SUMMARY
Cranial placodes are embryonic structures essential for sensory and endocrine organ development. Human placode development has remained largely inaccessible despite the serious medical conditions caused by the dysfunction of placode-derived tissues. Here, we demonstrate the efficient derivation of cranial placodes from human pluripotent stem cells. Timed removal of the BMP inhibitor Noggin, a component of the dual-SMAD inhibition strategy of neural induction, triggers placode induction at the expense of CNS fates. Concomitant inhibition of FGF signaling disrupts placode derivation and induces surface ectoderm. Further fate specification at the pre-placode stage enables the selective generation of placode-derived trigeminal ganglia capable of in vivo engraftment, mature lens fibers and anterior pituitary hormone-producing cells that upon transplantation produce human GH and ACTH in vivo. Our results establish a powerful experimental platform to study human cranial placode development and set the stage for the development of human cell-based therapies in sensory and endocrine disease.
INTRODUCTION
Cranial placodes give rise to cells of the sensory organs, including the optic lens, the nasal epithelium, otic structures, the adenohypophysis, and a subset of cranial nerves such as the trigeminal ganglia. During development, sensory placodes are formed at the interface of the non-neural ectoderm and neural plate, surrounding the anterior portion of the future central nervous system (CNS) (Figure S1A). Defects in placode development cause a wide spectrum of human congenital malformations ranging from blindness and deafness to hormone imbalance or loss of smell (Abdelhak et al., 1997; Baker and Bronner-Fraser, 2001; Ruf et al., 2004). To date, cranial placode development has been characterized in model organisms, including the frog, zebrafish, chicken and to a lesser extent, the mouse (Baker and Bronner-Fraser, 2001; Bhattacharyya and Bronner-Fraser, 2004; Schlosser, 2006). However, human placode development has remained largely unexplored due to lack of access to early human tissue and specific placode markers.
Human pluripotent stem cells (hPSCs), including human embryonic (hESCs) and human induced pluripotent stem cells (hiPSCs) have the potential to self-renew, while retaining a very broad differentiation potential. Over the last few years, protocols have been developed for directing the fate of hESCs into specific cell lineages. The derivation of CNS cells was among the first hESC differentiation protocols developed in the field (Reubinoff et al., 2001; Zhang et al., 2001). The differentiation of hESCs into cells of the peripheral nervous system has also been achieved (Lee et al., 2007; Menendez et al., 2011). In contrast to the successful derivation and application of defined CNS and NC derived cell types, there has been limited success on modeling cranial placode development in hPSCs.
Recently, we developed a neural induction strategy based on the concomitant inhibition of the Bone Morphogenetic Protein (BMP) and TGFβ/Activin/Nodal signaling pathways (dual-SMAD inhibition (dSMADi), (Chambers et al., 2009)). Exposure to Noggin (N) and SB431542 (SB) leads to the synchronized, rapid and efficient differentiation of hPSCs into CNS fates. Here, we report that de-repression of endogenous BMP signaling during dSMADi is sufficient for the selective induction of human cranial placodes. Using the novel placode induction protocol (PIP) > 70% of all cells adopt a SIX1+ cranial placode precursor fate by day 11 of differentiation. We further identify a pre-placodal lineage competent to differentiate into selective placode fates including trigeminal sensory neurons, mature lens fibers and hormone-producing anterior pituitary cells. Trigeminal sensory neurons are characterized by marker expression, electrophysiology and by transplantation into the developing chick embryo and the adult mouse CNS. Finally, we report the derivation of human pituitary cells producing GH and ACTH hormones in vitro and in vivo.
RESULTS
De-repression of endogenous BMP signaling induces placode at the expense of neuroectoderm
To address whether the dSMADi protocol is suitable for the derivation of placodal cells, we first defined a set of appropriate placode markers. Based on studies in model organisms, we hypothesized that members of the SIX, EYA, and DLX family of transcription factors (Baker and Bronner-Fraser, 2001; Schlosser, 2006) mark human placode fate. Within the ectodermal lineage, SIX1 is placode-specific, marking both the early pre-placodal region and the various specific placodes (Schlosser, 2006). Based on studies in the chick embryo, placode induction relies on a complex interplay of FGF, BMP and WNT signals during early ectodermal patterning in vivo (Litsiou et al., 2005). Activity of BMPs within the ectoderm is thought to be particularly critical in allocating fates. A model has been proposed initially whereby high levels of signaling promote an epidermal fate, moderate levels induce placodes, intermediate levels specify NC and a complete absence of BMP activity is required for neural plate formation (Wilson et al., 1997). More recent studies have revised the original model by confirming an early role for BMP signaling in establishing placode competence (Kwon et al., 2010) while the subsequent stage was shown to require BMP-inhibition rather than BMP activation (Ahrens and Schlosser, 2005; Kwon et al., 2010; Litsiou et al., 2005)
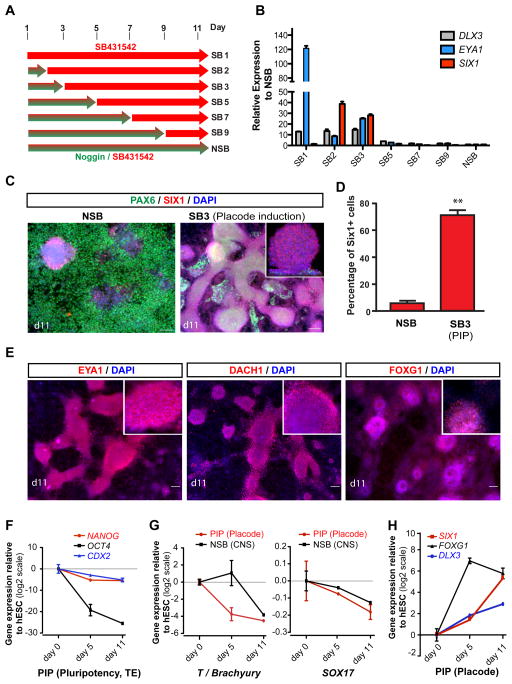
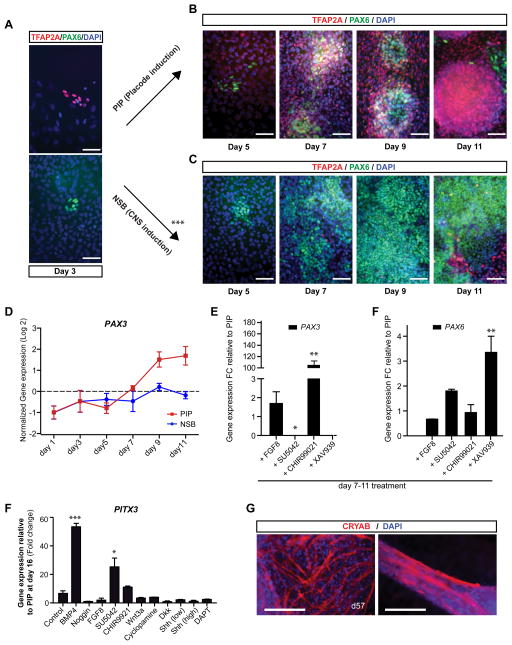
To test whether early BMP exposure promotes the derivation of SIX1+ placodal cells, we exposed SB (the TGFβ inhibitor) treated hESCs to various concentrations of BMP4. However, addition of BMP4 in the presence of SB caused a dramatic morphological change and triggered induction of CDX2 (Figure S1B, C), similar to the BMP-mediated induction of trophectoderm-like lineages reported previously (Xu et al., 2002). We next tested whether timed withdrawal of the BMP inhibitor Noggin during N-SB differentiation could induce placodal fates via de-repressing endogenous BMP signaling. We performed a time course analysis during which we removed Noggin at different time points of the N-SB protocol (Figure 1A). Gene expression analysis at day 11 revealed a robust induction of DLX3, SIX1 and EYA1 (Figure 1B) upon withdrawal of Noggin at day 2 or 3 of differentiation. In contrast, Noggin withdrawal at day 1 of differentiation led to the induction of EYA1 in the absence of SIX1 expression and triggered morphological changes as well as CDX2 expression, suggesting trophectodermal differentiation (though CDX2 and EYA1 can also be expressed in hESC-derived mesodermal lineages (Bernardo et al., 2011)). Our data indicate that EYA1 is expressed in both trophectodermal and placodal lineages, and that co-expression with SIX1 is required to define placodal lineage. Immunocytochemical analysis of hESC progeny at day 11 of differentiation demonstrated that Noggin withdrawal at day 3 (PIP conditions) induced a switch from 82% PAX6+ neuroectodermal cells under N-SB conditions to 71% SIX1+ putative placode precursor cells under PIP (Figure 1C, 1D, S1D). SIX1+ clusters expressed other placodal markers such as EYA1, DACH1 and FOXG1 (BF1) (Figure 1E). DACH1 is also expressed in anterior neuroectodermal cells (Elkabetz et al., 2008) marking neural rosettes while in PIP treated cultures DACH1 marks placodal clusters (Figure S1E). Temporal analysis of gene expression under PIP conditions revealed rapid downregulation of pluripotency markers (OCT4, NANOG), as well as markers of trophectoderm (CDX2) (Figure 1F), mesoderm (T) and endoderm (SOX17) (Figure 1G). SIX1 expression in placode was confirmed in human primary tissue (Carnegie Stage 15, ~ 5.5 weeks p.c.; data not shown). SIX1 is also expressed in precursors of skeletal muscle, thymus and kidney cells. However, we did not detect expression of skeletal muscle (MYOD), endoderm (SOX17), or mesoderm (Brachyury (T)) markers during PIP confirming that the hESC-derived SIX1+ cells are of placode identity. We also observed very few (< 1%) NC lineage cells under PIP conditions based on SOX10 expression by immunocytochemistry and SOX10::GFP (Chambers et al., 2012; Mica et al., 2013) reporter line expression (Figure S1F). Induction of cranial placode markers was observed by day 5 with FOXG1 preceding expression of SIX1 and DLX3 (Figure 1H). The PIP protocol was validated in multiple hESC and hiPSC lines (Figure S1G, H).
Figure 1. Derivation of Six1+ placodal precursors using a modified dual-SMAD inhibition protocol (see also Figure S1).
A) Schematic illustration of timed Noggin withdrawal paradigm to determine temporal requirement for endogenous BMP signaling during placode specification. The protocol is based on modifying the Noggin + SB431542 (NSB) protocol developed for CNS induction (Chambers et al., 2009). B) Relative induction of placodal markers comparing modified NSB protocol (various time points of Noggin withdrawal) to N-SB treatment maintained throughout differentiation (NSB condition). Data represent fold changes of mRNA expression measured by qRT-PCR at day 11. C) Immunocytochemical analyses of SIX1 and PAX6 expression at day 11 of differentiation. Inset shows a confocal section to demonstrate SIX1 expression within clusters. Scale bars correspond to 50 μm. D) Quantification of the percentage of Six1+ cells generated under modified N-SB (SB3 = placode induction (PIP) protocol) versus N-SB condition. E) Immunocytochemical analysis of placodal markers, EYA1, DACH1, and FOXG1 in placodal clusters. Insets show higher magnification images for respective marker. Scale bars correspond to 50 μm. F–H) Temporal analysis of gene expression in PIP versus N-SB protocol. Values are normalized to the expression observed in undifferentiated hESCs. F) Loss of expression of pluripotency (NANOG, OCT4/POU5F1) and trophectodermal (TE) markers; G) Lack of expression of mesodermal marker T; H) of non-neural fates and induction of placodal fates by monitoring time course expression of pluripotency markers (NANOG, OCT4), trophectoderm (CDX2), mesoderm (T), endoderm (SOX17), and placodal markers (DLX3, SIX1, FOXG1). Error bar represents SD. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared with control N-SB condition (n = 3 independent experiments).
To further validate the identity of hESC-derived placodal precursors, we made use of a conserved Eya1 enhancer element (Ishihara et al., 2008). We readily observed GFP expression following nucleofection of the enhancer in olfactory placode but not in age-matched midbrain cultures (Figure S1I–J) confirming specificity. In hESC-derived placode we observed an 8-fold increase in the percentage of cells with enhancer activity in PIP versus N-SB (Figure S1K).
Microarray analysis reveals novel human placode progenitor gene expression
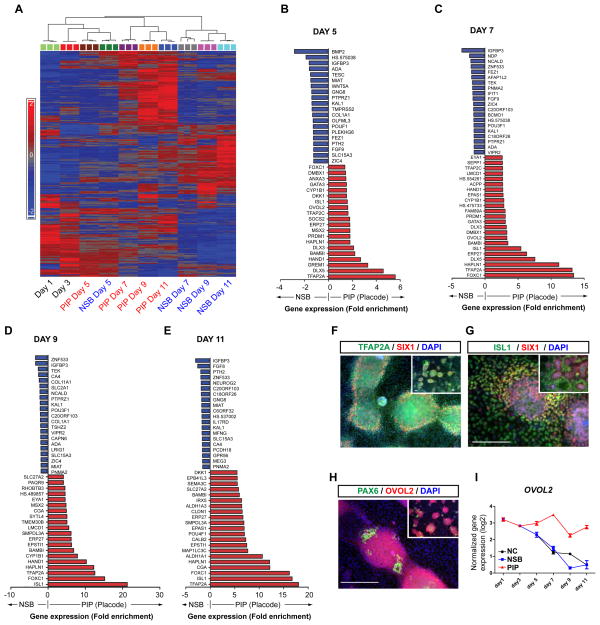
We next performed temporal transcriptome analysis to establish an unbiased molecular assessment of the in vitro placode induction process. RNA was collected at five time points in triplicates (day 1, 3, 5, 7, and 11) in control N-SB versus PIP treated cultures (Figure 2A–E; all raw data are available on GEO: http://www.ncbi.nlm.nih.gov/geo/: Accession # pending. Prior to microarray analysis, the quality of each sample was verified for expression of a panel of placode markers (SIX1, DLX3, EYA1) and the absence of other lineage markers (FOXA2, (endoderm), SOX17 (endoderm), MYOD (skeletal muscle), CDX2 (trophoblast), and T (mesoderm). Cluster and principal component analyses showed a temporal segregation of the transcriptome data in PIP versus N-SB treated cells by day 7 of differentiation (Figure 2A, S2A). Transcriptome data also defined a set of genes that distinguish placodal from neuroectodermal fate (Table S1, 2).
Figure 2. Temporal global gene expression profiles during hESC-derived placode specification (see also Figure S2).
A) Clustering of the differentially regulated genes during PIP versus N-SB protocol. B–E) The top twenty most significant up (red) and downregulated (blue) genes in PIP versus N-SB protocol by fold change at day 5, 7, 9 and 11. F–G) Confirmation of ISL1 and TFAP2A expression at the protein level under PIP conditions (day 11). H) Confirmation of OVOL2 expression by immunocytochemistry (day 11). I) Time course analysis of OVOL2 gene expression during PIP, N-SB and neural crest (NC) protocol. Scale bars in F, G and H correspond to 50 μm.
To gain insight into specific genes differentially expressed during placode induction, we performed pairwise comparisons for each differentiation time point (Figure 2B–E). Among the most highly enriched transcripts under PIP condition were known placode markers such as GATA3, DLX5, DLX3, TFAP2A, and TFAP2C. SIX1 was significantly upregulated in the microarray analysis by day 9 of differentiation, though it was not among the 20 most differentially regulated genes. Differential expression for these and additional genes was verified by qRT-PCR (Figure S2B). We also observed significant transcriptional changes in WNT and BMP pathway components such as an increase in the WNT pathway inhibitor DKK-1 and BMP antagonists, such as GREMLIN-1 and BAMBI (Figure 2B–D), which are known transcriptional targets of BMP signaling (Grotewold et al., 2001). Interestingly, we also noticed induction of ISL1, a well known marker for sensory neurons, motoneurons, heart progenitors and pancreatic islet cells (Hunter and Rhodes, 2005). Based on the early onset of ISL1 expression and the absence of mesodermal fates, we surmised that under PIP conditions ISL1 represents an early human placode marker. ISL1 was one of the first makers induced during PIP differentiation. Similar to TFAP2A and SIX1 (Figure 2F), ISL1 protein remained expressed in the majority of cells by day 11 of differentiation partially co-localizing with SIX1 (Figure 2G). Another novel placode marker identified by our microarray expression data is OVOL2, a member of the Ovo family of zinc-finger transcription factors. OVOL2 was enriched during PIP by day 5 of differentiation and placode clusters showed strong immunoreactivity for OVOL2 (Figure 2H). In contrast, OVOL2 was downregulated under conditions promoting CNS (N-SB) or neural crest (N-SB/CHIR (Chambers et al., 2012; Mica et al., 2013)) fates (Figure 2I).
We identified additional differentially expressed genes during anterior placode specification, including FOXC1 and HAPNL1 (also known as CRTL1). FOXC1 marks the lens placode during chick development (Bailey et al., 2006) but may also be expressed at the pre-placode stage (Sasaki and Hogan, 1993). HAPNL1 was one of the most differentially expressed genes in PIP and was previously shown to be expressed in the surface ectoderm and chick neural plate border (Colas and Schoenwolf, 2003). Gene ontology (GO) analysis, using DAVID (http://david.abcc.ncifcrf.gov/; (Dennis et al., 2003)) indicated that transcripts highly enriched in PIP versus N-SB conditions at day 7 and day 9 of differentiation are associated with sensory organ development, BMP and WNT pathways as wells as ectoderm and epidermis development (Figure S2C). The total number of differentially expressed genes increased by day 7 of differentiation (Figure S2D, E) matching the proposed time frame for placode commitment. In summary, our molecular analyses present the first roadmap of human placode development and an important resource for future functional studies.
Early treatment with FGF-inhibitor suppresses placode and induces surface ectoderm fate
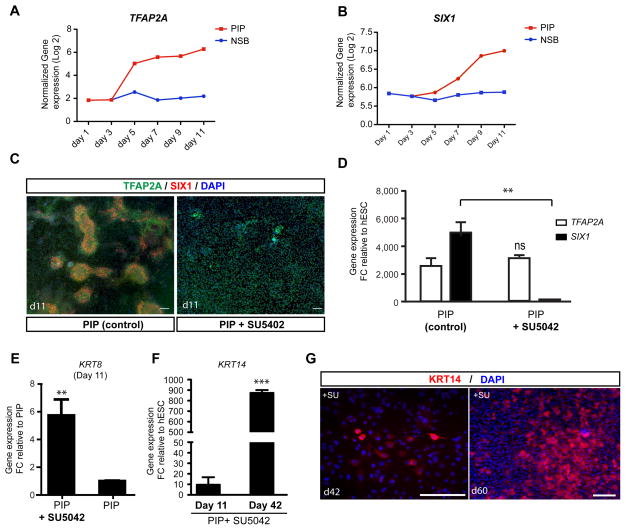
Lineage studies in several vertebrate species indicate that cranial placode precursors originate from the surface ectoderm and are induced by signals emanating from the adjacent neuroectoderm (Pieper et al., 2012), rather than being of neural origin. However, no such data are available for human development. The putative timing of human placode induction (~ day 20 p.c. (O’Rahilly, 1987)) makes this a stage largely inaccessible to experimental manipulations. Gene expression data during PIP indicate that TFAP2A is one of the earliest upregulated and differentially expressed genes (Figure 2B–E). During mouse development early expression of the orthologous gene, Tcfap2a (E7.5) is restricted to the future surface ectoderm (Arkell and Beddington, 1997).
The induction of TFAP2A was first detected at day 5 of PIP while absent in N-SB protocol (Figure 3A) suggesting that the period between day 3 and 5 of PIP corresponds to the commitment towards surface ectoderm. SIX1 was induced two days later than TFAP2A (Figure 3A, B) further pointing to a surface ectoderm intermediate. In vivo genetic studies indicate that high levels of BMPs in the absence of FGF signaling promote epidermal fate whereas placodal cells are established at reduced levels of BMPs upon activation of FGFs (Kudoh et al., 2004; Litsiou et al., 2005). Therefore, we tested whether blocking endogenous FGF signaling under PIP conditions disrupts placode induction and triggers epidermal fate. Exposure to SU5402, a small molecule inhibiting FGF signaling, from day 3–11 of differentiation (Figure S3A) suppressed the emergence of SIX1+ clusters while maintaining TFAP2A expression (Figure 3C). Quantification of SIX1 and TFAP2A gene expression at day 11 confirmed a complete loss of placode marker expression (Figure 3D), while TFAP2A expression was maintained. The ectodermal precursor identity of TFAP2A+ cells was supported by the expression of the epidermal precursor marker KRT8 in PIP + SU5402 treated cultures (Figure 3E). Furthermore, long-term cultures (day 42–60) showed robust induction of the mature keratinocyte marker KRT14 (Figure 3F) and formed E-CADHERIN positive patches with KRT14-immunoreactive cells at the periphery (Figure S3B). Analysis of the proliferative capacity of the epidermal precursors showed KI67 expression primarily in E-CADHERIN+/KRT14− cells. Cell proliferation decreased by day 60 concomitant with an increased percentage of cells expressing KRT14 (Figure 3G, S3C). In addition to the requirement for endogenous FGF signaling, we observed low WNT and BMP levels as other important parameters for the transition from surface ectoderm (day3) to early placode fate. Exposure to high concentrations of CHIR or BMP4 suppressed placode at the expense of NC or putative trophectoderm, respectively (Figure S3D, E).
Figure 3. FGF signaling determines placodal versus non-neural ectoderm/epidermal fate (see also Figure S3).
A–B) Time course of the induction of TFAP2A and SIX1 under PIP conditions. C) TFAP2A (green) is expressed in placodes and surface ectoderm, while SIX1 (red) is only expressed in placode cells. Blocking PIP via pharmacological inhibition of endogenous FGF signaling, blocks the formation of SIX1+ cells. D) Quantification of the loss of SIX1 gene expression following treatment with the FGF inhibitor SU5402. E) Expression of early epidermal marker KRT8. Data represent fold changes of mRNA expression by qRT-PCR at day 11 compared to PIP condition. F) Expression of late epidermal marker KRT14 during keratinocyte differentiation. Data represent fold changes of mRNA expression by qRT-PCR at day 11 and day 42 compared to hESC. G) Long-term SU5402 treated cultures are expressing KRT 14 protein suggesting epidermal/keratinocyte fate. Error bar represents SD. Error bar represents SD. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared with control PIP condition (n = 3 independent experiments). Scale bars in C and G correspond to 50 μm.
Placode precursors efficiently differentiate into trigeminal-type sensory neurons
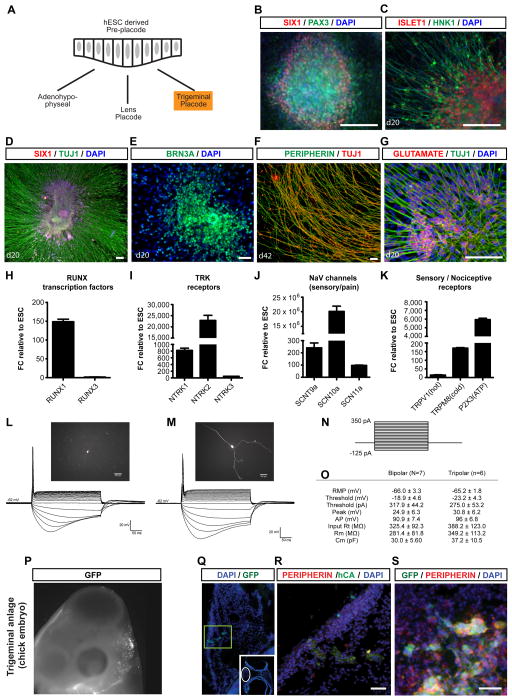
Cranial placodes give rise to a broad range of specialized cell types, including hormone-producing cells of the anterior pituitary gland, structural cells such as lens fibers in the eye and sensory neurons including trigeminal neurons (Figure 4A). Placodes can be characterized by the expression of specific PAX genes (McCauley and Bronner-Fraser, 2002). We observed that under standard PIP conditions most SIX1+ clusters co-expressed PAX3 (Figure 4B) suggesting ophthalmic trigeminal placode identity (McCabe et al., 2004; Stark et al., 1997). The spontaneous generation of HNK1+ (Metcalfe et al., 1990) cells with neuronal morphologies and co-expression of ISL1 confirmed peripheral sensory neuron identity (Figure 4C). To further ascertain the placode origin of the sensory neurons under PIP conditions, we assessed co-expression of neuronal markers with SIX1 (Figure 4D, S4A). Lack of SOX10 expression during PIP-based sensory neuron differentiation ruled out a neural crest origin. By day 20 of differentiation (7 days after replating), the cells formed ganglia-like structures with neurons extending long, radial processes and with nuclear expression of BRN3A (Figure 4D, E), a sensory neuron marker. Most neurons retained ISL1 expression by day 42 of differentiation and acquired expression of the peripheral neuron marker Peripherin (Figure 4F, S4B). Immunocytochemical analysis for neurotransmitter phenotypes revealed expression of Glutamate (Figure 4G) but lack of expression of TH and GABA (data not shown). These data are compatible with the generation of glutamatergic trigeminal sensory neurons. Gene expression analysis showed induction of RUNX1 (Figure 4H, S4C) and RET (Figure S4D) indicating sensory/nociceptive lineage. Expression of TRK receptors (Figure 4I, S4E) including NTRK1 and NTRK2 points to the presence of both nociceptive and non-nociceptive sensory neurons (Figure S4E–G). Diagnostic markers of nociceptive neuron identity include expression of specific sodium channels (SCN9A, SCN10A, SCN11A; Figure 4J) as well as classic pain receptors such as the capsaicin receptor (TRPV1), the receptor for cold sensation (TRPM8) and the P2X3 receptor critical for sensation of inflammatory pain mediated by ATP (Figure 4K). Gene expression analysis in long-term trigeminal neuronal cultures (day 55) showed sustained expression of RET1 and RUNX1 (Figure S4C, D).
Figure 4. Derivation, characterization and transplantation of hESC-derived trigeminal type sensory neurons (see also Figure S4 and Figure S5).
A) Schematic representation of various placodes derived during development for pre-placode cells including hormone producing cells (pituitary placode), structural cells such as lens fibers (lens placode) and sensory neurons (trigeminal placode). Trigeminal placode fate (highlighted in orange) appears to be a default under PIP conditions. B) PAX3, a trigeminal placode marker is expressed in SIX1+ placode clusters at the placode stage at day 11. C) Placode clusters by default rapidly yield cells expressing sensory neuron makers such as ISL1 and HNK1. D) Immunocytochemical analysis at day 20 of differentiation demonstrates that SIX1+ placodal clusters efficiently yield large numbers of TUJ1 positive neurons that initially retain SIX1 expression. E) Sensory neuron identity is further confirmed by expression of BRN3A in the majority of neurons derived from SIX1+ clusters. F) At day 42 of differentiation, neurons show increased expression of Peripherin and decreased levels of TUJ1 staining suggesting adoption of a more mature peripheral neuron fate. G) Trigeminal neurons stain for glutamate. H) Expression of RUNX factors, I) TRK receptors, J, K) nociceptor-specific channels and receptors in trigeminal neurons responsible for hot (TRPV1) and cold (TRPM8) sensation and for inflammatory pain (P2X3; n = 3 independent experiments). L, M) Examples of single cell patch clamp electrophysiological analysis in hESC-derived trigeminal-type neurons at day 53 of differentiation. N) The intensity of the stimulus was started from −125 pA and increased by 25 pA until a single action potential was observed. Steps shown correspond to 25 pA increments. O) Summary of quantitative electrophysiological parameters showed comparable patterns for both bipolar and tripolar type neurons. P–S) Transplantation into trigeminal anlage in chick embryo. P) GFP labeled cells and graft morphology 2 days after transplantation. Q) Low power image shows GFP+ fiber bundles: GFP (green) and DAPI (blue). Insert, section at midbrain level shows GFP+ fiber bundles adjacent to trigeminal anlage. R) The human fiber bundles express human cytoplasmic marker (green) and the sensory neuron marker, Peripherin (red). S) GFP+/PERIPHERIN+ cells bodies are arranged in ganglia-like clusters in vivo. All scale bars correspond to 50 μm.
Functional analysis of placode-derived sensory neurons was performed by whole-cell patch-clamp recordings (Figure S4G). Neurons were filled with Lucifer yellow from the recording pipette exhibiting either bipolar or tripolar morphologies (Figure 4L, M). Responses were measured to a series of hyperpolarizing and depolarizing pulses (Figure 4N). The hESC – derived neurons produced single action potentials at a threshold depolarization matching the functional properties reported for primary embryonic trigeminal neurons (Grigaliunas et al., 2002). The average resting membrane potential (RMP) was −65.6± 6.7 (Figure 4L, M). Passive membrane and action potential properties were comparable between bipolar and tripolar sensory neurons as summarized in (Figure 4O) suggesting that two morphologically distinct populations do not reflect functionally distinct subgroups. We further assessed the robustness of the trigeminal sensory neuron induction protocol across multiple hESC and hiPSC lines. We observed comparable percentages of ISL1 and BRN3A expressing neurons across lines (Figure S4H).
In vivo analysis of hESC-derived trigeminal neurons in the developing chick embryo and adult mouse CNS
To assess the in vivo properties of hESC-derived trigeminal placode precursors, PIP-induced neuronal clusters, derived from a constitutively GFP expressing hESC line (Figure S5A, B), were injected into the developing chick embryo targeting the early trigeminal anlage at H&H stage 10–12 (Figure S5C). Human cells were identified based on GFP expression and use of human specific antibodies against cytoplasmic antigen (hCA). Two days after in ovo transplantation surviving GFP+ cells were found dispersed in the area of the endogenous chick trigeminal ganglion (Figure 4P). We observed extensive GFP+ human fiber bundles co-expressing hCA and Peripherin (Figure 4Q, R). In contrast, no hCA or Peripherin expression was detected in the neural tube of the embryo (Figure S5D). The in vivo fiber outgrowth 2 days after transplantation was reminiscent of the extensive in vitro fiber outgrowth of replated trigeminal neuron clusters (Figure S5A). Peripherin expression in vivo (Figure 4S) confirmed the peripheral neuron identity of the grafted cells.
We next addressed whether hESC-derived trigeminal neurons can engraft in the adult mouse CNS and project towards their physiological target. The trigeminal nuclei in the brainstem receive afferent innervation from the trigeminal sensory ganglion that is relayed to the contralateral thalamus. The pons was selected as site for transplantation, as it is surgically accessible and located within proximity of the trigeminal brain stem nuclei that receive afferent input from the trigeminal ganglia. Hence, GFP+ human trigeminal neuron clusters were injected into adult NOD/SCID mice via stereotactic surgery (see Material and Methods). Histological analysis 4 weeks after transplantation showed survival of GFP+ human cell graft in the ventral pons (Figure S5E). While GFP+ cell bodies remained tightly clustered at injection site, GFP+ fibers showed extensive projections into the host brain (N= 6) including the endogenous trigeminal nuclei (Figure S5F). Expression of BRN3A confirmed the sensory neuron identity of the cells (Figure S5G). Graft-derived human fiber bundles (hNCAM+ and GFP+) were observed emanating from the graft core (Figure S5H). These data demonstrate in vivo survival of trigeminal placode derivatives, differentiation along sensory neuron lineage and the establishment of axonal projections towards relevant endogenous targets in the embryonic chick and adult mouse brain.
Identification of a putative pre-placode stage
Our results indicate that current PIP conditions efficiently induce ophthalmic trigeminal placode fates. To investigate whether other placodal fates can be generated using modified PIP conditions, we first addressed the presence of putative pre-placode cells in our culture system. During vertebrate development, the pre-placode is characterized as the developmental anlage containing precursor cells competent to respond to signals determining placode identity (Martin and Groves, 2006). Pre-placodal cells in various model organisms have been shown to express Six1 and to co-express markers of both ectodermal and neural fate. However, the development of a human pre-placode remains unexplored. We performed a time-course co-expression analysis for TFAP2A (early ectodermal marker) and PAX6 (early neuroectoderm marker (Zhang et al., 2010)). During the first three days of differentiation only a few sparse patches of PAX6 or TFAP2A expressing cells were observed without evidence of co-expression (Figure 5A). At day 5, two days following Noggin withdrawal (PIP), there was an increase in the number of TFAP2A+ cells (Figure 5B) under PIP conditions and a concomitant loss of TFAP2A+ cells in N-SB (Figure 5C). At day 7, N-SB conditions yielded PAX6+ cells devoid of TFAP2A expression (Figure 5C) while PIP treated cultures showed extensive co-expression of TFAP2A and PAX6, indicating pre-placode identity (Figure 5B). The emergence of TFAP2A/PAX6 double positive cells coincided with the onset of SIX1 gene expression and the emergence of SIX1+ clusters around day 7 of differentiation (Figure 3B, S6A). By day 11, placode clusters were negative for PAX6 but retained expression of TFAP2A (Figure 5B) suggesting that early anterior PAX6+ pre-placode cells give rise to PAX6-negative posterior placode populations enriched for PAX3+. This model was further supported by gene expression data showing a robust PAX3 increase from day 7–11 of PIP (Figure 5D). Treatment with inhibitors of WNT or FGF signaling from day 7–11 of PIP suppressed PAX3 induction (Figure 5E) while maintaining PAX6 (Figure 5F). Our results indicate that endogenous signals contribute to the transition from an anterior PAX6+ pre-placode to a posterior PAX3+ placode lineage. The small number of TFAP2A cells in day 11 N-SB cultures (Figure 5C) likely represent neural crest precursors (Chambers et al., 2009) that lack PAX6 expression. Under PIP conditions the percentage of contaminating SOX10+ NC cells at day 11 was < 1% (Figure S1F).
Figure 5. Identification of putative pre-placode and directed differentiation towards human lens placode lineage (see also Figure S6).
A) Immunocytochemical analysis for TFAP2A and PAX6 at day 3 of differentiation. B) Time course analysis at day 5, 7, 9 and 11 of PIP differentiation show co-expression of TFAP2A and PAX6 at days 7 and 9 of differentiation. C) Time course analysis at day 5, 7, 9 and 11 of N-SB differentiation shows lack of TFAP2A expression but expression of PAX6 in CNS neuroectodermal cells. Scale bars in A–C correspond to 25 μm. D) Temporal analysis of PAX3 gene expression during PIP versus NSB protocol. E) PAX3 expression levels following treatment (days 7–11) with activators or inhibitor of FGF (FGF8, SU5402) and WNT signaling (CHIR99021, XAV939) during PIP. F) PAX6 expression levels using same treatment as in (E). F) Results of a four signaling pathway screen (modulators of BMP, FGF, WNT and SHH signaling added at days 7–11 of PIP). Induction of the lens placode marker PITX3 by qRT-PCR was observed upon treatment with activators of BMP signaling (BMP4) or inhibitors of FGF signaling (SU5402). G) Modified placode cultures differentiating into Crystalline+ cells with mature lens fiber morphologies by day 57 of differentiation. Scale bars in (G) correspond to 50 μm. Error bar represents SD. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared with control PIP condition (n = 3 independent experiments).
Treatment with FGF-inhibitor SU5402 at pre-placode stage induces lens fates
We next tested whether putative pre-placode cells, can be coaxed into specific placode fates other then trigeminal neurons. The spontaneous appearance of lens precursors (lentoid bodies) from primate and hESCs has been previously reported (Ooto et al., 2003; Zhang et al., 2010), although the lineage origin and inducing signals remained unexplored in those studies.
We tested the impact of four developmental signaling pathways on lens placode specification using activators and inhibitors of BMP, FGF, WNT, and Hedgehog signaling. To quantify the induction of lens placode fate, we monitored expression of the lens precursor marker PITX3 at day 16 of differentiation (Figure S6B). PITX3 expression was significantly induced in the presence of recombinant BMP4 or upon exposure to the FGF-inhibitory molecule, SU5402 (Figure 5F). A role for BMPs and FGFs has been previously proposed in developmental studies in the chick (Sjödal et al., 2007). Further differentiation revealed strong induction of αβcrystalline and the formation of mature lens fiber structures by day 57 (Figure 5G, left panel). The characteristic layering of lens fibers (Figure 5G, right panel) mimicked the structural properties of developing lens in vivo.
Treatment with SHH at pre-placode stage induces anterior pituitary cells
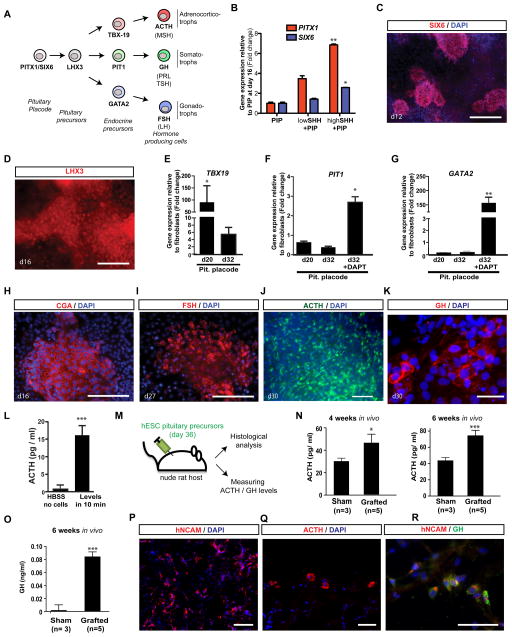
We next explored whether pre-placodal cells can be differentiated into anterior pituitary placode and recreate the various pituitary precursors and hormone producing cell types (Figure 6A). We observed that treatment with agonists for SHH signaling (Figure S7A) at the pre-placode stage (day 7–11) induced expression of the oral ectoderm marker SIX6 and PITX1 (Figure 6B), master regulators of pituitary gland development (Tremblay et al., 1998). Induction of PITX1 and SIX6 at transcript and protein levels was dependent on SHH dose (Figure 6B, C). Furthermore, SHH treatment triggered the expression of the definitive pituitary precursor marker LHX3 (Figure 6D).
Figure 6. Specification and functional characterization of hormone-producing pituitary placode derivatives (see also Figure S7).
A) Schematic illustration of normal pituitary lineage development in vivo (Tabar, 2011). Examples of hormone producing cells generated using our modified PIP are listed in bold (ACTH, GH, FSH). B) Treatment with SHH from day 7–11 of PIP differentiation induced PITX1 and SIX6 expression as assessed by qRT-PCR. PITX1 and SIX6 mark the pituitary anlage. Low SHH: 20ng/ml C25II SHH; high SHH: 100ng/ml C25II SHH + 1μM puromorphamine. C) Immunocytochemical analysis showed expression of SIX6 at the protein level in a subset of clusters in the presence of SHH treatment. D) Immunocytochemical analysis for expression of LHX3 at day 16 (upon SHH treatment). E–G) Induction of defined endocrine precursor lineages: E) TBX19 expression was highly induced by day 20. F) PIT1 expression at day 20 and 32 of differentiation (see Figure S7A for treatment paradigm). G) GATA2 expression at day 20 and 32 of differentiation. H–L) Immunocytochemical evidence of hormone production H) CGA expression was readily detected by day 16 in SHH-treated PIP cultures. I) FSH, was expressed by day 27. J) ACTH expression was most abundant in SHH-treated PIP culture by day 30 of differentiation. K) GH expression at day 30 of differentiation. L) ELISA measurement of in vitro hormone production after 10 min exposure in HBSS. M) Schematic illustration of transplantation paradigm in nude rat host. N) ACTH plasma levels in grafted adult nude rats and sham-grafted controls at 4 and 6 weeks after transplantation. O) GH plasma levels using a human specific ELISA (6 weeks after transplantation). P–R) Histological analysis 6 weeks after transplantation: P) Robust survival of hNCAM+ human cells. Q) ACTH expressing and (R) GH expressing cells in vivo. Scale bars are: 100 μm in (C, P), 50 μm in (D, H, I, J, Q, R, S) and 10 μm in (K). Error bar represents SEM (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared with controls PIP condition in (B), hESC in (E, F, G; n = 3 independent experiments), HBSS without cells in (L), and plasma samples from matrigel-only injected (Sham) animals in (N, O; n=3 and n=5 animals).
Endocrine cells of the anterior pituitary gland are derived from three main precursors lineages (Scully and Rosenfeld, 2002). Following PIP + SHH treatment, we observed robust induction of TBX19 (Figure 6E), specific to precursors giving rise to ACTH and MSH producing cells. Induction of PIT1 and GATA2 precursor lineages was less efficient but was increased following treatment with the γ-secretase inhibitor DAPT (Figure 6F, G). Immunocytochemistry for pituitary hormones showed expression of CGA by 16 days while protein expression of FSH, ACTH and GH was first observed by day 25–30 of differentiation (Figure 6H–K). The induction of ACTH+ cells was particularly efficient (Figure 6J) and in vitro release of ACTH hormone could readily be detected by ELISA (Figure 6L).
We next tested whether hESC-derived pituitary cells are capable of in vivo survival and function in mouse and rat xenograft models (Figure 6M, S7B, S7C). Subcutaneous injection of GFP-marked, hESC-derived pituitary precursors (day 16 of differentiation) into adult male NOD/SCID mice demonstrated survival of GSU and FSH cells in vivo (Figure S7D, S7E). Longer-term survival studies in nude male rats (n=8; 4–6 weeks post grafting) showed significant increases in serum ACTH levels (Figure 6N) as measured during early morning hours (low point of endogenous ACTH expression during diurnal cycle). We also measured levels of human GH in vivo using an ELISA assay that selectively detects human but not mouse GH (Figure 6O). Transplantation into rat hosts allowed for repeated blood draws and showed a consistent increase of ACTH and GH levels in grafted as compared to sham injected (matrigel-only) animals. Finally, histological analysis demonstrated in average 0.46 ± 0.015 million surviving hNCAM+ cells (Figure 6P) at 6 weeks after transplantation. About 10% of the surviving human cells expressed ACTH (Figure 6Q) and 6% of the cells were immunoreactive for GH (Figure 6R). The ability to derive functional hormone producing pituitary cells from hESCs via modified PIP conditions is particularly intriguing given previous work in mouse ESCs suggesting the need for complex co-culture systems to induce pituitary lineages (Suga et al., 2011). In vivo survival and production of graft-derived ACTH and GH suggest translational potential for patients suffering from genetic, surgical or radiation induced hypopituitarism (Tabar, 2011).
DISCUSSION
An experimental platform for the efficient generation of human cranial placodes in vitro
The study of human cranial placode development has been challenging due to the lack of a tractable experimental system, the inaccessibility of this transient structure during early human development and the absence of validated human placode markers. Our data resolve those major challenges and establish a versatile platform for the study of human placode and ectoderm development (Figure 7). Previous studies have observed the emergence of certain placode derivatives such as lens (Ooto et al., 2003; Zhang et al., 2010) or otic placode derived cells (Chen et al., 2012; Oshima et al., 2010). However, the mechanisms of placode induction were not addressed, and no general model of placode specification has emerged from those studies. Very recent studies have shown some promise in directing pluripotent stem cells towards placode fates (Leung et al., 2013; Mengarelli and Barberi, 2013; Shi et al., 2007). However, no functional human placode derivatives have been reported under those conditions. In contrast, studies in mouse ESCs have successfully derived functional otic (Koehler et al., 2013) and pituitary (Suga et al., 2011) placode derivatives.
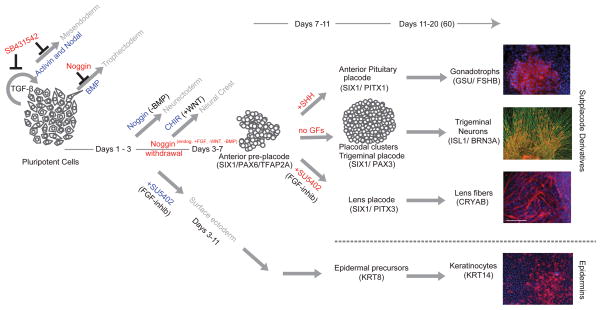
Figure 7. Proposed model for the derivation of human placodes from hPSCs.
Placode induction is dependent on de-repression of BMP signaling using dSMADi protocol. Continuous BMP repression induced CNS fates, de-repression of BMP in combination with the inhibition of FGF signaling by SU5402 triggers epidermal fates. Pre-placodal cells can be further patterned towards specific placode fates by modulating FGF, BMP or SHH signaling at day 7 of differentiation leading to functional lens, trigeminal neuron and anterior pituitary derivatives.
A developmental question of particular interest is the origin of placode cells. The prevalent hypothesis is that placode tissue originates from non-neural ectoderm upon response to inductive signals from the adjacent neural tissue (Schlosser, 2006). Our data indicate that human placode development similarly originates from non-neural ectoderm based on the result that TFAP2A expression precedes the induction of SIX1 while TFAP2A is suppressed during N-SB induction. BMP-based induction of TFAP2A and GATA3 may be critical in establishing non-neural ectoderm lineage competent for placode induction (Kwon et al., 2010). The ability to block placode induction at the expense of non-neural ectoderm by inhibition of FGF signaling further supports a non-neural ectoderm origin and suggests that endogenous FGF signals may be the neural signal responsible for the placode default in PIP. Our findings further delineate the time point of developmental commitment to placodal fate by day 7, given that treatment of cells at day 7 of differentiation (pre-placode)induced a switch between various placode fates but did not affect the ratio of cells of placode versus non-neural ectoderm fate.
Previous work described FOXG1 and DACH1 as markers of anterior neuroectoderm and neural rosette stage cells during hPSC differentiation (Chambers et al., 2009; Elkabetz et al., 2008). Our study reports a small percentage of SIX1+ cells that emerge spontaneously under N-SB conditions. Therefore, differentiation studies aimed at generating CNS lineages should address whether contaminating placodal tissues are present in hPSC derived neural cultures. Those spontaneously emerging placodal cells are likely the source of lentoid bodies and other placode derivatives observed in past neural differentiation studies. Our gene expression data define a broad set of placode markers valuable for future studies. For example, OVOL2 has been shown to be expressed in the mouse epiblast and surface ectoderm, and loss of Ovol2 leads to early embryonic lethality (Mackay et al., 2006). Here we define OVOL2 as a human pre-placode marker. FOXC1 and ISL1 are additional transcription factors specifically expressed at the pre-placode stage. Our data provide a framework for defining transcriptional networks that distinguish cranial placode identity from early CNS, neural crest and surface ectoderm identity.
Specification towards functional hESC-derived placode derivatives
The current study defines a transient pre-placode population competent to adopt various specific placode identities. The emergence of a pre-placodal region has been described during Xenopus and zebrafish development (Bailey et al., 2006; Martin and Groves, 2006; Schlosser, 2006). Our data indicate that PIP initially yields a PAX6+/SIX1+ anterior pre-placode population that spontaneously adopts a more posterior, PAX3+ ophthalmic trigeminal fate upon further differentiation, likely due to the caudalizing effects of endogenous FGF and WNT signals. Exposure to SHH or suppression of FGF signaling at day 7 of differentiation directs the putative pre-placode precursors into anterior pituitary and lens placodes fates. Lens placode has been suggested as the default state during chick placode development with FGF8 being necessary and sufficient to specify olfactory placode fate (Bailey et al., 2006). Elegant studies in mouse ESCs (Koehler et al., 2013; Suga et al., 2011) have shown the feasibility of generating hormone-producing pituitary cells and otic sensory neurons respectively using sophisticated 3D culture systems. Our results demonstrate that modified PIP conditions yield human pituitary precursors efficiently without the need for complex 3D culture conditions. While we observe a bias in generating preferentially TBX19-related pituitary lineages including ACTH producing cells, all three major precursor lineages (GATA-2, TBX19, and PIT1 lineages, Figure 6A) could be derived. Therefore it is likely that further optimization of the protocol will provide selective access to individual pituitary hormone lineages. Our current in vivo studies are limited to a maximum engraftment period of three months, subcutaneous injections into the flank of adult murine hosts, and use of animals (both mouse and rat) with normal pituitary function. Future experiments should include longer-term survival studies and an in-depth analysis of graft integration into the hypothalamic-pituitary axis, possibly via orthotopic graft placement into the hypothalamus or the sella. Finally, in vivo hormone function will need to be validated in animal models of pituitary dysfunction.
We present data on the specification of trigeminal, lens and anterior pituitary placode lineages. However, preliminary evidence indicates that other placode fates are accessible as well using modified PIP conditions. For example, exposure of pre-placode cells to FGF8 enriches for ASCL1 expression compatible with olfactory placode fate (Balmer and LaMantia, 2005). Exposure to caudalizing cues such as WNT3A leads to the induction of a population of cells co-expressing SIX1 and SOX10 compatible of otic placode fates. Those conditions, while requiring further optimization, support the notion that PIP represents a universal platform for cranial placode fate specification.
One of the most immediate applications of PIP-based differentiation is the derivation of trigeminal sensory neurons. Given our recent success in deriving neural crest-derived nociceptive neurons (Chambers et al., 2012), it will be intriguing to compare the developmental and functional features of neural crest versus placode-derived nociceptors. Trigeminal neurons are involved in several pain syndromes such as trigeminal nerve palsy, trigeminal neuralgia and migraine pain (Love and Coakham, 2001). Therefore, the ability to generate large numbers of trigeminal neurons will be particularly useful for modeling human nociception and for the development of cell-based drug screens in pain research. Another important application will be modeling Herpes simplex encephalitis using human iPSCs (Lafaille et al., 2012). HSV-1 is a virus that specifically persists in a latent form within the trigeminal ganglia (Barnett et al., 1994). The ability to derive trigeminal neurons from patient-specific iPSCs should address whether defects in the control of viral latency contributes to Herpes simplex encephalitis. The robust in vivo survival of trigeminal placode precursors raises the possibility for developing future regenerative approaches with the goal of nerve repair following mechanical, radiation or chemotherapy-induced damage. Finally, the potentially largest impact on regenerative medicine may come from deriving functional hormone producing cells. Hypopituitarism is a common consequence of congenital defects, head injury or therapeutic intervention in patients with pituitary tumors or patients receiving radiation therapy (Tabar, 2011). While replacement hormones can be given to normalize resting serum levels in patients, the financial, logistic and medical costs for such life-long treatments are considerable. Furthermore, hormone replacement therapy does not allow for dynamic release in response to circadian rhythms, or rapid adjustments to physiological changes in the environment or stressful challenges. Therefore, the ability to generate large numbers of functional ACTH and GH producing cells launches the possibility of long-term therapeutic cell replacement strategies in pediatric and adult patients for restoring endocrine function.
EXPERIMENTAL PROCEDURES
Cell culture
hESCs (WA-09; passages 35–45), hiPSC lines (iPS-14, iPS-27; passages 20–30), and I6 were maintained at undifferentiated state and differentiated towards CNS lineages using dual-SMAD inhibition protocol described previously. For placode induction protocol (PIP), Noggin was removed at day3 of differentiation. In some experiments, BMP-4, Noggin, DKK-1, FGF8, SU5402, Wnt-3a, DAPT, CHIR99021, Cyclopamine, Sonic Hedgehog (SHH) and Purmorphamine were added as detailed in Supplementary methods. For differentiation towards trigeminal sensory fate placode clusters were maintained in N2 medium supplemented with ascorbic acid and BDNF. Pituitary fate was induced by exposure to SHH and Puromorphamine from day 7–11 of PIP followed by treatment with DAPT to promote PIT1+ fate.
Cell characterization
qRT-PCR data were normalized to HPRT and are based on 4–6 technical replicates from at least 3 independent experiments. Global gene expression analysis was performed by the MSKCC genomics core according to the specification of the manufacturer (Illumina Human-6 oligonucleotide arrays). Detailed information on the use of primary antibodies for immunocytochemistry and flow analysis and on the electrophysiological analyses is presented in extended supplementary methods.
Animal studies
Animal studies were done in accordance with protocols approved by our institutional Animal Care and Use Committee and following NIH guidelines. Hormone producing cells were injected subcutaneously into adult male NOD-SCID IL2Rgc mice and adult 8 male nude rats. Blood was collected at 4 – 6 weeks after the transplantation followed by ELISA analysis for determining hormone levels. Chick transplantation studies were performed at HH Stage 9–10 and embryos were harvested at HH Stage 20. Injections into pons of adult NOD-SCID IL2Rgc mice were performed by stereotactic surgery.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 5.0b (GraphPad Software). All data were derived from at least 3 independent experiments. Asterisks mark experimental groups that were significantly different from control groups by a two-tailed Students t-test, or by ANOVA followed by Dunnett test to compare control against multiple independent treatment groups. Data are presented as mean ± SEM unless indicated otherwise.
Supplementary Material
HIGHLIGHTS.
Timed withdrawal of BMP inhibitor is sufficient to induce placode fates from hESCs
Timed FGF inhibition suppresses placode fate and induce epidermal lineage
Placode-derived trigeminal neurons are functional in vitro and engraft in vivo
Pituitary placode-derived cells are capable of hormone release in vitro and in vivo
Acknowledgments
This work was supported in part through grants from the Starr Foundation, NINDS grant NS072381 and NYSTEM contract C026447. We are grateful to E. Lai for BF1 antibody, A.S. McNeilly for GSU antibody, R. Vinagolu for FSH antibody and K. Kawakami for Eya1::GFP enhancer element. We also would like to thank H. Ford for advice on SIX1 antibody, M. Hashimi for advice on microarray analysis, M. Tomishima & J. Tchieu for critical review of the manuscript and M. Bronner for help in designing the chick in vivo transplantation studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nature genetics. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Developmental biology. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Arkell R, Beddington RS. BMP-7 influences pattern and growth of the developing hindbrain of mouse embryos. Development. 1997;124:1–12. doi: 10.1242/dev.124.1.1. [DOI] [PubMed] [Google Scholar]
- Bailey AP, Bhattacharyya S, Bronner-Fraser M, Streit A. Lens specification is the ground state of all sensory placodes, from which FGF promotes olfactory identity. Dev Cell. 2006;11:505–517. doi: 10.1016/j.devcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Balmer CW, LaMantia AS. Noses and neurons: induction, morphogenesis, and neuronal differentiation in the peripheral olfactory pathway. Developmental dynamics: an official publication of the American Association of Anatomists. 2005;234:464–481. doi: 10.1002/dvdy.20582. [DOI] [PubMed] [Google Scholar]
- Barnett EM, Jacobsen G, Evans G, Cassell M, Perlman S. Herpes simplex encephalitis in the temporal cortex and limbic system after trigeminal nerve inoculation. J Infect Dis. 1994;169:782–786. doi: 10.1093/infdis/169.4.782. [DOI] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Bronner-Fraser M. Hierarchy of regulatory events in sensory placode development. Curr Opin Genet Dev. 2004;14:520–526. doi: 10.1016/j.gde.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jongkamonwiwat N, Abbas L, Eshtan SJ, Johnson SL, Kuhn S, Milo M, Thurlow JK, Andrews PW, Marcotti W, et al. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature. 2012;490:278–282. doi: 10.1038/nature11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas JF, Schoenwolf GC. Localization of cartilage linking protein 1 during primary neurulation in the chick embryo. Brain Res Dev Brain Res. 2003;141:141–148. doi: 10.1016/s0165-3806(03)00011-7. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology 2003. 2003;4:34, 3. [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigaliunas A, Bradley RM, MacCallum DK, Mistretta CM. Distinctive neurophysiological properties of embryonic trigeminal and geniculate neurons in culture. Journal of Neurophysiology. 2002;88:2058–2074. doi: 10.1152/jn.2002.88.4.2058. [DOI] [PubMed] [Google Scholar]
- Grotewold L, Plum M, Dildrop R, Peters T, Rüther U. Bambi is coexpressed with Bmp-4 during mouse embryogenesis. Mech Dev. 2001;100:327–330. doi: 10.1016/s0925-4773(00)00524-4. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Rhodes SJ. LIM-homeodomain genes in mammalian development and human disease. Mol Biol Rep. 2005;32:67–77. doi: 10.1007/s11033-004-7657-z. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Sato S, Ikeda K, Yajima H, Kawakami K. Multiple evolutionarily conserved enhancers control expression of Eya1. Dev Dyn. 2008;237:3142–3156. doi: 10.1002/dvdy.21716. [DOI] [PubMed] [Google Scholar]
- Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013 doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh T, Concha ML, Houart C, Dawid IB, Wilson SW. Combinatorial Fgf and Bmp signalling patterns the gastrula ectoderm into prospective neural and epidermal domains. Development. 2004;131:3581–3592. doi: 10.1242/dev.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Bhat N, Sweet EM, Cornell RA, Riley BB. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010;6:e1001133. doi: 10.1371/journal.pgen.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaille FG, Pessach IM, Zhang SY, Ciancanelli MJ, Herman M, Abhyankar A, Ying SY, Keros S, Goldstein PA, Mostoslavsky G, et al. Impaired intrinsic IFN-α/β immunity to HSV-1 in human iPSC-derived UNC-93B- and TLR3-deficient central nervous system cells. Nature. 2012;491:769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Kim H, Elkabetz Y, Al Shamy G, Panagiotakos G, Barberi T, Tabar V, Studer L. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- Leung AW, Kent Morest D, Li JY. Differential BMP signaling controls formation and differentiation of multipotent preplacodal ectoderm progenitors from human embryonic stem cells. Dev Biol. 2013;379:208–220. doi: 10.1016/j.ydbio.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsiou A, Hanson S, Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051–4062. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- Love S, Coakham HB. Trigeminal neuralgia: pathology and pathogenesis. Brain. 2001;124:2347–2360. doi: 10.1093/brain/124.12.2347. [DOI] [PubMed] [Google Scholar]
- Mackay DR, Hu M, Li B, Rhéaume C, Dai X. The mouse Ovol2 gene is required for cranial neural tube development. Dev Biol. 2006;291:38–52. doi: 10.1016/j.ydbio.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2006;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- McCabe KL, Manzo A, Gammill LS, Bronner-Fraser M. Discovery of genes implicated in placode formation. Dev Biol. 2004;274:462–477. doi: 10.1016/j.ydbio.2004.07.012. [DOI] [PubMed] [Google Scholar]
- McCauley DW, Bronner-Fraser M. Conservation of Pax gene expression in ectodermal placodes of the lamprey. Gene. 2002;287:129–139. doi: 10.1016/s0378-1119(01)00894-0. [DOI] [PubMed] [Google Scholar]
- Menendez L, Yatskievych TA, Antin PB, Dalton S. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19240–19245. doi: 10.1073/pnas.1113746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengarelli I, Barberi T. Derivation of multiple cranial tissues and isolation of lens epithelium-like cells from human embryonic stem cells. Stem Cells Transl Med. 2013;2:94–106. doi: 10.5966/sctm.2012-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe WK, Myers PZ, Trevarrow B, Bass MB, Kimmel CB. Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development. 1990;110:491–504. doi: 10.1242/dev.110.2.491. [DOI] [PubMed] [Google Scholar]
- Mica Y, Lee G, Chambers SM, Tomishima MJ, Studer L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013;3:1140–1152. doi: 10.1016/j.celrep.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rahilly RMF. Developmental stages in human embryos. Vol. 637. Washington, D.C: Carnegie Institution of Washington; 1987. [Google Scholar]
- Ooto S, Haruta M, Honda Y, Kawasaki H, Sasai Y, Takahashi M. Induction of the differentiation of lentoids from primate embryonic stem cells. Invest Ophthalmol Vis Sci. 2003;44:2689–2693. doi: 10.1167/iovs.02-1168. [DOI] [PubMed] [Google Scholar]
- Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper M, Ahrens K, Rink E, Peter A, Schlosser G. Differential distribution of competence for panplacodal and neural crest induction to non-neural and neural ectoderm. Development. 2012;139:1175–1187. doi: 10.1242/dev.074468. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Itsykson P, Turetsky T, Pera MF, Reinhartz E, Itzik A, Ben-Hur T. Neural progenitors from human embryonic stem cells. Nature biotechnology. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Jr, et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235. doi: 10.1126/science.1062736. [DOI] [PubMed] [Google Scholar]
- Shi F, Corrales CE, Liberman MC, Edge AS. BMP4 induction of sensory neurons from human embryonic stem cells and reinnervation of sensory epithelium. Eur J Neurosci. 2007;26:3016–3023. doi: 10.1111/j.1460-9568.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- Sjödal M, Edlund T, Gunhaga L. Time of exposure to BMP signals plays a key role in the specification of the olfactory and lens placodes ex vivo. Dev Cell. 2007;13:141–149. doi: 10.1016/j.devcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Stark MR, Sechrist J, Bronner-Fraser M, Marcelle C. Neural tube-ectoderm interactions are required for trigeminal placode formation. Development. 1997;124:4287–4295. doi: 10.1242/dev.124.21.4287. [DOI] [PubMed] [Google Scholar]
- Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, Takata N, Wataya T, Muguruma K, Miyoshi H, et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature. 2011;480:57–62. doi: 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- Tabar V. Making a pituitary gland in a dish. Cell Stem Cell. 2011;9:490–491. doi: 10.1016/j.stem.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JJ, Lanctot C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Lagna G, Suzuki A, Hemmati-Brivanlou A. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development. 1997;124:3177–3184. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li X, Ayala M, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.