Background: The light-harvesting complex (LHC) motif is an amino acid consensus sequence found in various thylakoid proteins.
Results: Modification and replacement of the transmembrane domain encompassing the LHC motif of a plant protein retains its membrane-anchoring function but impairs its protein-protein interaction.
Conclusion: This domain functions in membrane anchoring and complex formation.
Significance: This study provides new insights regarding the function of the LHC motif.
Keywords: Arabidopsis, Chloroplast, Isoprenoid, Plant Molecular Biology, Transgenic, Chlorophyll, Geranylgeranyl Reductase, Light-harvesting-like Protein
Abstract
The light-harvesting complex (LHC) constitutes the major light-harvesting antenna of photosynthetic eukaryotes. LHC contains a characteristic sequence motif, termed LHC motif, consisting of 25–30 mostly hydrophobic amino acids. This motif is shared by a number of transmembrane proteins from oxygenic photoautotrophs that are termed light-harvesting-like (LIL) proteins. To gain insights into the functions of LIL proteins and their LHC motifs, we functionally characterized a plant LIL protein, LIL3. This protein has been shown previously to stabilize geranylgeranyl reductase (GGR), a key enzyme in phytol biosynthesis. It is hypothesized that LIL3 functions to anchor GGR to membranes. First, we conjugated the transmembrane domain of LIL3 or that of ascorbate peroxidase to GGR and expressed these chimeric proteins in an Arabidopsis mutant lacking LIL3 protein. As a result, the transgenic plants restored phytol-synthesizing activity. These results indicate that GGR is active as long as it is anchored to membranes, even in the absence of LIL3. Subsequently, we addressed the question why the LHC motif is conserved in the LIL3 sequences. We modified the transmembrane domain of LIL3, which contains the LHC motif, by substituting its conserved amino acids (Glu-171, Asn-174, and Asp-189) with alanine. As a result, the Arabidopsis transgenic plants partly recovered the phytol-biosynthesizing activity. However, in these transgenic plants, the LIL3-GGR complexes were partially dissociated. Collectively, these results indicate that the LHC motif of LIL3 is involved in the complex formation of LIL3 and GGR, which might contribute to the GGR reaction.
Introduction
Protein domains represent functional units of protein sequences. Domains are often conserved during evolution. In some instances, specific regions of genomic DNA encoding domains have been duplicated, modified, and transferred within or between genomes. Genomic modifications such as these have contributed toward the evolution of proteins. From an analysis perspective, domains can be recognized through characterization of sequence footprints or “motifs.” Currently, numerous protein motifs have been identified by the analysis of protein sequences that are deposited in public sequence databases (1, 2). In addition to providing invaluable information regarding protein functions, these motifs also provide clues pertaining to protein evolution. The light-harvesting complex (LHC)2 motif, alternatively called the chlorophyll a/b motif, is a widely distributed motif among the genomes of oxygenic photosynthetic organisms (3–7) and was first identified in plants (8). The LHC motif plays essential roles in the LHC, providing ligands for the binding of chlorophyll and carotenoids, and also serves as a structural backbone for the LHC (9).
The LHC motif is not only identified in the protein sequences for the LHC, but it is also found in many other proteins from oxygenic photosynthetic organisms (1, 2, 6, 7). These proteins are collectively called light-harvesting-like (LIL) proteins (3–7). Unlike the LHC, LIL does not appear to be involved in light harvesting, although some of the LIL proteins appear to be at least temporarily associated with the photosynthetic apparatus (8, 10–13).
Both photosynthetic eukaryotes and cyanobacteria contain multiple LIL proteins. For example, the model cyanobacterium Synechocystis sp. PCC6803 contains five different types of LIL proteins (9, 14), whereas Arabidopsis contains at least eight different types (5). The LHC motif sequences are well conserved among all types of LIL proteins. On the contrary, sequences residing outside of the LHC motifs are not conserved among different types of LIL proteins, indicating that different types of LIL proteins have different functions (6, 7). Among various LIL proteins, ferrochelatase (FC) and LIL3 have clearly assigned functions. FC is responsible for the final step of heme biosynthesis, in which this enzyme inserts Fe2+ into protoporphyrin IX (15). LIL3 is involved in the stabilization of geranylgeranyl reductase (GGR), which is the enzyme responsible for phytol formation (16). A few other types of LIL proteins have been characterized, and their functions have been proposed. Cyanobacterial LIL proteins are hypothesized to be involved in chlorophyll turnover (17, 18) or stabilization of photosystem 1 (19). The ELIP protein, which is alternatively called LIL1, is reported to be functionally associated with seed germination (20). Both early light-inducible protein (ELIP) (13) and stress-enhanced protein (SEP) (21) are suggested to be involved in the response of plants to light stresses. Even though LIL proteins have been studied extensively, we still have a limited understanding regarding the functions of their LHC motifs. At least three specific functions have been postulated for the LHC motif: membrane anchoring, providing binding sites of tetrapyrroles, and facilitating protein-protein interactions (5, 22). The function of the LHC motif of cyanobacterial FC has been studied at both the in vivo and in vitro levels (22–24). Although removal of the LHC motif from the FC sequence did not affect its catalytic activity, it resulted in a dissociation of the dimeric FC into monomers (22) and in the increased turnover of the FC reaction (24). These results indicate that the LHC motif does not participate in the FC reaction. Instead, the LHC motif contributes to the formation of dimers, and it is possibly involved in the regulation of FC reactions by controlling monomer-dimer transitions (24). To the best of our knowledge, the function of the LHC motif still remains to be elucidated for other LIL proteins. As a result, it is not clear whether the LHC motifs of LIL proteins have common functions or not.
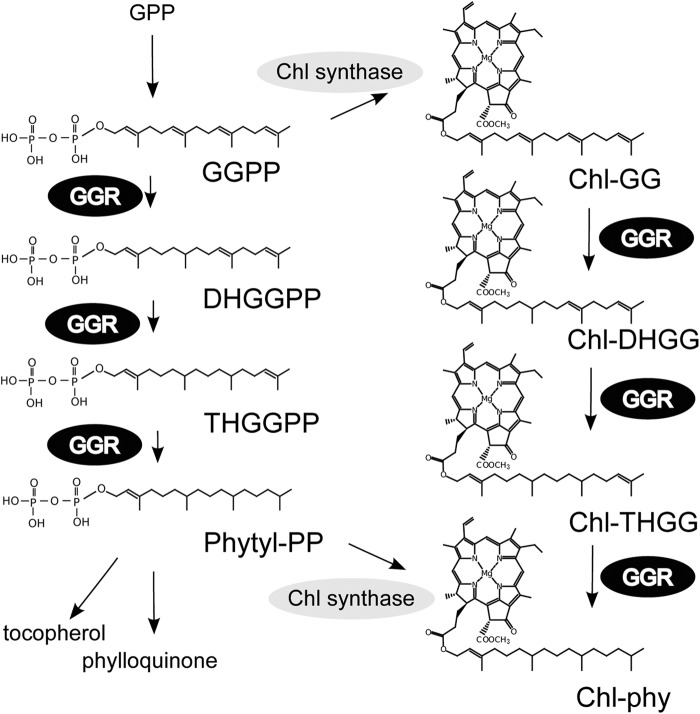
To gain a better understanding regarding the functions of the LHC motif, we functionally characterized the LHC motif with Arabidopsis LIL3 protein in this study. LIL3 is a chloroplastic protein that contains two putative transmembrane (TM) helices, one of which encompasses an LHC motif (5). LIL3 is shown to interact with GGR, which catalyzes the reduction of geranylgeranyl-diphosphate (GGPP) to form phytyl-diphosphate, which is a key reaction leading to α-tocopherol, chlorophyll (phytylated chlorophyll (Chl-phy)), and phylloquinone (Fig. 1). In addition, this enzyme catalyzes the reduction of the geranylgeranyl side chain of chlorophyll to phytol (25, 26). The Arabidopsis genome encodes two LIL3 isoforms, designated LIL3:1 and LIL3:2. Both isoforms are similar in sequence and exhibit 76% identity at the amino acid level. The LIL3 isoforms consist of the N-terminal region (120–130 amino acids), which is predicted to be soluble, and two TM helices (approximately 90 amino acids) at their C terminus, the first of which contains a well conserved LHC motif. In the Arabidopsis lil3:1/lil3:2 double mutant lacking both LIL3 isoforms, GGR levels are reduced significantly (16). Accordingly, this mutant accumulates unusual chlorophyll species containing incompletely saturated side chains (see Fig. 1 for formulas), including geranylgeranylated chlorophyll (Chl-GG), dihydrogeranylgeranylated chlorophyll (Chl-DHGG), and tetrahydrogeranylgeranylated chlorophyll (Chl-THGG). In contrast, WT plants almost exclusively synthesize Chl-phy (16). Because a knockout mutation of a single gene encoding either LIL3:1 or LIL3:2 only leads to a slight reduction in GGR levels (16), it is most likely that LIL3:1 and LIL3:2 are functionally redundant. A yeast two-hybrid assay and blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis demonstrate that LIL3 and GGR interact with each other (16). Taken together, it has been shown that LIL3 stabilizes GGR by a direct interaction.
FIGURE 1.
Schematic of the metabolic pathways in which GGR is involved. GGR catalyzes six different reactions, as indicated in this illustration. The structure of chlorophyll a is representatively depicted. GPP, geranyl-diphosphate; DHGGPP, dihydrogeranylgeranyl-diphosphate; THGGPP, tetrahydrogeranylgeranyl-diphosphate; Phytyl-PP, phytyl-diphosphate.
To analyze the function of an LHC motif, it would be desirable to use an experimental system in which a functional impairment of an LHC motif is readily detectable. However, it has not always been possible to observe dysfunction of LIL proteins in LIL-deficient mutants (21). For other LIL proteins, a complex and sophisticated experimental system is required to observe the effects of LIL protein dysfunction (15, 20, 27, 28). On the contrary, a defect in the function of LIL3 is easily detectable by observing GGR levels and Chl-phy levels (16). Thus, it is expected that Arabidopsis lil3 mutants are capable of serving as an ideal system to enable the functional characterization of an LHC motif. Here, we describe the functional analysis of the LIL3 protein and its LHC motif using the Arabidopsis lil3 mutant as a model system. It should be noted that, for this study, we utilized a transgenic approach in plants rather than adopting the yeast split ubiquitin system used in our previous study (16). This approach was chosen because the plant system is expected to provide more physiologically relevant functional information regarding the function-structure relationship of LIL3 and its LHC motif. Such information pertaining to the functionality of LIL3 is difficult to obtain with the yeast system.
In this study, we introduced a modification in the LIL3 sequence and assessed its effects by overexpressing the modified LIL3 gene in the lil3:1/lil3:2 double mutant. Furthermore, we constructed a chimeric protein by conjugating GGR with the membrane-spanning domain of LIL3 (we designated this domain as TMLIL3). For comparison, we also fused the membrane-spanning domain of thylakoid-bound ascorbate peroxidase (we designated this region as TMAPX), which we consider a non-LIL protein representative, to GGR. If the LIL3 protein functions to anchor GGR to membranes, a fusion of a membrane-spanning domain to GGR should functionally complement the lack of LIL3. The results suggest that anchoring GGR to membranes fulfills the function of LIL3. Furthermore, we found that an alteration of the LHC motif results in partial dissociation of GGR-LIL3 complexes and a substantial reduction of GGR activity. Taken together, we propose that the TM domain of LIL3 possesses a dual function to anchor GGR to membranes and to also oligomerize this enzyme.
EXPERIMENTAL PROCEDURES
Plants
The Arabidopsis lil3:1/lil3:2 mutant was isolated from a Ds transposon-tagged mutant pool of the Nossen ecotype (16). Plants were grown at 23 °C under continuous light conditions (∼80 μmol m−2 s−1) as described previously (16). Counting from the top, the fourth whorl of the leaves was harvested 4 weeks after germination for the extraction of pigments, RNA, and protein.
Chlorophyll Measurement
Chlorophyll was extracted from frozen leaves with cold acetone and subjected to HPLC analysis as described previously (16).
Construction of Arabidopsis Transgenic Plants
For the construction of Arabidopsis transgenic plants overexpressing FLAG-tagged LIL3:2, cDNA for the full-length LIL3:2 was PCR-amplified with a specific primer set that was designed to incorporate a FLAG tag (DYKDDDDK) coding sequence into the PCR product. Likewise, for the construction of Arabidopsis transgenic plants expressing the amino acid substituted LIL3:2 sequence in which three conserved amino acid residues were substituted with alanine (E171A, N174A, and D189A), GAA-to-GCA, AAT-to-GCT, and GAT-to-GCT substitutions were incorporated by an overlapping PCR method. The modified LIL3:2 cDNA fragments were cloned into the pGreenII-0229 vector (30) together with the cauliflower mosaic virus 35S promoter, the ω sequence, and the nopaline synthase terminator. For the construction of GGR-TMLIL3- or GGR-TMAPX-expressing plants, sequences encompassing the full-length GGR sequence (1–1401 of the At1g74470 sequence) and its own 5′ UTR (−83 to −1) and 3′ UTR (1402–1562) were amplified separately by PCR. The amplified fragments were subsequently fused with another DNA fragment coding for the transmembrane domain of LIL3:2 (corresponding to nucleotides 499–774, which were counted from the initiation codon) or that for the transmembrane domain of ascorbate peroxidase (corresponding to nucleotides 1105–1278) by an overlapping PCR method. The amplified PCR fragments were then cloned into the pENTR4-dual vector (Invitrogen), and the sequences were subsequently transferred to the pEarleyGate100 vector (31) by the Gateway recombination system (Invitrogen). The transfer DNA region of the aforementioned binary plasmids were introduced into the lil3:1/lil3:2 double mutant (16) by Agrobacterium-mediated transformation, with the exception of the LIL3:2-FLAG-expressing plasmid. This plasmid was introduced into the lil3:2 mutant (16) of Arabidopsis.
Immunoblotting
For immunoblotting, total leaf proteins were extracted from the fourth whorl of 4-week-old plants with extraction buffer containing 50 mm Tris-HCl (pH 8.0), 1.5% DTT, 12% sucrose, and 2% lithium dodecyl sulfate. Ten microliters of extracts, corresponding to 1 mg of leaf material, were separated on a 14% polyacrylamide gel and subsequently electroblotted to a PVDF membrane. LIL3 protein and GGR protein were detected with an anti-GGR antiserum using the ECL-Plus immunodetection kit according to the instructions of the manufacturer (GE Healthcare).
Immunoprecipitation
A single selected transgenic line expressing LIL3:2-FLAG was used for chloroplast isolation. Thylakoid membranes were isolated according to the method of Salvi et al. (32). Standard procedures for the purification of the LIL3 complex containing LIL3:2-FLAG were followed. Specifically, isolated chloroplasts from ∼120 plants were solubilized in buffer containing 1% α-dodecyl maltoside, 10 mm MOPS, and 4 mm MgCl2. Solubilized proteins were then incubated with FLAG antibody-linked beads (Tamagawa Seiki Co. Ltd.) using a rotator for 2 h at 4 °C. The beads were washed with the same buffer and then incubated with DYKDDDDK peptide-containing buffer (Wako Chemicals) for 30 min.
BN-PAGE Analysis
For BN-PAGE analysis, thylakoid membranes were isolated from the leaves (containing 3–8 μg of chlorophyll) of 4-week-old plants. Leaves were homogenized by mortar and pestle in leaf grinding medium containing 0.45 m sorbitol, 20 mm Tricine/KOH (pH 8.4), 10 mm EDTA, 10 mm NaHCO3, and 0.1% (w/v) BSA. The homogenate was initially centrifuged at 1000 × g at 4 °C for 1 min. The supernatant was then recovered and centrifuged again at 3000 × g at 4 °C for 5 min. The pellet was washed twice with the washing medium containing 0.3 m sorbitol, 20 mm Tricine/KOH (pH 7.6), 5 mm MgCl2, and 2.5 mm EDTA. The green pellet was suspended in the solubilization buffer containing 50 mm imidazole-HCl (pH 7.0), 20% glycerol, 5 mm 6-aminocaproic acid, and 1 mm EDTA and mixed with an equal volume of 2% (w/v) α-dodecyl maltoside solution. Solubilized membrane proteins were then separated by 5–14% acrylamide gradient gels according to the method of Wittig et al. (33). SDS-PAGE was performed as described previously (34).
Alignment of Amino Acid Sequences
Amino acid sequences of representative LIL3 proteins and those of Arabidopsis Lhcb1.1 were aligned by the MUSCLE software developed by Edgar (35) with its default parameters. The accession numbers of the amino acid sequences are as follows: LIL3:1 (Arabidopsis, NP_567532), LIL3:2 (Arabidopsis, NP_199522), Brassica1 (Brassica oleracea, ABD64919.1), Brassica2 (B. oleracea, ABD65018.1), Oryza1 (Oryza sativa, BAG92255.1, Oryza2 (O. sativa, BAD08011.1), Chlamydomonas (Chlamydomonas reinhardtii, XP_001699421.1), Physcomitrella1 (Physcomitrella patens, XP_001756081.1), Physcomitrella2 (P. patens, XP_001782647.1), and Lhcb1.1 (Arabidopsis thaliana, NP_564340.1).
RESULTS
Isolation of GGR-LIL3 Complexes
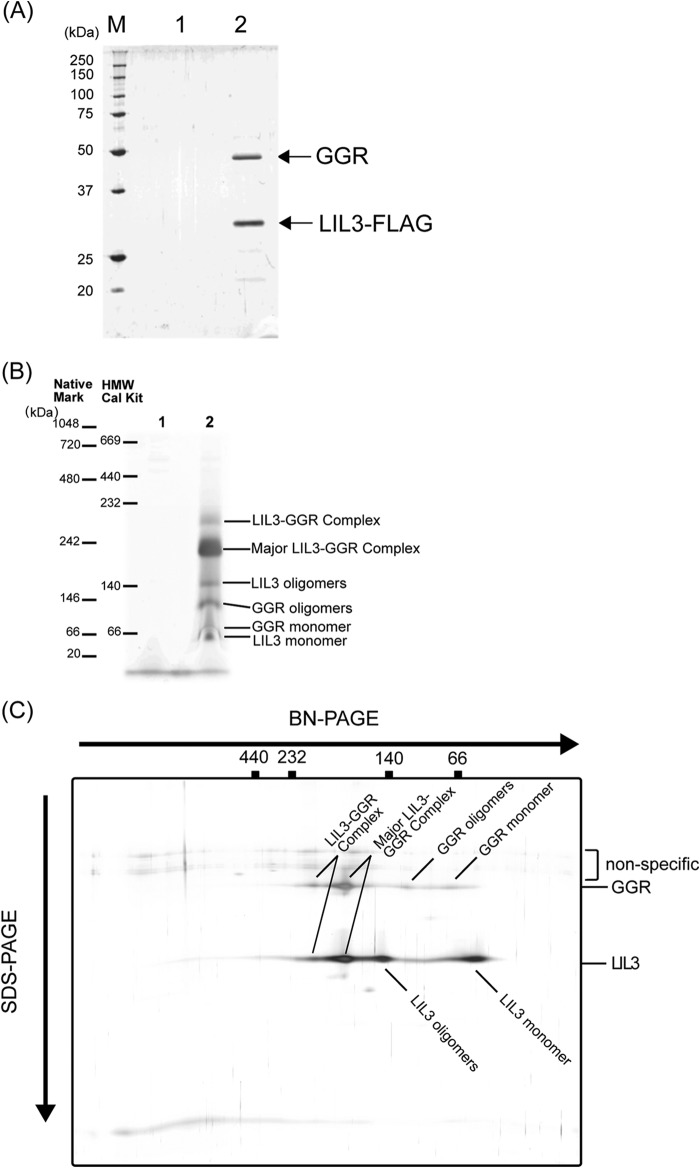
In our previous study, we showed that LIL3 interacts with GGR, forming a few protein complexes whose sizes are estimated to range between 160–250 kDa (16). To obtain further insight into the function of LIL3, we analyzed the protein compositions of the LIL3-GGR complexes. We expressed a FLAG-tagged LIL3:2 protein in the Arabidopsis lil3:2 mutant and purified protein complexes using anti-FLAG antibody-conjugated magnetic beads. Isolated LIL3-GGR complexes were analyzed by BN-PAGE and SDS-PAGE (Fig. 2). The isolated LIL3:2-containing complex was subsequently resolved by SDS-PAGE, which yielded two major bands (Fig. 2A). Mass spectroscopic analyses determined that these bands represent LIL3:1 and LIL3:2, both of which are estimated to be 30 kDa, and GGR (47 kDa). The results indicate that the major constituents of the isolated fraction are heteromeric complexes consisting of LIL3:1, LIL3:2, and GGR. The isolated complexes were separated into several major bands in the BN-PAGE analysis (Fig. 2B). To assign each band, we analyzed the isolated complexes with two-dimensional BN-PAGE/SDS-PAGE analyses (Fig. 2C). The protein profile was in good accordance with those obtained by immunoblotting analysis of the whole cell extracts, which were probed with anti-GGR and anti-LIL3 antibodies (16). Collectively, these data indicate that the isolated complexes reflect the protein compositions of the prepurified LIL3-GGR complexes.
FIGURE 2.
A, SDS-PAGE analysis of the purified LIL3-GGR complexes. M, molecular weight marker; 1, protein profile of purified WT extracts as a negative control; 2, protein profile of the purified extracts from the LIL3-FLAG-expressing plant. B, BN-PAGE analysis of the purified LIL3-GGR complexes. Two molecular weight markers (NativeMark from Invitrogen and the HMW calibration kit (HMW Cal Kit) from GE Healthcare) were run for comparison. 1, protein profile of purified WT extracts as a negative control; 2, protein profile of the purified extracts from the LIL3-FLAG-expressing plant. C, two-dimensional BN-PAGE/SDS-PAGE analysis of the purified LIL3-GGR complexes. For BN-PAGE analysis, the HMW calibration kit was used for estimation of the molecular weights, and gels were stained with a silver staining kit (Thermo Fisher Scientific Inc.).
In the BN-PAGE analysis, we ran two molecular weight markers, NativeMark unstained protein standard (Invitrogen, catalog no. LC0725) and HMW native marker kit (GE Healthcare, catalog no. 17-0445-01), alongside the isolated protein complexes on the BN-PAGE gel (Fig. 2B). Because of the nature of native gel electrophoresis, the migration of protein complexes is largely influenced by the surface charges and the structure of protein complexes. As a result, the migration patterns of protein complexes in a native PAGE gel do not always correlate with the molecular weight of these complexes. In our analysis, the two representative molecular weight markers showed significant differences in their migration patterns. The major protein complex was estimated to be ∼230 kDa, according to the NativeMark unstained protein standard. On the other hand, the same complex was estimated to be 190 kDa, according to the HMW native marker kit.
To gain further information regarding the composition of the LIL3-GGR complexes, the isolated LIL3-GGR complexes were analyzed by a combination of BN-PAGE and SDS-PAGE (Fig. 2C). Both of the largest LIL3-GGR and the major (second-largest) LIL3-GGR complexes appear to be solely composed of LIL3 and GGR proteins. On the contrary, the third-largest band seems to consist of LIL3, whereas the fourth-largest band seems to only consist of GGR. It is likely that the protein that we assigned as GGR oligomers and monomers was dissociated from the LIL3-GGR complexes during electrophoresis because the protein was purified using the anti-FLAG antibody-conjugated beads, which interact with the FLAG tag conjugated with LIL3. On the other hand, it is not clear whether the protein entities to which we assigned LIL3 oligomers and LIL3 monomers are present in vivo. It should be noted that nonspecific signals were detected above GGR signals in our two-dimensional analysis. It is likely that these signals were the result of human keratin contamination. Taken together, these results indicate that the major LIL3-GGR complexes are composed of the oligomers of LIL3 and GGR.
Subsequently, we analyzed the intraorganelle localization of LIL3 and GGR. Isolated chloroplasts were further separated by sucrose density gradient centrifugation into three different fractions enriched for thylakoid membranes, envelope membranes, and stromal components, respectively. Immunoblotting analyses revealed that LIL3 was exclusively detected in the thylakoid membrane fraction (Fig. 3). Likewise, the majority of GGR was detected in the same fraction (Fig. 3). A trace amount of GGR was also detected in the envelope fraction. It is possible that our envelope fraction contained a small amount of thylakoid membranes, which might have resulted in the trace detection of GGR and LHCB1 (a thylakoid marker) in this fraction. Considering that the envelope membrane only constitutes a minor fraction of total chloroplast membranes, which is estimated to be only 2% of the total membrane fraction (36), it is concluded that a vast majority of GGR is localized in thylakoid membranes.
FIGURE 3.
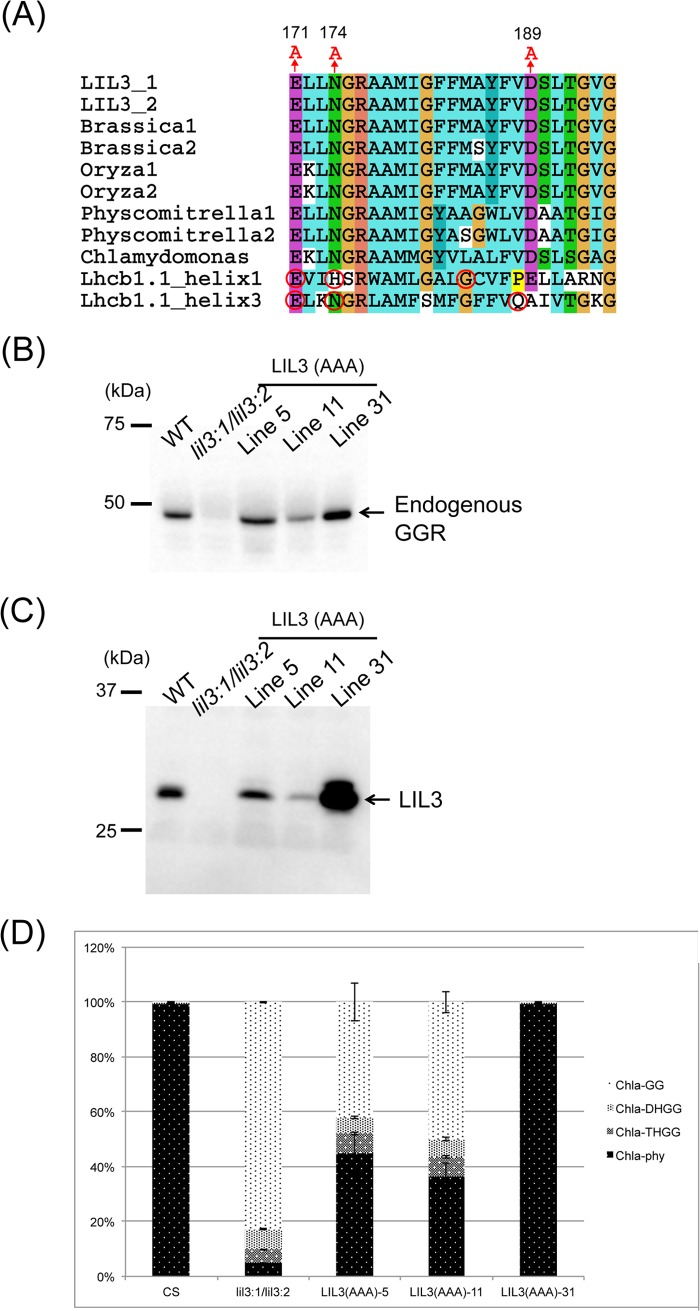
Suborganellar localization of LIL3 and GGR. Chloroplasts were isolated from 4-week-old WT leaves and fractionated into stroma, envelope membranes, and thylakoid membranes. 10 μg of protein was loaded per lane for the analyses with anti-LIL3 and anti-GGR antibodies. 2 μg of protein was loaded per lane for the analyses with the other antibodies. Anti-Rubisco antibody (stroma marker), anti-Tic110 antibody (envelope marker), anti-Lhcb1 antibody (thylakoid marker), anti-LIL3 antibody, and anti-GGR antibody were incubated with protein blots and subsequently detected with a Western Lightning Plus enhanced chemiluminescence kit (PerkinElmer Life Sciences).
Membrane-anchored GGR Is Stabilized in the Absence of LIL3
Although LIL3 has been shown to be necessary for the stabilization of GGR (16), the exact functional role of LIL3 was still not known. Considering that GGR lacks predictable hydrophobic domains (25) although it is mainly localized on thylakoid membranes (Fig. 3), we hypothesized that LIL3 anchors GGR to membranes and that this anchoring stabilizes GGR. If this hypothesis were correct, a direct fusion of a TM domain to GGR would substitute the function of LIL3. To assess this hypothesis, we fused the putative TM domain of LIL3 (hereafter referred to as TMLIL3), to GGR and constitutively expressed this chimeric protein (hereafter referred to as GGR-TMLIL3) in the Arabidopsis lil3:1/lil3:2 double mutant lacking LIL3:1 and LIL3:2 (see Fig. 4 for an overview of the experimental scheme). The GGR-TMLIL3-expressing lines exhibited enhanced growth and greener leaves in comparison with the lil3:1/lil3:2 double mutant (Fig. 5). These data clearly indicated that GGR-TMLIL3 is capable of complementing the phenotype of the mutant.
FIGURE 4.
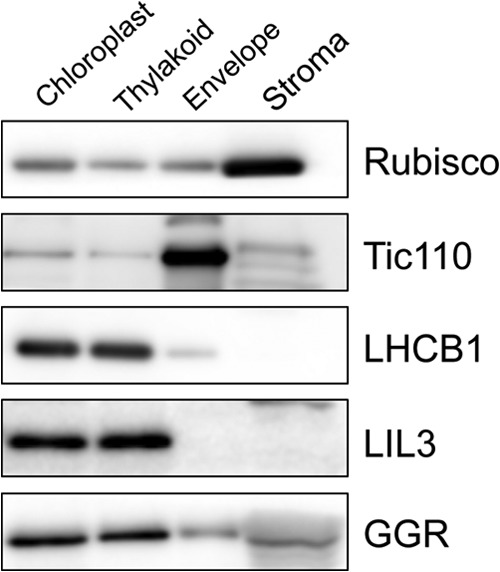
Schematic of the postulated structure of the modified GGR or LIL3 protein. The depicted stoichiometries of each subunit are hypothetical. The large globular structure colored in purple represents GGR, and the small globular structure colored in yellow depicts the N-terminal domain of LIL3. Yellow or green cylindrical structures represent TMLIL3 and TMAPX, respectively. The LHC motif in TMLIL3 is shown in orange. A, postulated structure of the native LIL3-GGR complex. The structure is shown as a dimer, but it could be a trimer or other type of oligomer. B, postulated structure of GGR-TMLIL3. C, postulated structure of GGR-TMAPX. D, structure of the postulated GGR-LIL3(TMAPX) complex. E, postulated structure of the LIL3(AAA)-GGR complex, which appears to be much smaller than the native LIL3-GGR complexes.
FIGURE 5.

Appearance of 4-week-old Arabidopsis transgenic plants expressing GGR-TMLIL3, GGR-TMAPX, LIL3(TMAPX), and LIL3(AAA) together with the WT and lil3:1/lil3:2 plants. Three independent transgenic lines are presented for each construct.
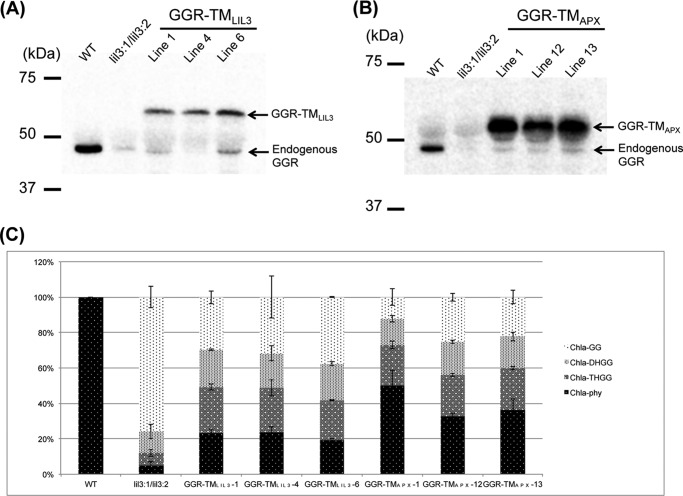
To further assess the phytol-synthesizing activity in the GGR-TMLIL3-expressing lines, we analyzed the compositions of Chl species that are conjugated with incompletely reduced side chains (Chl-GG, Chl-DHGG, and Chl-THGG) and that with the completely reduced side chain (Chl-phy). (We found that the compositions of the side chains for Chl a were well correlated with those for Chl b in all plants used in this study. Therefore, we only show the results of Chl a for simplicity.) WT plants almost exclusively synthesize Chl-phy, whereas the Chl-phy level is less than 5% of the total Chl level in the lil3:1/lil3:2 double mutant (Fig. 6C). Instead, the major Chl species in this mutant is Chl-GG, which constitutes nearly 80% of the total Chl (Fig. 6C). The Chl-phy levels in the GGR-TMLIL3-overexpressing lines have been recovered to ∼20% of the total Chl a level (Fig. 6C). Concomitantly, the levels of Chl-GG were substantially reduced to 30% of total Chl in the GGR-TMLIL3-overexpressing lines. These results indicate that the phytol-synthesizing activity is partially restored in the GGR-TMLIL3-overexpressing lines. Immunoblotting analysis showed that the GGR-TMLIL3 lines accumulate a substantial level of this chimeric protein, which is approximately half of the wild-type GGR levels (Fig. 6A). These results show that conjugating the TMLIL3 domain increases the stability of GGR.
FIGURE 6.
A, immunoblotting analysis of GGR-TMLIL3 levels in three independent GGR-TMLIL3-expressing lines. B, immunoblot analysis of GGR-TMAPX levels in three GGR-TMAPX-expressing lines. C, the compositions of Chl a derivatives in the GGR-TMLIL3 and GGR-TMAPX lines. Error bars represent S.D. n = 4.
To test whether the TMLIL3 domain stabilizes GGR by anchoring or through another specific function, we fused the membrane-spanning domain of ascorbate peroxidase (APX), which we referred to as TMAPX, to the C terminus of GGR and overexpressed them under the control of the 35S promoter of cauliflower mosaic virus in the lil3:1/lil3:2 double mutant (see Fig. 4). It has been shown previously that TMAPX is not involved in the catalytic mechanism of APX and just functions to tether the catalytic domain of APX to thylakoid membranes (37). If TMAPX stabilizes GGR in a similar manner as TMLIL3, it would indicate that anchoring GGR to membranes stabilizes GGR. We found that the growth of the GGR-TMAPX-expressing lines was recovered and that their leaves become greener when compared with the lil3:1/lil3:2 double mutant (Fig. 5). In addition, the proportion of Chl-phy was higher (∼40%) in these lines than in the lil3:1/lil3:2 double mutant (∼5%) (Fig. 6C). In three independent GGR-TMAPX-expressing lines, GGR-TMAPX levels were significantly increased when compared with GGR levels in the lil3 double mutant (Fig. 6B). These results show that TMAPX also stabilizes GGR, indicating that anchoring GGR to membranes increases the stability of GGR. Taken together, these results demonstrate that the function of LIL3 includes the anchoring of GGR to membranes.
The stabilizing effect of TMAPX appears to be greater in the GGR-TMAPX lines than that of TMLIL3 in the GGR-TMLIL3-expressing lines (Fig. 6, A and B). One possible explanation is that TMAPX has less interference with the structure of GGR because TMAPX contains a single membrane-spanning domain, whereas TMLIL3 contains two. As a result, it is possible that TMAPX might have a more compact structure.
It should be noted that the GGR-TMLIL3- and GGR-TMAPX-expressing plants also accumulate a low level of endogenous GGR protein, which is equivalent to that of the double mutant (Fig. 6, A and B). These results indicate that overexpression of modified GGR-TMLIL3 or GGR-TMAPX does not affect the stability of endogenous GGR. Thus, we conclude that the chimeric proteins (GGR-TMLIL3 or GGR-TMAPX) contribute to the recovery of Chl-phy synthesizing ability in the GGR-TMLIL3- or GGR-TMAPX-expressing lines.
Substitution of the LHC Motif with the TMAPX Domain in the LIL3 Protein Stabilizes Endogenous GGR
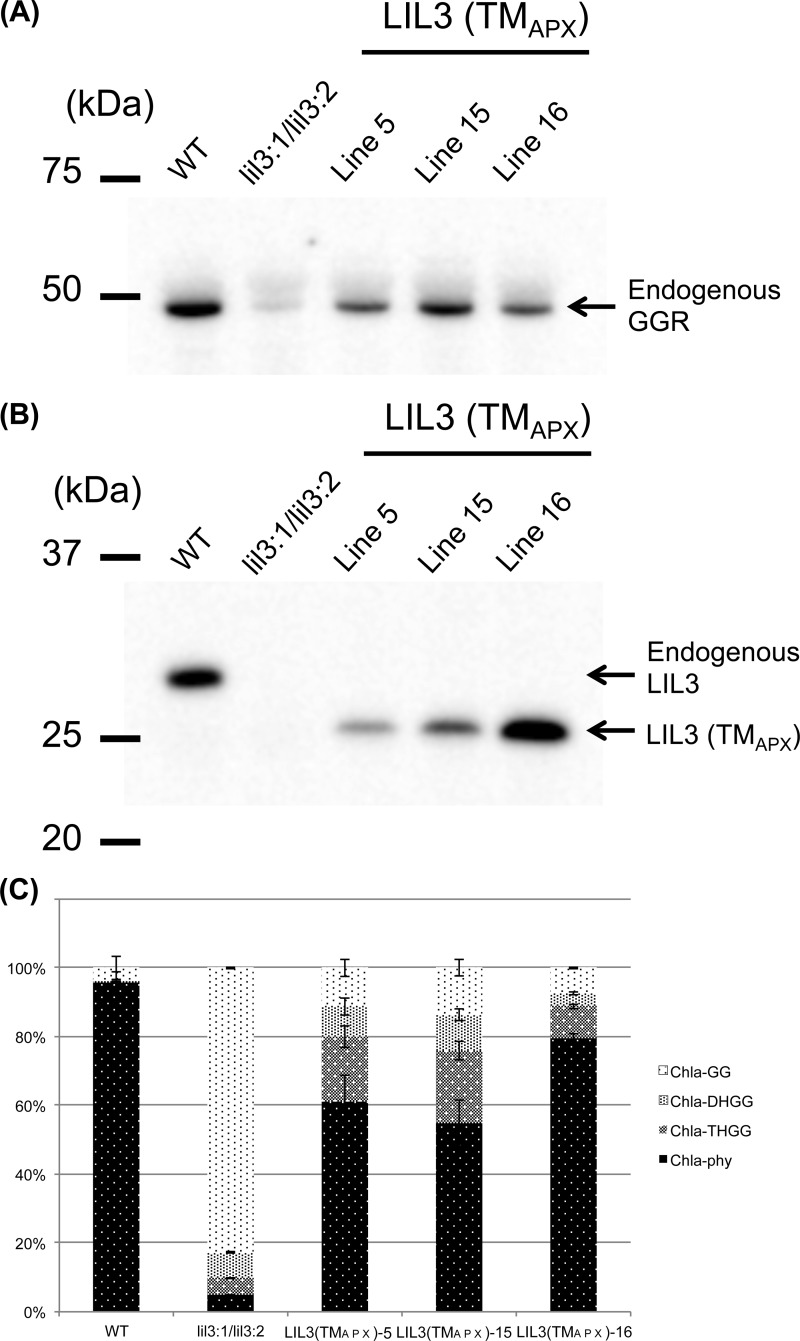
If LIL3 solely functions to anchor GGR to thylakoid membranes, TMLIL3 could be replaced with another membrane-spanning domain without compromising the function of LIL3. We tested this hypothesis by replacing TMLIL3 with TMAPX in the LIL3:2 sequence and overexpressed this chimeric protein in the lil3:1/lil3:2 double mutant (see Fig. 4). These transgenic lines are termed LIL3(TMAPX) lines, and we analyzed three independent LIL3(TMAPX) lines in this study. These lines also recovered their growth, which was almost undistinguishable from the WT (Fig. 5). Consistent with the recovery of plant growth, these plants accumulated a substantial amount of Chl-phy, which accounts for ∼60–80% of total Chl content. In contrast, the amount of Chl-GG decreased from 80% of total Chl content of the lil3 double mutant to ∼10% of the total Chl content in the LIL3(TMAPX) lines (Fig. 7). These results demonstrate that the LIL3(TMAPX) lines substantially recovered phytol-biosynthesizing activity. Among the characterized transgenic lines, the levels of LIL3(TMAPX) protein varied (Fig. 7B). Specifically, the level was much lower in lines 5 and 16 of the LIL3(TMAPX)-expressing lines compared with that of the WT. It appears that the level of LIL3(TMAPX) is not well correlated with that of GGR in line 16. Nevertheless, accumulation of LIL3(TMAPX) increased the stability of endogenous GGR in the LIL3(TMAPX)-overexpressing lines (Fig. 7A), compared with the lil3 double mutant. These results show that the TMLIL3 domain can be replaced by another membrane-anchoring domain that lacks an LHC motif, indicating that membrane anchoring is a primary function of LIL3. On the contrary, if we consider the observation that GGR levels are still less in the LIL3(TMAPX)-expressing plants compared with the WT (Fig. 7A), this would suggest that the function of LIL3 is not completely fulfilled with TMAPX. It is possible that TMAPX interferes with the correct folding of LIL3. Alternatively, TMLIL3 may have an extra function to enhance the function of LIL3.
FIGURE 7.
A, immunoblot analysis of GGR levels in three independent LIL3(TMAPX)-expressing lines. B, immunoblot analysis of LIL3(TMAPX) levels in three independent LIL3(TMAPX)-expressing lines. C, the compositions of Chl a derivatives in the LIL3(TMAPX) lines. Error bars represent S.D. n = 4.
Amino Acid Substitution in the LHC Motif of LIL3 Protein Partly Compromised Complex Formation and GGR Accumulation
To gain further insight into the function of TMLIL3, which includes the LHC motif, we modified the sequence of the LHC motif within the LIL3:2 protein. Three conserved amino acid residues (Gln-171, Asn-174, and Asp-189) of the LHC motif within the LIL3:2 sequence were all substituted with alanine (A) residues (Fig. 8A). Two of these amino acids (Gln-171 and Asn-174) were selected because they correspond to the conserved chlorophyll-binding amino acid residues in the solved structure of LHC (9, 39). Asp-189 was chosen because it is another positively charged residue within the LHC motif of LIL3:2, and we speculated that it might potentially interact with a Chl molecule. We named the amino acid substituted version of the LIL3:2 protein LIL3(AAA), and three independent LIL3(AAA)-expressing transgenic Arabidopsis lines were selected for subsequent functional characterization. Among them, line 5 accumulates LIL3 protein to a similar level to that of the WT, whereas line 11 shows a lower LIL3 level, and line 31 shows a higher LIL3 level (Fig. 8C). It is possible that the varied levels of LIL3(AAA) within three independent lines might be due to a variation in transgene expression, which is often caused by a positional effect of transgene insertion into the Arabidopsis genome. The growth of line 31 was recovered, and it was undistinguishable from the WT, whereas lines 5 and 11 were still smaller than the WT (Fig. 5). However, both of these lines grew better than the lil3:1/lil3:2 double mutant (Fig. 5). Consistent with the recovery of the growth, line 31 accumulated Chl-phy levels that were comparable with the WT, whereas the other two LIL3(AAA)-expressing lines had less than the WT (Fig. 8D). The endogenous GGR levels were correlated with the Chl-phy levels in the LIL3(AAA) lines (Fig. 8B).
FIGURE 8.
A, amino acid alignment of representative LIL3 and LHC sequences. The numbers above the alignment indicate the amino acid positions in the Arabidopsis LIL3:2 sequence. Chlorophyll-binding ligands in helices 1 and 3 of Lhcb1 are shown with red circles. B, immunoblot analysis of GGR levels in three independent LIL3(AAA)-expressing lines. C, immunoblot analysis of LIL3(AAA) levels in three independent LIL3(AAA)-expressing lines. D, the compositions of Chl a derivatives in the LIL3(AAA) lines. Error bars represent S.D. n = 4.
Although line 31 recovered phytol-synthesizing activity with the LIL3 and GGR levels that greatly exceeded the WT levels, line 5 accumulated LIL3 and GGR proteins at similar levels as those observed for the WT. It is important to note that line 5 did not recover phytol-synthesizing activity to a similar level as the WT (Fig. 8D). These results indicate that targeted amino acid substitution in the LHC motif partly compromises the function of LIL3. Although these data do not exclude the possibility that the amino acid modification exerted conformational changes on other parts of LIL3, we think this is unlikely because the experiments with LIL3(TMAPX) indicated that the conformation of the TM domain and that of the other part of LIL3 were independent from each other. In the case of cyanobacterial FC, removal of the LHC motif from the FC sequence resulted in the dissociation of the FC dimer into monomers (23). The results led us to hypothesize that the modification of the LHC motif in TMLIL3 also affects the oligomerization of LIL3-GGR complexes. Therefore, we analyzed the LIL3-GGR complexes in the LIL3(AAA) lines by BN-PAGE, which was followed by SDS-PAGE and immunoblotting. We found that the larger LIL3-GGR complex, which was estimated to be ∼170 kDa with the HMW native marker kit, almost disappeared in the LIL3(AAA) line. On the other hand, smaller complexes that were estimated to be less than 100 kDa (Fig. 9) were observed to increase in the LIL3(AAA) line. Although the exact molecular sizes and subunit compositions of these LIL3-GGR complexes are not known at this time, modification of the conserved amino acids of the LHC motif was shown to result in partial dissociation of the LIL3-GGR complex. We hypothesize that the observed LIL3-GGR oligomers in the LIL3(AAA) lines reflect the in vivo functional status of these proteins in thylakoid membranes, and it is not due to an aggregation of these proteins during BN-PAGE analysis. It is because our analysis of the Chl-phy levels in the LIL3(AAA) lines shows that these lines substantially recovered their phytol-synthesizing activity, indicating that LIL3-GGR is (at least partly) functional (Fig. 8D). However, it is still possible that LIL3 and GGR form an even larger complex and that they are partly dissociated to form the observed complexes during the BN-PAGE analysis. In either case, these results indicate that the LHC motif of LIL3 functions to facilitate stable oligomerization of the proteins.
FIGURE 9.
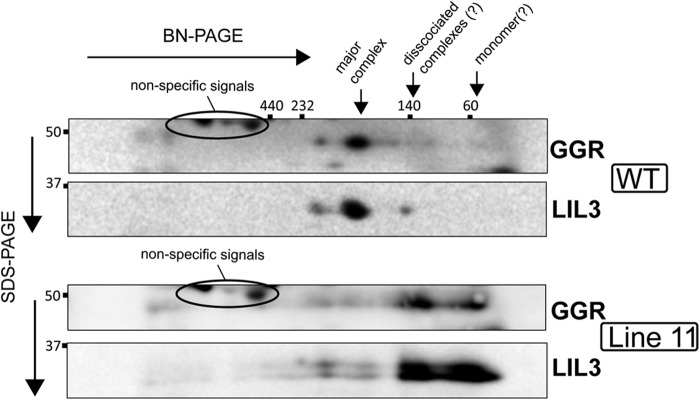
Complex formation of GGR and LIL3 in the WT and LIL3(AAA) lines resolved by BN-PAGE/SDS-PAGE. Blots were detected with anti-GGR and anti-LIL3 antibodies.
DISCUSSION
Our previous study confirmed that LIL3 is required for the stabilization of GGR in Arabidopsis (16). This finding was unexpected because a few characterized GGR enzymes from prokaryotes and plants have been shown to perform their reactions without requiring the presence of additional proteins (25, 40). The exclusive distribution of LIL3 to the green lineage of photosynthetic eukaryotes also raises the question why only Viridiplantae GGR requires LIL3 (6, 7). Considering that GGRs from various organisms are known to accept multiple substrates, such as GGPP, Chl-GG, or 2,3-di-O-geranylgeranylglyceryl phosphate (25, 40), it is reasonable to consider that LIL3 is involved in the supply of certain substrates to the GGR reactions in Viridiplantae. In particular, it is interesting to consider that LIL3 holds the tetrapyrrole groups of the substrates of the GGR reaction with its LHC motif to provide them to GGR. However, findings from this study do not support this model. Transgenic Arabidopsis plants expressing LIL3(TMAPX) are still able to synthesize phytylated chlorophyll to significant levels (Fig. 7), demonstrating that the tetrapyrrole-binding ability (if the LHC motif of LIL3 possesses such an ability) is dispensable for GGR reactions. Instead, it is shown that fusion of the TMLIL3 or TMAPX to GGR stabilizes GGR in the absence of intact LIL3, indicating that membrane anchoring is essential for the stabilization of GGR. Taken together, we suggest that the primary function of LIL3 is to tether GGR to membranes, and this function is specifically fulfilled by the TM domain, which spans the LHC motif.
On the other hand, we hypothesize that LIL3 conveys another function to support the GGR reactions. The Chl-phy levels in the LIL3(TMAPX) and LIL3(AAA) lines are still lower than in the WT, with the exception of line 31 of LIL3(AAA), which accumulates a massive amount of LIL3(AAA) protein and synthesizes nearly 100% of Chl-phy (Fig. 8). These results indicate that modification of the TM(LIL3) domain compromises the function of LIL3. One simple explanation for this observation is that the modification of the TMLIL3 domain hampers its interaction with GGR and, thus, destabilizes GGR. However, this explanation does not seem likely because similar protein levels for GGR are observed in the LIL3(AAA) lines in relative comparison to the WT, and, furthermore, a substantial amount of GGR and LIL3(AAA) appear to interact with each other to form a smaller complex (Fig. 9). Instead, it is shown that the amino acid substitution in the TMLIL3 domain dissociates the large LIL3-GGR complexes into smaller complexes (Fig. 9). These results indicate that the LHC motif of LIL3 facilitates the formation of the native LIL3-GGR complexes, whose size is estimated to be 160 kDa or above. In good accordance with these results, it is shown that the TM domains containing the LHC motif have been shown to be essential in oligomerization of other LIL or LHC proteins. The TM domain of cyanobacterial FC, which contains an LHC motif, is functionally involved in the dimerization of FC in vitro (22). Two transmembrane helices of LHC, both of which contain an LHC motif, are also known to interact with each other to hold a symmetrical structural backbone of LHC (38). Furthermore, it is suggested that two pairs of cyanobacterial LIL proteins, ScpB/E and ScpC/D, form heterodimers (28). Taking these observations into account, it is possible that one fundamental function of LHC motifs is to facilitate the oligomerization of proteins.
Although GGR accumulates to the WT level in line 5 of LIL3(AAA)-overexpressing plants, the Chl-phy levels do not show a similar level of recovery as compared with the WT. These results indicate that the formation of the large LIL3-GGR complex is essential for the optimal activity of GGR. It is possible to explain these observations by assuming that the formation of the large LIL3-GGR complex facilitates the substrate shuttling between the active sites of each GGR subunit, which may enable efficient consecutive reduction of GGPP to phytyl-PP or of Chl-GG to Chl-phy. A substrate shuttling mechanism has been extensively studied with fatty acid synthase (41). This enzyme catalyzes the consecutive elongation of fatty acid chains by shuttling reaction substrates between the symmetrically positioned identical subunits of the enzyme complex (41). A similar substrate-shuttling mechanism has been postulated for the reaction mechanism of uroporphyrinogen III decarboxylase, which catalyzes the consecutive reduction of the four side chains of uroporphyrinogen III to form coproporphyrinogen III (42). In our study, most of the transgenic plants overexpressing the modified versions of LIL3 were found to accumulate intermediate substrates of the reactions (Chl-DHGG and Chl-THGG) (Figs. 7 and 8). It is possible that the modified LIL3-GGR complexes cannot efficiently shuttle the intermediate reaction substrates (such as DHGGPP or THGGPP) between active sites. As a result, these modified complexes would release these intermediates into the environment. A substrate shuttling model of the GGR reaction may serve as a basis for future analysis of the GGR reaction mechanism.
Amino acid substitution in the conserved amino acid residues within the LHC motif of LIL3 results in a partial dissociation of the large LIL3-GGR complex into a smaller complex. These results do not necessarily indicate that these conserved residues bind chlorophyll or other pigments. Moreover, we did not detect any pigment binding to the isolated LIL3-GGR complexes.3 These results are consistent with the observation of cyanobacterial FC, in which pigment binding was not detected by fluorescence resonance energy transfer (24). On the contrary, other types of cyanobacterial LIL protein, ScpB, ScpC, ScpD, and ScpE, have been shown to bind chlorophyll in vitro (28). In addition, one of the plant LIL proteins, ELIP, has been reported to bind photosynthetic pigments (38). The cyanobacterial LIL proteins mentioned above and ELIP are suggested to associate with the photosynthetic apparatus (10–12) or light-harvesting complexes (13). It is possible that a specific set of LIL proteins, including ScpB-E proteins and ELIP, gain a pigment-binding ability. On the contrary, thylakoid localization and interaction with other proteins are apparently common functions of LIL proteins (10, 12, 13, 21, 29). As a result, it is intriguing to consider that an ancestral protein containing an LHC motif is a membrane-localized protein with a superior ability to interact with other proteins and that this protein may have gained a pigment binding ability to evolve to LHC.
In conclusion, we performed a functional analysis of the LIL3 protein and demonstrated that the TMLIL3 domain is not essential in the GGR reaction. Instead, by analyzing GGR-TMLIL3- and GGR-TMAPX-overexpressing plants, we have shown that the membrane-anchoring function of the TMLIL3 domain is important for stabilizing GGR. Furthermore, we hypothesize that LIL3 facilitates the formation of the multimeric LIL3-GGR complex, which appears to more efficiently perform reactions that function to catalyze the reduction of GGPP. Taken together, this study provides novel insight into the function of LIL3 and its TM domain containing a characteristic LHC motif. Collectively, these findings have increased our understanding of the function of the LHC motif, which is ubiquitous among oxygenic photosynthetic organisms.
Acknowledgment
We thank Sachiko Tanaka for technical assistance.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan, by Grant-in-Aid for Scientific Research 23570042, and by a leadership research grant from the Institute of Low Temperature Science, Hokkaido University (to R. T.).
Unpublished results from Kaori Takahashi and Ryouichi Tanaka.
- LHC
- light-harvesting complex
- LIL
- light-harvesting-like
- FC
- ferrochelatase
- GGR
- geranylgeranyl reductase
- TM
- transmembrane
- GGPP
- geranylgeranyl-diphosphate
- Chl-phy
- phytylated chlorophyll
- Chl-GG
- geranylgeranylated chlorophyll
- Chl-DHGG
- dihydrogeranylgeranylated chlorophyll
- Chl-THGG
- tetrahydrogeranylgeranylated chlorophyll
- BN-PAGE
- blue native PAGE
- APX
- ascorbate peroxidase
- Chl
- chlorophyll
- ELIP
- early light-inducible protein.
REFERENCES
- 1. Sonnhammer E. L., Eddy S. R., Birney E., Bateman A., Durbin R. (1998) Pfam. Multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26, 320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Green B., Pichersky E. (1994) Hypothesis for the evolution of three-helix Chl a/b and Chl a/c light-harvesting antenna proteins from two-helix and four-helix ancestors. Photosynth. Res. 39, 149–162 [DOI] [PubMed] [Google Scholar]
- 4. Green B., Kühlbrandt W. (1995) Sequence conservation of light-harvesting and stress-response proteins in relation to the three-dimensional molecular-structure of LhcII. Photosynth. Res. 44, 139–148 [DOI] [PubMed] [Google Scholar]
- 5. Jansson S. (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240 [DOI] [PubMed] [Google Scholar]
- 6. Engelken J., Brinkmann H., Adamska I. (2010) Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol. Biol. 10, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neilson J. A., Durnford D. G. (2010) Evolutionary distribution of light-harvesting complex-like proteins in photosynthetic eukaryotes. Genome 53, 68–78 [DOI] [PubMed] [Google Scholar]
- 8. Green B. R., Pichersky E., Kloppstech K. (1991) Chlorophyll a/b-binding proteins. An extended family. Trends Biochem. Sci 16, 181–186 [DOI] [PubMed] [Google Scholar]
- 9. Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 [DOI] [PubMed] [Google Scholar]
- 10. Kufryk G., Hernandez-Prieto M. A., Kieselbach T., Miranda H., Vermaas W., Funk C. (2008) Association of small CAB-like proteins (SCPs) of Synechocystis sp. PCC 6803 with photosystem II. Photosynth. Res. 95, 135–145 [DOI] [PubMed] [Google Scholar]
- 11. Yao D., Kieselbach T., Komenda J., Promnares K., Prieto M. A., Tichy M., Vermaas W., Funk C. (2007) Localization of the small CAB-like proteins in photosystem II. J. Biol. Chem. 282, 267–276 [DOI] [PubMed] [Google Scholar]
- 12. Promnares K., Komenda J., Bumba L., Nebesarova J., Vacha F., Tichy M. (2006) Cyanobacterial small chlorophyll-binding protein ScpD (HliB) is located on the periphery of photosystem II in the vicinity of PsbH and CP47 subunits. J. Biol. Chem. 281, 32705–32713 [DOI] [PubMed] [Google Scholar]
- 13. Heddad M., Norén H., Reiser V., Dunaeva M., Andersson B., Adamska I. (2006) Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiol. 142, 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Funk C., Lindström V., Vermaas W. (1999) Small cab-like proteins: relatives to the chlorophyll a/b binding proteins in cyanobacteria. in The Chloroplast: From Molecular Biology to Biotechnology, pp. 103–106, Springer, Dordrecht, The Netherlands [Google Scholar]
- 15. Funk C., Vermaas W. (1999) A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38, 9397–9404 [DOI] [PubMed] [Google Scholar]
- 16. Tanaka R., Rothbart M., Oka S., Takabayashi A., Takahashi K., Shibata M., Myouga F., Motohashi R., Shinozaki K., Grimm B., Tanaka A. (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 16721–16725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vavilin D., Yao D., Vermaas W. (2007) Small Cab-like proteins retard degradation of photosystem II-associated chlorophyll in Synechocystis sp. PCC 6803. Kinetic analysis of pigment labeling with 15N and 13C. J. Biol. Chem. 282, 37660–37668 [DOI] [PubMed] [Google Scholar]
- 18. Hernandez-Prieto M. A., Tibiletti T., Abasova L., Kirilovsky D., Vass I., Funk C. (2011) The small CAB-like proteins of the cyanobacterium Synechocystis sp. PCC 6803. Their involvement in chlorophyll biogenesis for photosystem II. Biochim. Biophys. Acta 1807, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 19. Wang Q., Jantaro S., Lu B., Majeed W., Bailey M., He Q. (2008) The high light-inducible polypeptides stabilize trimeric photosystem I complex under high light conditions in Synechocystis PCC 6803. Plant Physiol. 147, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rizza A., Boccaccini A., Lopez-Vidriero I., Costantino P., Vittorioso P. (2011) Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytol. 190, 896–905 [DOI] [PubMed] [Google Scholar]
- 21. Heddad M., Adamska I. (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc. Natl. Acad. Sci. U.S.A. 97, 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sobotka R., Tichy M., Wilde A., Hunter C. N. (2011) Functional assignments for the carboxyl-terminal domains of the ferrochelatase from Synechocystis PCC 6803. The CAB domain plays a regulatory role, and region II is essential for catalysis. Plant Physiol. 155, 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sobotka R., McLean S., Zuberova M., Hunter C. N., Tichy M. (2008) The C-terminal extension of ferrochelatase is critical for enzyme activity and for functioning of the tetrapyrrole pathway in Synechocystis strain PCC 6803. J. Bacteriol. 190, 2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Storm P., Tibiletti T., Hall M., Funk C. (2013) Refolding and enzyme kinetic studies on the ferrochelatase of the cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE 8, e55569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keller Y., Bouvier F., d'Harlingue A., Camara B. (1998) Metabolic compartmentation of plastid prenyllipid biosynthesis. Evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 251, 413–417 [DOI] [PubMed] [Google Scholar]
- 26. Tanaka R., Oster U., Kruse E., Rudiger W., Grimm B. (1999) Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 120, 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rossini S., Casazza A. P., Engelmann E. C., Havaux M., Jennings R. C., Soave C. (2006) Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol. 141, 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Storm P., Hernandez-Prieto M. A., Eggink L. L., Hoober J. K., Funk C. (2008) The small CAB-like proteins of Synechocystis sp. PCC 6803 bind chlorophyll. In vitro pigment reconstitution studies on one-helix light-harvesting-like proteins. Photosynth. Res. 98, 479–488 [DOI] [PubMed] [Google Scholar]
- 29. Adamska I. (2001) Stable insertion of the early light-induced proteins into etioplast membranes requires chlorophyll a. J. Biol. Chem. 276, 8582–8587 [DOI] [PubMed] [Google Scholar]
- 30. Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M. (2000) pGreen. A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 [DOI] [PubMed] [Google Scholar]
- 31. Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., Pikaard C. S. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 [DOI] [PubMed] [Google Scholar]
- 32. Salvi D., Rolland N., Joyard J., Ferro M. (2008) Methods in Molecular Biology, pp. 19–36, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 33. Wittig I., Braun H.-P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 34. Kunugi M., Takabayashi A., Tanaka A. (2013) Evolutionary changes in chlorophyllide a oxygenase (CAO) structure contribute to the acquisition of a new light-harvesting complex in Micromonas. J. Biol. Chem. 288, 19330–19341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salvi D., Moyet L., Seigneurin-Berny D., Ferro M., Joyard J., Rolland N. (2011) Preparation of envelope membrane fractions from Arabidopsis chloroplasts for proteomic analysis and other studies. Methods Mol. Biol. 775, 189–206 [DOI] [PubMed] [Google Scholar]
- 37. Yoshimura K., Yabuta Y., Tamoi M., Ishikawa T., Shigeoka S. (1999) Alternatively spliced mRNA variants of chloroplast ascorbate peroxidase isoenzymes in spinach leaves. Biochem. J. 338, 41–48 [PMC free article] [PubMed] [Google Scholar]
- 38. Adamska I., Roobol-Bóza M., Lindahl M., Andersson B. (1999) Isolation of pigment-binding early light-inducible proteins from pea. Eur. J. Biochem. 260, 453–460 [DOI] [PubMed] [Google Scholar]
- 39. Barros T., Royant A., Standfuss J., Dreuw A., Kühlbrandt W. (2009) Crystal structure of plant light-harvesting complex shows the active, energy-transmitting state. EMBO J. 28, 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sasaki D., Fujihashi M., Iwata Y., Murakami M., Yoshimura T., Hemmi H., Miki K. (2011) Structure and mutation analysis of archaeal geranylgeranyl reductase. J. Mol. Biol. 409, 543–557 [DOI] [PubMed] [Google Scholar]
- 41. Jenni S., Leibundgut M., Boehringer D., Frick C., Mikolásek B., Ban N. (2007) Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 316, 254–261 [DOI] [PubMed] [Google Scholar]
- 42. Martins B. M. (2001) Crystal structure and substrate binding modeling of the uroporphyrinogen-III decarboxylase from Nicotiana tabacum. Implications for the catalytic mechanism. J. Biol. Chem. 276, 44108–44116 [DOI] [PubMed] [Google Scholar]