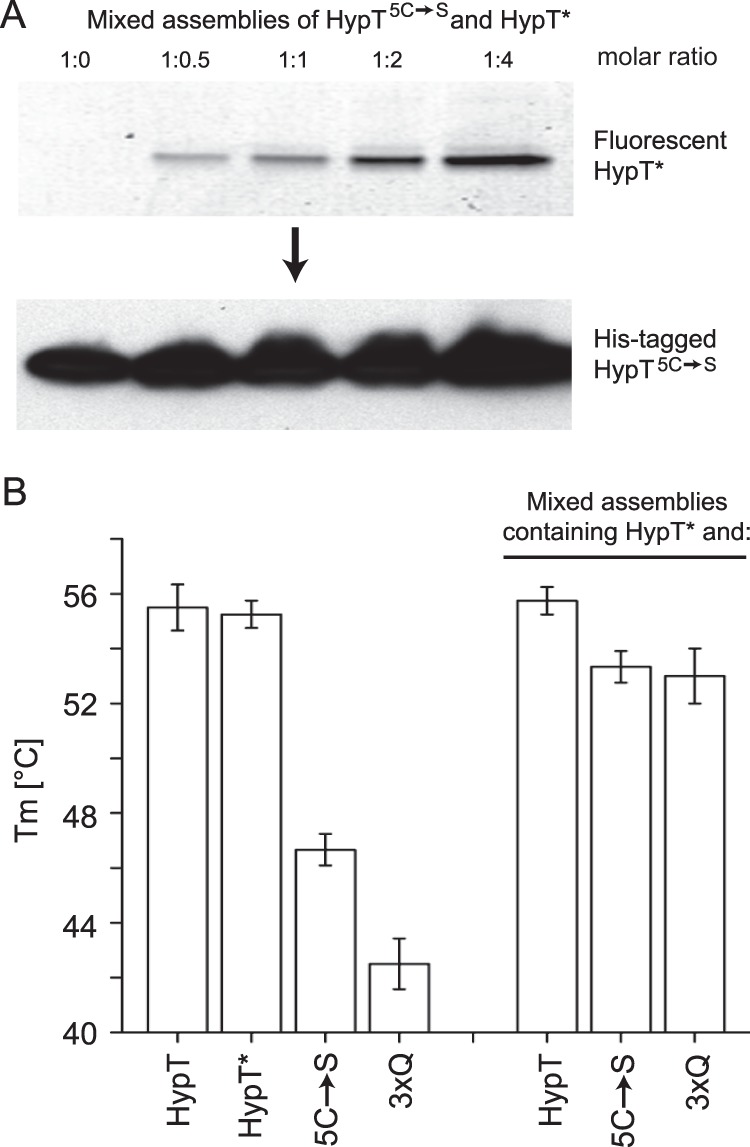
Analysis of mixed assemblies of various HypT subunits. A, mixed assemblies of HypT5C→S and HypT* subunits were generated at the indicated molar ratios. Samples were separated on nonreducing SDS gels, fluorescence was analyzed to detect HypT* (upper panel), and the His tag of HypT5C→S was detected by Western blotting using His tag-specific antibodies (lower panel). B, the thermal stability of HypT, HypT*, HypT5C→S, HypTM123,206,230Q, and different mixed assemblies was analyzed, and midpoints of thermal transition were determined. Note that wild-type HypT subunits stabilized the thermally unstable HypT5C→S and HypTM123,206,230Q subunits. Shown is the mean ± S.D. of at least three independent experiments.
We first incubated HypT* and His-tagged HypT proteins separately with 0.3 m l-arginine to dissociate them into dimers. We then mixed HypT*and one His-tagged species, each present as dimers, in various ratios and eventually diluted the samples to obtain a final concentration of 0.05 m l-arginine to induce reassociation. Thus, the smallest identical assembly was a dimer. Assemblies were purified using a HisTrap column and then used for further analysis. To test whether or not mixed assemblies were generated, samples eluted from the HisTrap column were separated by SDS-PAGE, fluorescence was then analyzed to detect HypT*, and His-tagged HypT species were additionally detected by Western blotting using antibodies specific to the His tag. When titrating HypT* to HypT5C→S, the fluorescence intensity increased with increasing proportions of HypT*. Simultaneously, similar amounts of HypT5C→S were detected in all samples, confirming that mixed assemblies were generated (Fig. 3A). Importantly, no fluorescence was detected in the sample containing only HypT5C→S (Fig. 3A, left lane), and the sample containing only HypT* did not bind to the HisTrap column (data not shown), demonstrating specific purification of the His-tagged protein from the HisTrap column.
As a further readout for the generation of mixed assemblies, we analyzed the thermal stability of HypT* and the individual His-tagged HypT mutants and compared it to that of the generated mixed assemblies (HypT* + HypT, HypT* + HypT5C→S, and HypT* + HypTM123,206,230Q). We used the thermal stability assay, which utilizes the fluorescent dye SYPRO Orange, which binds to exposed hydrophobic surfaces upon unfolding of proteins, thus exhibiting increased fluorescence (23). From these data, the midpoint of thermal transition (Tm) can be calculated, which represents the thermal stability of the protein. HypT and HypT* showed a similar thermal stability, with Tm = 55.5 ± 0.8 °C and 55.3 ± 0.5 °C, respectively (Fig. 3B). In contrast, HypT5C→S and HypTM123,206,230Q showed a lower thermal stability than HypT, with Tm = 46.7 ± 0.6 °C and 42.5 ± 0.9 °C, respectively (Fig. 3B). In contrast, the mixed assemblies demonstrated a similar thermal stability as HypT, with Tm = 55.8 ± 0.5 °C (HypT* + HypT), 53.3 ± 0.6 °C (HypT* + HypT5C→S), and 53.0 ± 1.0 °C (HypT* + HypTM123,206,230Q) (Fig. 3B). Of note, these values are ∼4–5 °C lower than the values observed by CD analysis (HypT, 60 °C (9); HypT5C→S, 50.5 °C (12); and HypTM123,206,230Q, 47.0 °C (data not shown)), yet both methods show a striking difference in the thermal stability of HypT and the mutants. We conclude that mixed assemblies of HypT and HypT mutants can be generated and that the HypT mutants are stabilized in a mixed assembly with HypT.
What Are the Effects of Arginine and NaCl on HypT?
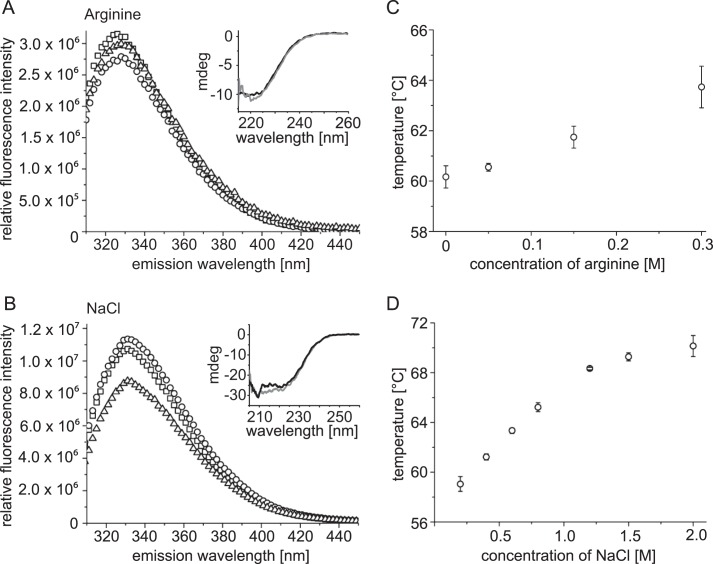
To further elucidate the effect of arginine and NaCl on HypT, we analyzed the secondary structure by CD and the conformational state by intrinsic tryptophan fluorescence. Both assays were performed upon the addition of dl-arginine (0.2–0.3 m) and NaCl (0.8–1.5 m), respectively, to induce dissociation of HypT. The secondary structure of HypT was not significantly altered either by dl-arginine (Fig. 4A, inset) or by NaCl (Fig. 4B, inset). The intrinsic tryptophan fluorescence of HypT was only slightly altered by both additives (Fig. 4, A and B). Given that the intrinsic fluorescence intensity changed only marginally without a significant wavelength shift, this indicates that the hydrophobic environment of the fluorescent tryptophan residues did not change upon dl-arginine or NaCl addition. These results show that the overall structure (i.e. secondary and tertiary structures) of HypT does not significantly change upon dissociation of HypT into small oligomers.
FIGURE 4.
Effect of dl-arginine and NaCl on structure and stability of HypT. The tertiary structure of HypT was analyzed by intrinsic tryptophan fluorescence in the presence of 0 m (squares), 0.2 m (circles), and 0.3 m (triangles) dl-arginine (A) and in the presence of 0.4 m (squares), 0.8 m (circles), and 1.5 m (triangles) NaCl (B). Insets, the secondary structure was analyzed by CD in the presence of 0 m (black line) and 0.3 m (gray line) dl-arginine (A) and 0.4 m (black line) and 0.8 m (gray line) NaCl (B). Shown are the results of one representative experiment. The thermal stability of HypT was determined by CD in buffers containing the indicated concentrations of dl-arginine (C) and NaCl (D). Shown is the mean ± S.E. of at least two independent experiments. mdeg, millidegrees.
Arginine and NaCl are known to stabilize proteins. Consequently, we analyzed the thermal stability of HypT in buffers containing various concentrations of additives by CD. HypT in standard buffer showed Tm = 59.9 ± 0.7 °C (compared with 55 °C as determined by the thermal stability assay; see above). The addition of dl-arginine induced a small increase in thermal stability, with Tm = 63.7 ± 0.8 °C (0.3 m dl-arginine) (Fig. 4C). A more pronounced effect on the thermal stability of HypT was observed for NaCl; the Tm increased to 70.1 ± 0.8 °C (2 m NaCl) (Fig. 4D). This shows that arginine and NaCl increase the thermal stability of HypT.
NaCl Stabilizes HypT against HOCl-induced Aggregation
Next, we tested the additive-dependent stability of HypT against HOCl-induced aggregation. HOCl is known to cause oxidative unfolding, leading to protein aggregation on a proteome-wide scale (6). HOCl also activates HypT in vitro and in vivo via reversible oxidation of three methionine residues to methionine sulfoxide (10). In vitro activation of HypT requires that cysteine residues are reversibly modified with glutathione before HOCl treatment, and it requires a 6:1 HOCl:HypT molar ratio. No activation is observed at lower ratios, and aggregation of HypT occurs at higher ratios (10). We hypothesized that if NaCl increases the thermal stability of HypT, it may also improve HypT stability against unfolding caused by HOCl. We used NaCl for this assay because it is chemically inert and does not react with HOCl, in contrast to arginine, whose side chain guanidinium group is a target for HOCl (32). HypT was incubated with NaCl at a final concentration of 0.4 m, at which it is present as dodecamer (standard condition), and 1.5 m, at which HypT is present as tetramer. Dodecameric HypT and tetrameric HypT were treated with different HOCl:HypT ratios (Fig. 5). Samples were centrifuged, soluble HypT was analyzed by SDS-PAGE, and band intensities were quantified. At a 12:1 HOCl:HypT molar ratio, HypT in 0.4 m NaCl aggregated significantly (Fig. 5, squares), whereas HypT in 1.5 m NaCl remained highly soluble (Fig. 5, triangles). With increasing HOCl:HypT molar ratios (up to 95:1), HypT in 0.4 m NaCl aggregated almost completely, and HypT in 1.5 m HOCl was still ∼50% soluble (Fig. 5). Only at HOCl:HypT molar ratios above 100:1 did both proteins aggregate completely. Thus, arginine and NaCl seem to have two distinct effects on HypT: they destabilize the quaternary structure, leading to dissociation of the dodecamer, and concomitantly stabilize the small oligomers.
FIGURE 5.
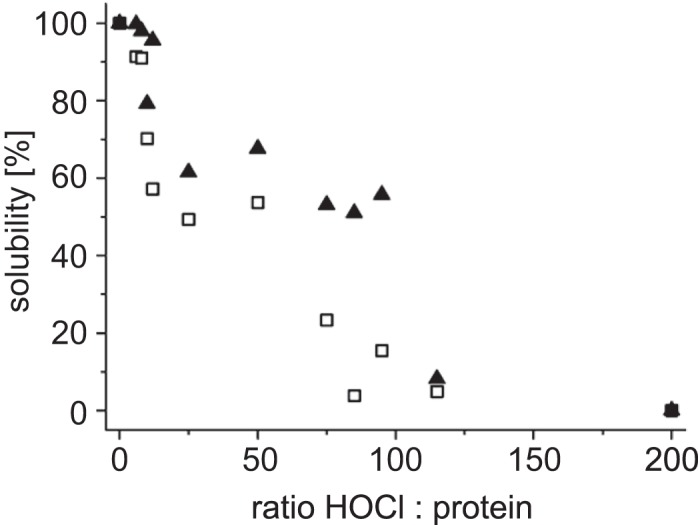
Stability of HypT against HOCl-induced aggregation. HypT in 0.4 m (squares) and 1.5 m (triangles) NaCl was incubated with the indicated molar ratios of HOCl to protein, and soluble and aggregated proteins were analyzed on SDS gels. Band intensities were quantified.
Correlation between the Oligomeric State and HypT Activation
As mentioned above, HypT is activated in standard buffer at a 6:1 HOCl:HypT molar ratio (10). We asked whether the oligomerization state of HypT influences its activation by HOCl. To address this question, we treated S-glutathionylated HypT (HypTGSSG) either in 0.4 m NaCl (dodecameric HypT) with a 6:1 HOCl:HypT molar ratio (10) or in 1.5 m NaCl (tetrameric HypT) with different HOCl ratios (Fig. 6). Samples were removed after different time points, and HOCl was then quenched by the addition of tris(2-carboxyethyl)phosphine, which also removed the glutathione moiety from HypT. Subsequently, samples were buffer-exchanged to standard buffer with a final concentration of 0.4 m NaCl. DNA-binding activity was analyzed by fluorescence anisotropy and compared under the different incubation conditions. We used reduced HypT (inactive, no HOCl treatment) and the constitutively active HypTM123,206,230Q mutant as controls, which showed anisotropy values of r = 0.030 and r = 0.070, representing low and high DNA-binding activities, respectively (10). Of note, the DNA-binding activity of HypT that was dissociated and reassociated but not treated with HOCl was higher (r = 0.046) than that of reduced HypT (r = 0.030). This was surprising because HypT treated as such should reform the dodecamer upon reassociation and show similar DNA binding as the original HypT. This suggests that dissociation into tetramers may cause slight changes in or close to the DNA-binding domain that are too small to be detected by CD or intrinsic tryptophan fluorescence but that are significant enough to form a more DNA binding-competent state.
FIGURE 6.
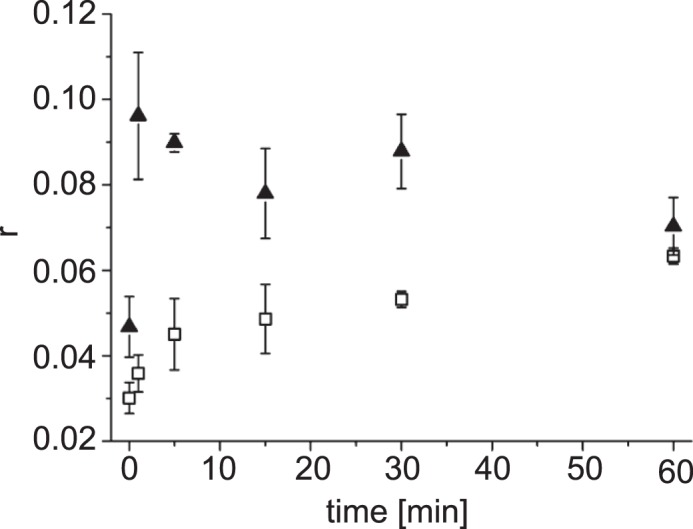
Time-dependent activation of HypT at different NaCl concentrations in vitro. HypTGSSG in 0.4 m NaCl was treated with a 6:1 molar ratio of HOCl to HypT (squares), or HypTGSSG was first dissociated with 1.5 m NaCl and treated with a 1:1 HOCl molar ratio (triangles), and reassociation was then induced. Samples were taken at the indicated time points and reduced, and DNA binding was analyzed by fluorescence anisotropy. The control sample (time point 0) was not treated with HOCl. Shown is the mean ± S.D. of at least three independent experiments.
Activation of HypT dodecamers (i.e. without prior dissociation) at a 6:1 HOCl:HypT molar ratio proceeded with a rate constant of ∼30 min−1. The DNA-binding activity reached its maximum after 1 h of incubation (Fig. 6, squares). Surprisingly, HypT that was dissociated into tetramers prior to the addition of HOCl was activated already at a 1:1 molar ratio to HOCl (Fig. 6, triangles). Higher HOCl ratios did not result in HypT activation, most likely because HOCl attacks other amino acids in HypT or the glutathione moiety (32), thus preventing DNA binding. Furthermore, activation of HypT tetramers at a 1:1 HOCl molar ratio was extremely fast and was complete within 1 min (Fig. 6, triangles). Given that activated HypT forms tetramers (10), such HOCl-treated tetramers, i.e. active HypT, also likely remain in this oligomerization state and are immediately available for DNA binding. Therefore, activation at stoichiometric amounts of HOCl makes HypT a very potent HOCl sensor. We conclude that HOCl activation of HypT tetramers occurs quickly, which supports our previous results that the active form of HypT is a tetramer in vivo (9) and in vitro (10).
DISCUSSION
Arginine and NaCl are widely used as chemical additives to refold proteins and stabilize them against aggregation (17, 19). Here, we used these additives as a tool to study HypT dissociation in a DNA-independent manner. Both additives were shown to dissociate the HypT dodecamer into small oligomers representing dimers and tetramers. Given that dissociation was achieved so far only in the presence of DNA (9), a DNA-independent dissociation now allows dissection of dissociation and DNA binding or activation. The observed effects of additives were nonspecific. Whereas DNA was negatively charged, arginine was positively charged. Thus, dissociation of HypT dodecamers does not depend on the charge of the component used for triggering dissociation. Furthermore, l-arginine-induced dissociation has also been described for other proteins such as α-crystallin, whose dissociation of the oligomer simultaneously enhances its chaperone-like activity (23). In addition, l-arginine is well known to stabilize proteins and folding intermediates and to reduce the aggregation propensity of numerous proteins (e.g. Refs. 19 and 20). Also, NaCl stabilizes proteins (e.g. Refs. 26 and 27), thus supporting the observation of increased thermal and chemical stability of HypT upon the addition of NaCl or l-arginine. However, salt ions can also promote aggregation (29) and fibrillation of proteins such as HypF, insulin, and prion proteins (28, 33–35).
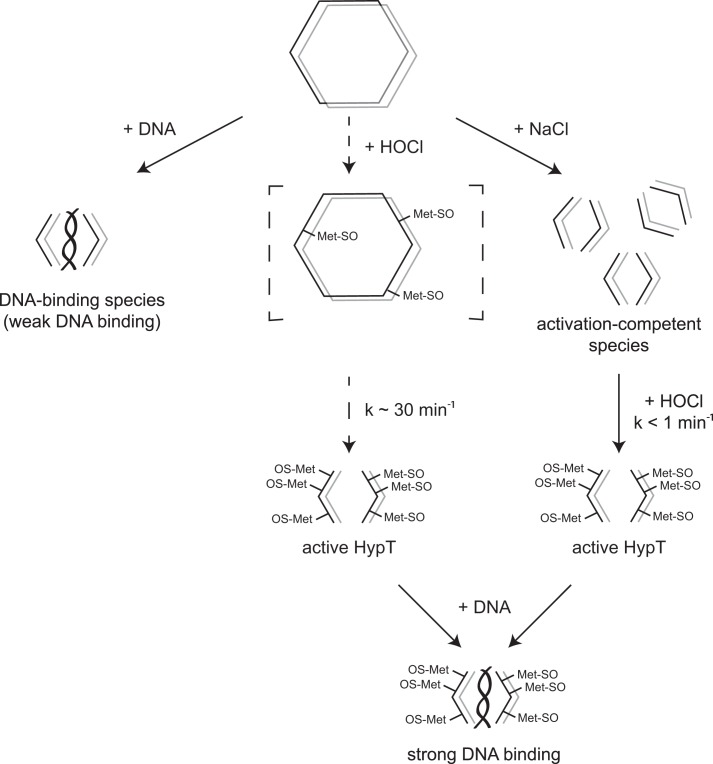
The DNA-independent dissociation of HypT allowed us to analyze two effects in vitro: the activation of HypT by HOCl as well as the reassociation of different HypT species and formation of mixed assemblies of various subunits. The dissociation of HypT is reversible, therefore enabling HypT to shift from one oligomerization state to another. HypT can be present as a storage form, i.e. dodecamer, when not required in an active state (Fig. 7). If conditions change such that active HypT is required, it can form the small species from existing dodecamers, which can be activated by HOCl very quickly. Thus, HypT exists in different states: the inactive dodecamer, the DNA binding-competent small oligomers, the activation-competent small oligomers, and the active species, i.e. oxidized tetramers (Fig. 7). At present, it is not clear whether HOCl treatment of dodecameric HypT first triggers dissociation and then methionine oxidation or if the order is reversed (Fig. 7, middle).
FIGURE 7.
Model of HypT activation. Purified HypT is present as a dodecamer and depicted as two hexamer rings stacked on top of each other. Left, in the presence of DNA, dodecamers dissociate into dimers and tetramers, generating the DNA-binding species with weak DNA-binding activity. Middle, HOCl treatment of dodecamers leads to oxidation of methionine residues and concomitant dissociation of dodecamers into tetramers. This reaction proceeds fairly slowly (k ∼ 30 min−1) and is completed within 1 h. As a result, active HypT tetramers are formed. Right, dissociation of dodecamers into tetramers can be triggered by additives such as NaCl, thus generating activation-competent tetramers. Subsequent treatment with HOCl quickly forms active HypT (k < 1 min−1). Active HypT tetramers show strong DNA-binding activity.
When we analyzed the mixed assemblies, we observed that HypT mutants with decreased thermal stability were stabilized by the presence of wild-type HypT species in the assembly. Given that the Tm values of the HypT variants are comparable when obtained by thermal stability assay after dissociation and reassociation as described in this study and CD analysis as described previously (9, 12), this indicates that the dissociation and reassociation procedure itself does not influence the thermal stability of HypT and mutants. Thus, the mixed reassociation between HypT mutants and wild-type HypT leads to an increase in thermal stability, showing that the effect is derived from wild-type HypT. Considering that the smallest identical assembly is the dimer, one could assume that wild-type HypT forms more tightly packed tetramers, maybe due to more extensive stabilizing dimer-dimer contacts, which stabilize any kind of assembly containing wild-type HypT.
Changes in oligomerization as part of an activation mechanism are not unique to HypT but are described for a number of proteins, including the LysR-type transcriptional regulator CrgA (16), the bacterial DNA-binding protein Dps (36), several small heat shock proteins (see below) that function as molecular chaperones, the Bacillus subtilis AAA+ protein ClpC (37), as well as caspases and procaspases (38). CrgA forms octameric ring-like structures that, upon binding to DNA, build a double octameric species, indicating that DNA stabilizes the larger assembly (16). Dps forms spherical dodecamers that form large biocrystals in the presence of DNA in vitro and in vivo (36). Except for small heat shock proteins, the other examples also form larger oligomers in the course of activation. In contrast, some small heat shock proteins undergo dissociation into smaller species as part of the activation mechanism. The ubiquitous small heat shock proteins form large oligomeric assemblies and protect substrate proteins from aggregation (for review, see Refs. 39 and 40). The trigger for activation may be increased temperatures and/or phosphorylation. These triggers can induce destabilization and remodeling of the large oligomer, thus causing formation of smaller oligomers with increased chaperone activity (e.g. Hsp26 (41), Hsp16.9 (42), Hsp27 (43), and αB-crystallin (44)). However, dissociation into smaller oligomers is not always required for efficient substrate binding, indicating that increased conformational flexibility may itself be sufficient for chaperone activity (e.g. HSP18.1 (45) and Hsp26 (46)).
At this point, it should be noted that the in vitro situation differs from the in vivo situation for HypT in wild-type E. coli cells. The salt composition and concentration in the E. coli cytosol are largely different from our in vitro buffer composition. Whereas the cytosol contains ∼300 mm combined sodium and potassium ions (47) in addition to many other salts and macromolecules (48), our buffer contains merely 10 mm sodium phosphate and 0.4 m NaCl (plus potential additives). Previous in vitro experiments showed that the binding of RNA polymerase and lac repressor to DNA is strongly inhibited in buffers containing high salt concentrations (49, 50). In contrast, such dependence was not observed in vivo at similarly high intracellular concentrations of salt (51). Richey et al. (51) suggested that not only is the salt concentration important but that also relatively larger numbers of nonspecific DNA-binding sites in in vivo compared with in vitro conditions could account for the observed difference between the in vitro and in vivo results. Likewise, the enzyme activity may be different if analyzed in a standard in vitro buffer compared with a buffer that represents the in vivo situation. van Eunen et al. (52) analyzed the kinetic parameters of glycolytic and fermentative enzymes of Saccharomyces cerevisiae in a buffer comprising the composition of the yeast cytosol. They showed that some enzymes had largely different activity in the in vivo buffer compared with the enzyme-specific optimum in vitro buffer (52). Furthermore, HypT dodecamers are the main species under strong overexpression conditions, whereas small oligomers, as typical for LysR-type transcriptional regulators, prevail in wild-type cells (9). Small oligomeric species are also increased upon HOCl stress of mildly hypT-overexpressing cells (9). HypT activation upon HOCl treatment of overproducing cells leads to maximum gene regulation within 10 min (9, 10). Taking into account that HypT activation is very quick when starting with small oligomers, this indicates that activation of HypT in wild-type cells is unequally faster. Taking further into account that activation requires only equimolar amounts of HOCl, this makes HypT a very sensitive HOCl probe, being perfectly equipped to immediately recognize its activating reactive oxygen species and become activated.
Acknowledgments
We thank Drs. Hauke Lilie and Stefan Gleiter and members of the Winter and Buchner laboratories for discussion.
Footnotes
This work was supported by the Elitenetzwerk Bayern (to A. D. and K. M. G.) and the Emmy-Noether Program of the Deutsche Forschungsgemeinschaft (DFG; to J. W.).
REFERENCES
- 1. Ha E. M., Oh C. T., Bae Y. S., Lee W. J. (2005) A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 [DOI] [PubMed] [Google Scholar]
- 2. Winterbourn C. C., Hampton M. B., Livesey J. H., Kettle A. J. (2006) Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome. Implications for microbial killing. J. Biol. Chem. 281, 39860–39869 [DOI] [PubMed] [Google Scholar]
- 3. Davies M. J. (2005) The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109 [DOI] [PubMed] [Google Scholar]
- 4. Gray M. J., Wholey W. Y., Jakob U. (2013) Bacterial responses to reactive chlorine species. Annu. Rev. Microbiol. 67, 141–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosen H., Klebanoff S. J., Wang Y., Brot N., Heinecke J. W., Fu X. (2009) Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc. Natl. Acad. Sci. U.S.A. 106, 18686–18691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winter J., Ilbert M., Graf P. C., Ozcelik D., Jakob U. (2008) Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gray M. J., Wholey W. Y., Parker B. W., Kim M., Jakob U. (2013) NemR is a Bleach-sensing transcription factor. J. Biol. Chem. 288, 13789–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palm G. J., Khanh Chi B., Waack P., Gronau K., Becher D., Albrecht D., Hinrichs W., Read R. J., Antelmann H. (2012) Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR. Nucleic Acids Res. 40, 4178–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gebendorfer K. M., Drazic A., Le Y., Gundlach J., Bepperling A., Kastenmüller A., Ganzinger K. A., Braun N., Franzmann T. M., Winter J. (2012) Identification of hypochlorite-specific transcription factor from Escherichia coli. J. Biol. Chem. 287, 6892–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drazic A., Miura H., Peschek J., Le Y., Bach N. C., Kriehuber T., Winter J. (2013) Methionine oxidation activates a transcription factor in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 110, 9493–9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jakob U., Muse W., Eser M., Bardwell J. C. (1999) Chaperone activity with a redox switch. Cell 96, 341–352 [DOI] [PubMed] [Google Scholar]
- 12. Drazic A., Tsoutsoulopoulos A., Peschek J., Gundlach J., Krause M., Bach N. C., Gebendorfer K. M., Winter J. (2013) Role of cysteines in the stability and DNA-binding activity of the hypochlorite-specific transcription factor HypT. PLoS ONE 8, e75683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., Ryu S. E. (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105, 103–113 [DOI] [PubMed] [Google Scholar]
- 14. Lochowska A., Iwanicka-Nowicka R., Plochocka D., Hryniewicz M. M. (2001) Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276, 2098–2107 [DOI] [PubMed] [Google Scholar]
- 15. Bender R. A. (2010) A NAC for regulating metabolism, the nitrogen assimilation control protein (NAC) from Klebsiella pneumoniae. J. Bacteriol. 192, 4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sainsbury S., Lane L. A., Ren J., Gilbert R. J., Saunders N. J., Robinson C. V., Stuart D. I., Owens R. J. (2009) The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res. 37, 4545–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rudolph R., Lilie H. (1996) In vitro folding of inclusion body proteins. FASEB J. 10, 49–56 [PubMed] [Google Scholar]
- 18. Greene R. F., Jr., Pace C. N. (1974) Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, α-chymotrypsin, and β-lactoglobulin. J. Biol. Chem. 249, 5388–5393 [PubMed] [Google Scholar]
- 19. Lange C., Rudolph R. (2009) Suppression of protein aggregation by l-arginine. Curr. Pharm. Biotechnol. 10, 408–414 [DOI] [PubMed] [Google Scholar]
- 20. Winter J., Lilie H., Rudolph R. (2002) Renaturation of human proinsulin–a study on refolding and conversion to insulin. Anal. Biochem. 310, 148–155 [DOI] [PubMed] [Google Scholar]
- 21. Arakawa T., Ejima D., Tsumoto K., Obeyama N., Tanaka Y., Kita Y., Timasheff S. N. (2007) Suppression of protein interactions by arginine, a proposed mechanism of the arginine effects. Biophys. Chem. 127, 1–8 [DOI] [PubMed] [Google Scholar]
- 22. Lyutova E. M., Kasakov A. S., Gurvits B. Y. (2007) Effects of arginine on kinetics of protein aggregation light scattering and tubidimetry techniques. Biotechnol. Prog. 23, 1411–1416 [DOI] [PubMed] [Google Scholar]
- 23. Srinivas V., Raman B., Rao K. S., Ramakrishna T., Rao C. M. (2003) Structural perturbation and enhancement of the chaperone-like activity of α-crystallin by arginine hydrochloride. Protein Sci. 12, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dominy B. N., Perl D., Schmid F. X., Brooks C. L., 3rd (2002) The effects of ionic strength on protein stability: the cold shock protein family. J. Mol. Biol. 319, 541–554 [DOI] [PubMed] [Google Scholar]
- 25. Wright D. B., Banks D. D., Lohman J. R., Hilsenbeck J. L., Gloss L. M. (2002) The effect of salts on the activity and stability of Escherichia coli and Haloferax volcanii dihydrofolate reductases. J. Mol. Biol. 323, 327–344 [DOI] [PubMed] [Google Scholar]
- 26. Mao Y. J., Sheng X. R., Pan X. M. (2007) The effects of NaCl concentration and pH on the stability of hyperthermophilic protein Ssh10b. BMC Biochem. 8, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beauchamp D. L., Khajehpour M. (2012) Studying salt effects on protein stability using ribonuclease T1 as a model system. Biophys. Chem. 161, 29–38 [DOI] [PubMed] [Google Scholar]
- 28. Campioni S., Mannini B., López-Alonso J. P., Shalova I. N., Penco A., Mulvihill E., Laurents D. V., Relini A., Chiti F. (2012) Salt anions promote the conversion of HypF-N into amyloid-like oligomers and modulate the structure of the oligomers and the monomeric precursor state. J. Mol. Biol. 424, 132–149 [DOI] [PubMed] [Google Scholar]
- 29. Baldwin R. L. (1996) How Hofmeister ion interactions affect protein stability. Biophys. J. 71, 2056–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Cremer P. S. (2006) Interactions between macromolecules and ions: the Hofmeister series. Curr. Opin. Chem. Biol. 10, 658–663 [DOI] [PubMed] [Google Scholar]
- 31. Ericsson U. B., Hallberg B. M., Detitta G. T., Dekker N., Nordlund P. (2006) Thermofluor-based stability of proteins. Anal. Biochem. 357, 289–298 [DOI] [PubMed] [Google Scholar]
- 32. Pattison D. I., Davies M. J. (2001) Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem. Res. Toxicol. 14, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 33. Nielsen L., Khurana R., Coats A., Frokjaer S., Brange J., Vyas S., Uversky V. N., Fink A. L. (2001) Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry 40, 6036–6046 [DOI] [PubMed] [Google Scholar]
- 34. Jain S., Udgaonkar J. B. (2010) Salt-induced modulation of the pathway of amyloid fibril formation by the mouse prion protein. Biochemistry 49, 7615–7624 [DOI] [PubMed] [Google Scholar]
- 35. Yeh V., Broering J. M., Romanyuk A., Chen B., Chernoff Y. O., Bommarius A. S. (2010) The Hofmeister effect on amyloid formation using yeast prion protein. Protein Sci. 19, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolf S. G., Frenkiel D., Arad T., Finkel S. E., Kolter R., Minsky A. (1999) DNA protection by stress-induced biocrystallization. Nature 400, 83–85 [DOI] [PubMed] [Google Scholar]
- 37. Kirstein J., Schlothauer T., Dougan D. A., Lilie H., Tischendorf G., Mogk A., Bukau B., Turgay K. (2006) Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 25, 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang D. W., Ditsworth D., Liu H., Srinivasula S. M., Alnemri E. S., Yang X. (2003) Oligomerization is a general mechanism for the activation of apoptosis initiator and inflammatory procaspases. J. Biol. Chem. 278, 16466–16469 [DOI] [PubMed] [Google Scholar]
- 39. Richter K., Haslbeck M., Buchner J. (2010) The heat shock response: life on the verge of death. Mol. Cell 40, 253–266 [DOI] [PubMed] [Google Scholar]
- 40. Winter J., Jakob U. (2004) Beyond transcription–new mechanisms for the regulation of molecular chaperones. Crit. Rev. Biochem. Mol. Biol. 39, 297–317 [DOI] [PubMed] [Google Scholar]
- 41. Haslbeck M., Walke S., Stromer T., Ehrnsperger M., White H. E., Chen S., Saibil H. R., Buchner J. (1999) Hsp26, a temperature-regulated chaperone. EMBO J. 18, 6744–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Montfort R. L., Basha E., Friedrich K. L., Slingsby C., Vierling E. (2001) Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 8, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 43. Shashidharamurthy R., Koteiche H. A., Dong J., McHaourab H. S. (2005) Mechanism of chaperone function in small heat shock proteins. Dissociation of the HSP27 oligomer is required for recognition and binding of destabilized T4 lysozyme. J. Biol. Chem. 280, 5281–5289 [DOI] [PubMed] [Google Scholar]
- 44. Peschek J., Braun N., Rohrberg J., Back K. C., Kriehuber T., Kastenmüller A., Weinkauf S., Buchner J. (2013) Regulated structural transitions unleash the chaperone activity of αB-crystallin. Proc. Natl. Acad. Sci. U.S.A. 110, E3780–E3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stengel F., Baldwin A. J., Painter A. J., Jaya N., Basha E., Kay L. E., Vierling E., Robinson C. V., Benesch J. L. (2010) Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc. Natl. Acad. Sci. U.S.A. 107, 2007–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Franzmann T. M., Wühr M., Richter K., Walter S., Buchner J. (2005) The activation mechanism of Hsp26 does not require dissociation of the oligomer. J. Mol. Biol. 350, 1083–1093 [DOI] [PubMed] [Google Scholar]
- 47. Schultz S. G., Solomon A. K. (1961) Cation transport in Escherichia coli. I. Intracellular Na and K concentrations and net cation movement. J. Gen. Physiol. 45, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cossins B. P., Jacobson M. P., Guallar V. (2011) A new view of the bacterial cytosol environment. PLoS Comput. Biol. 7, e1002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roe J. H., Burgess R. R., Record M. T., Jr. (1984) Kinetics and mechanism of the interaction of Escherichia coli RNA polymerase with the λ PR promoter. J. Mol. Biol. 176, 495–522 [DOI] [PubMed] [Google Scholar]
- 50. Riggs A. D., Bourgeois S., Cohn M. (1970) The lac repressor-operator interaction. 3. Kinetic studies. J. Mol. Biol. 53, 401–417 [DOI] [PubMed] [Google Scholar]
- 51. Richey B., Cayley D. S., Mossing M. C., Kolka C., Anderson C. F., Farrar T. C., Record M. T., Jr. (1987) Variability of the intracellular ionic environment of Escherichia coli. Differences between in vitro and in vivo effects of ion concentrations on protein-DNA interactions and gene expression. J. Biol. Chem. 262, 7157–7164 [PubMed] [Google Scholar]
- 52. van Eunen K., Bouwman J., Daran-Lapujade P., Postmus J., Canelas A. B., Mensonides F. I., Orij R., Tuzun I., van den Brink J., Smits G. J., van Gulik W. M., Brul S., Heijnen J. J., de Winde J. H., de Mattos M. J., Kettner C., Nielsen J., Westerhoff H. V., Bakker B. M. (2010) Measuring enzyme activities under standardized in vivo-like conditions for systems biology. FEBS J. 277, 749–760 [DOI] [PubMed] [Google Scholar]