Abstract
Background
Leishmaniasis is a group of diseases that are created by intracellular parasites of Leishmania. Cutaneous leishmaniasis is considered as one of the health problems in some provinces of Iran.
Methods
In this study, a total of 178 Giemsa-stained slides from confirmed cases of cutaneous leishmaniasis were examined. The slides were prepared from the patients with cutaneous leishmaniasis that referred to health centers and infected during the epidemic of cutaneous leishmaniasis in Poldokhtar city, Lorestan Province, Iran in 2006.Genomic DNA from each slide was extracted. After DNA extraction, ITS-PCR was used.
Results
Out of 178 slides, 129 (72.47%) samples had a band in the range of 485 bp and 49 (27.53%) samples 626 bp that matched L. tropica and L. major standard samples, respectively.
Conclusion
This study showed that Leishmania DNA could be efficiently extracted and amplified even from old Giemsa-stained microscopic slides that were stored more than 6 yr. In this study was shown that both L. tropica and L. major species exist in Lorestan Province.
Keywords: Leishmania, Giemsa, PCR, Cutaneous leishmaniasis, Iran
Introduction
Leishmaniasis is a group of diseases that are created by intracellular parasites of Leishmania. Out of 30 species of the recognized Leishmania, about 10 species are important due to medical and veterinary (1). This disease is observed in three main forms of cutaneous, visceral and mucocutaneous leishmaniasis (2). Due to the importance of health in this disease, it has always been considered by the World Health Organization. Currently in 98 countries or territories leishmaniasis is endemic (3). Prevalence rate of leishmaniasis have been reported around 12 million cases (3–5).
Cutaneous leishmaniasis is considered as one of the health problems in some provinces of Iran and more than 20 thousand cases of this disease are reported annually (6). In Iran, Leishmania L. major is agent of zoonotic cutaneous leishmaniasis (ZCL), and L. tropica causes anthroponetic cutaneous leishmaniasis (ACL) (2, 7, 8).
Considering the biological properties of the parasite, reservoir and vector, the methods to combat these two forms of cutaneous leishmaniasis, are different. Therefore, identification of parasite species is important in the health planning (9).
For molecular studies, the different parts of Leishmania genome are used, one of them that has many applications is ribosomal gene used for ITS amplification fragment (10, 11). The advantages of these methods are that even with the low number of parasite, the infection is shown and parasite species identified (9, 10).
Cutaneous leishmaniasis is reported in several provinces of Iran and the species characterization in some of them was done by PCR method. For example, in endemic foci of south-eastern Iran was shown that L. tropica is the main species caused ACL and L. major is present in low level (12). A study in Qom Province were done by PCR and showed that L. major is the causative disease (13). In the studies on CL in Mashhad using ITS-PCR and PCR-RFLP, was determined that L. tropica was the dominant species (14, 15). Also in order to identify Leishmania species using Nested–PCR, the studies were performed in Shush city in Khuzestan Province and Shiraz city and reported that the predominant species was L. major (16, 17).
Lorestan Province is one of the endemic areas in Iran that both species of L. tropica and L. major have been reported in this province (18). Since the epidemic of cutaneous leishmaniasis happened in 2006 (19) and probable of its occurrence, to know which species are dominant in the epidemic is especially important.
This study was carried out for the first time to evaluate of PCR in order to detect of Leishmania DNA and molecular identification of Leishmania species in archived Giemsa-stained slides in Poldokhtar City, Iran.
Materials and Methods
The study was performed in Parasitology Department, Cellular and Molecular Research Center, Lorestan University of Medical Sciences, Iran.
A total of 178 Giemsa-stained slides from confirmed cases of cutaneous leishmaniasis were examined. The slides were prepared from the patients with cutaneous leishmaniasis that referred to health centers and infected during the epidemic of cutaneous leishmaniasis in Poldokhtar City, Lorestan Province, Iran in 2006.
DNA from each slide was extracted separately by AccuPrep® Genomic DNA Extraction Kit (Bioneer, South Korea) in accordance with the manufacturer's protocol. Extracted DNA was stored at - 20 °C until PCR amplification. The ribosomal internal transcribed spacer (ITS) was amplified with specific primers according to previous studies (8), LeishF (5‘- CAA CACGCCGCCTCCTCTCT -3‘), LeishR (5‘- CCTCTCTTTTTTCNCTGTGC-3‘). The PCR conditions consisted of one initial denaturing cycle at 94°C for 5 min, followed by 30 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 40 s and finally 1 cycle of 72° C for 5 min (8). At the end, PCR products were analyzed by 1.5% agarose gel electrophoresis.
Expected PCR products for L. major and L. tropica were 626 bp, 485 bp, respectively. Standard strains of L. major ((MRHO/IR/75/ER) and L. tropica (MHOM/IR/01/yaza) were used as positive controls.
Negative controls were used. In one of them was added distilled water instead of DNA. In another one extracted DNA was used that it was prepared from cutaneous lesion slide of patient with skin disease except CL which Leishmania was not found in lesion using microscopic exams, culture and PCR.
DNA sequencing
DNA for sequencing was prepared by the ITS-PCR.
Results
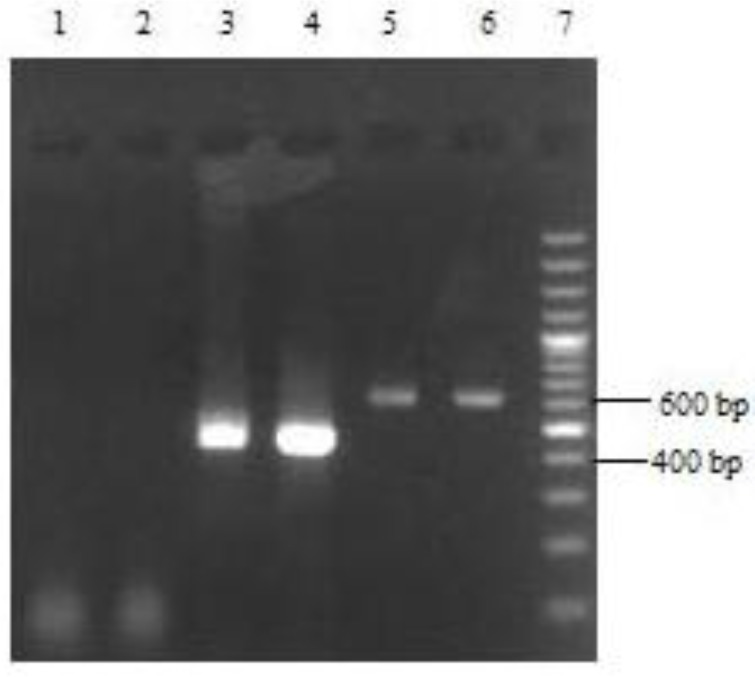
Out of 178 slides, 111 (62.36%) of whom were for male and 67 (37.64%) for female and also 14 (7.87%) slides were collected from urban and 164 (92.13%) from rural. Frequency distribution of cutaneous leishmaniasis base on age, number and location of the lesion were shown in Table 1 and Table 2, respectively. Electrophoresis patterns from each isolates were compared with reference strains of L. tropica and L. major. After DNA extraction and PCR performance, according to the pattern of electrophoresis, out of 178 slides, 129 (72.47%) samples had a band in the range of 485 bp and 49 (27.53%) samples 626 bp that matched L. tropica and L. major standard samples, respectively (Fig. 1). Number of Leishmania amastigotes on Giemsa-stained smears was shown in Table 3.
Table 1.
Frequency distribution of cutaneous leishmaniasis base on age in Poldokhtar City, Iran in 2006
| Age group (yr) | 0-9 | 10-19 | 20-29 | 30-39 | 40-49 | ≥ 50 | Total |
|---|---|---|---|---|---|---|---|
| Frequency | 26 | 46 | 37 | 29 | 19 | 21 | 178 |
Table 2.
Frequency distribution of cutaneous leishmaniasis base on the number and location of the lesion in Poldokhtar City, Iran in 2006
| Location | Number of the lesions | |||
|---|---|---|---|---|
| 1 | 2 | ≥3 | Total | |
| Face | 16 | 6 | 0 | 22 |
| Foot | 28 | 14 | 7 | 49 |
| Hand | 54 | 18 | 23 | 95 |
| Face & Hand | 0 | 5 | 3 | 8 |
| Face & Hand & Foot | 0 | 0 | 1 | 1 |
| Hand & Foot | 0 | 1 | 2 | 3 |
| Total | 98 | 44 | 36 | 178 |
Fig. 1.
1.5% agarose gel electrophoresis of PCR amplification for identification of Leishmania species on Giemsa-Stained slides
Lane 1, 2: Negative controls
Lane 3: L. major isolate
Lane 4: Reference strain of L. major
Lane 5: L. tropica isolate
Lane 2: Reference strain of L. tropica
Lane 7: 100bp DNA ladder marker
Table 3.
Leishmania amastigote numbers on Giemsa-Stained slides prepared from cutaneous leishmaniasis patients in Poldokhtar City, Iran in 2006
| Leishmania species | *Number of Amastigotes | Total | |||
|---|---|---|---|---|---|
| 1+ | 2+ | 3+ | 4+ | ||
| L. tropica | 25 | 31 | 46 | 27 | 129 |
| L. major | 11 | 28 | 4 | 6 | 49 |
| Total | 36 | 59 | 50 | 33 | 178 |
Grading of Leishmania Parasites was obtained by average parasite density using x10 eyepiece and x100 oil-immersion lens as follows: 4+ 1-10 parasites/fields/3+ 1-10 parasites/10 fields/2+ 1-10 parasites/100 fields/1+ 1-10 parasite/1000 field
Nucleotide sequence data reported in this article have been submitted to the GenBank database with accession numbers, L. tropica JX183382 and L. major JX183383.
Discussion
Identification of the epidemiological aspects of cutaneous leishmaniasis for control program is necessary. So it is especially imp-ortant to identify the parasite species, because different species may require distinct treatment regimens (20, 21).
Because all Leishmania species are very similar, identification of Leishmania species by microscopic examination of Giemsa-stained slides is not possible (22). In recent years molecular methods including PCR have been used for diagnosis of leishmaniasis (9, 10, 23–26). In endemic areas where more than one Leishmania species is present, diagnostic tools are required for detection and identification of parasites directly in samples (22).
ITS-PCR was used for diagnosis and characterization of Leishmania species on Giemsa-stained slides of the patients with cutaneous leishmaniasis referred to health centers that infected during the epidemic of cutaneous leishmaniasis in Poldokhtar City, Lorestan Province, Iran.
Similar studies were conducted in different parts of Iran and other countries for the diagnosis of cutaneous leishmaniasis by PCR Giemsa-stained slides. For example, in a study DNA was isolated from 92 Giemsa-stained smears that had been stored for up to 4 years and used for PCR-based diagnosis of Leishmania infection and demonstrated this method is effective (27). In another study, PCR-RFLP was conducted on 48 Giemsa-stained slides and reported that technique is an effective method (28). In a study using PCR was identified that archived Giemsa-stained slides were positive for CL which caused by L. tropica (29). A study in Brazil examined the ability of PCR to amplify Leishmania DNA from Giemsa-stained slides that prepared from American cutaneous leishmaniasis patients. The slides were stored for up to 36 years. The results showed that archived Giemsa-stained slides are a useful source of Leishmania DNA for performing clinical and epidemiological studies of leishmaniasis (30). A study on 102 Giemsa-stained slides by real-time PCR, sensitivity of this method was reported 98%. Archived slides that were stored more than 3 years can be use for Leishmania DNA extraction and amplification by real-time PCR (31). Another study was conducted to evaluate sensitivity and specificity of PCR in order to detect of Leishmania DNA in archived Giemsa-stained bone marrow slides for diagnosis of visceral leishmaniasis. The results showed this method is a suitable tool for confirming diagnosis in Kalaazar patients and useful in the diagnosis of complicated cases. Additionally, was announced Giemsa-stained slides are easily stored, do not require special storage conditions, can be easily posted to centers where PCR is available and making a super option for diagnosis in the field (32). The study was done on Giemsa-stained smears for Palestinian patients. DNA was extracted from each slide and subjected to PCR. PCR showed 87% sensitivity and 100% specificity. This study reported that Giemsa-stained slides are an easily usable sampling method for PCR (33). Our study similar to other studies was shown that PCR Giemsa-stained slide is effective. Identification of Leishmania species using ITS-PCR technique on Giemsa-stained slides is an accurate and useful method that can be used in other endemic areas of leishmaniasis in Iran. Also in this method, there is no need to parasite cultivation or injection into laboratory sensitive animals and identification of Leishmania species is possible directly by the slides stained. Additionally, Giemsa-stained slides are appropriate for field condition such as samples can be easily stored and sent to the diagnostic laboratory (22, 27, 32) and can be helpful when re-evaluating the diagnosis of controversial cases or in retrospective epidemiologic studies (22).
This study like previous studies showed that Leishmania DNA could be efficiently extracted and amplified even from old Giemsa-stained microscopic slides those were stored more than 6 years.
Since in 2005, only 9 cases of cutaneous leishmaniasis were reported in Poldokhtar City, and in 2006, 178 cases that is a sign of epidemic of this disease in this year (19).
According to our results, out of 178 slides examined, 129 (72.47%) slides were L. tropica and 49 (27.53%) L. major. In previous study that was done on identification of Leishmania species of 43 patients with cutaneous leishmaniasis in Poldokhtar City, the results showed that out of 43 patients, 6 (13.95%) were infected with L. major and 37 (86.05%) with L. tropica (18). To compare these two studies shows that there is a significant difference between the prevalence of each species in the two studies so that during the epidemic, L. major frequency had been more than expected due to its prevalence in the region (P<0.05).
One of the reasons of this epidemic could be the return of Lorestan troops from operational area of Ilam Province, one of the endemic area of cutaneous leishmaniasis in Iran, who deployed in silos around the city along with their belongings probably brought the Leishmania-infected vectors (19). Since the vectors of this parasite are found in abundance in the province so easily caused the spread of disease. Another likely reason could be an increase in human– vector contact. This is attributed to the development of villages and the spread of the human population into the habitats of the local vectors (34).
In this study, the disease was observed in all age groups but most cases were seen ranged in age from 10 to 40 year that could be due to more working these people outside of the home and the possibility of more contact with sandfly. In this study as previous study (18) was shown that both L. tropica and L. major species exist in the province. Since the reservoir of L. major is rodents, so their identification for the purpose of planning of disease control, especially in epidemics is important (35–37). As mentioned above, considering the biological properties of the parasite, reservoir and vector, methods of combating these two species are different, so an accurate and comprehensive planning in this regard should be designed (2).
Conclusion
The PCR procedure is a suitable tool for direct diagnosis and identification of Leishmania species from Giemsa-stained slides. Also this method is useful for retrospective epidemiologic studies.
Acknowledgment
Hereby, the authors appreciate deputy and colleagues of Research and Technology, deputy and colleagues of Razi Herbal Medicines Research Center, Lorestan University of Medical Sciences, for their great cooperation and all those who helped us in this research are also sincerely appreciated. The authors declare that there is no conflict of interest.
References
- 1.Bates PA. Transmission of Leishmania met acyclic promastigots by phlebotomine sand files. Int J Parasitol. 2007;37(10-3):1097–1106. doi: 10.1016/j.ijpara.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. Leishmaniasis: the global trend. 2009. http://www.who.into/neglected_disease/integrated-media_Leishmaniasis/en/index.html.
- 3.World Health Organization (WHO) Control of the leishmaniases; Geneva: 2010. pp. 1–187. Report of a Meeting of the WHO Expert Committee on the Control of Leishmaniases, WHO Technical Report Series 949. [Google Scholar]
- 4.Herwaldt BL, Magill AJ. Yellow book. 2012. Leishmaniasis, Cutaneous, Infectious diseases related to travel, Centers for Disease Control and Prevention, Traveler's health. Chapter. [Google Scholar]
- 5.Piscopo Tv, Mallia Azzopardi C. Leishmanias. Postqrad Med J. 2007;83(976):649–657. doi: 10.1136/pgmj.2006.047340corr1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoonoses Control Office. Ministry of Health and Medical Education, Iran: CDC; 2010. [Google Scholar]
- 7.Farahmand M, Nahrevanian H, Atashi Shirazi H, Naeimi S, Farzanehnejad Z. An overview of a diagnostic and epidemiologic reappraisal of cutaneous leishmaniasis in Iran. Braz J Infect Dis. 2011;15(1):17–21. [PubMed] [Google Scholar]
- 8.Rahbarian N, Mesgarian A, Mahmoudi Rad M, Hajaran H, Shahbazi F, Mesgarian Z, Taghipour N. Identification of Leishmania species isolated from human cutaneous leishmaniasis using PCR method. J Res Health Sci. 2009;9(2):48–51. [PubMed] [Google Scholar]
- 9.Singh S. New developments in diagnosis of Leishmaniasis. Indian J Med Res. 2006;123(3):311–30. [PubMed] [Google Scholar]
- 10.Tavares RG, Staggemeier R, Borges ALP, Rodrigues, Castelan LA, Vasconcelos J, et al. Molecular techniques for the study and diagnosis of parasite infection. J Venom Anim Toxins incl Trop Dis. 2011;17(3):239–248. [Google Scholar]
- 11.Banuls AL, Hide M, Prugnolle F. Leishmania and the Leishmaniases: A parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol. 2007;64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- 12.Sharifi F, Sharifi I, Zarean M, Hakimi Parizi M, Aflatoonian MR, Fasihi Harandi M, Zahmatkesh R, Mashayekhi M, Kermanizadeh AR. Spatial distribution and molecular identification of Leishmania species from endemic foci of south-eastern Iran. Iranian J Parasitol. 2012;7:45–52. [PMC free article] [PubMed] [Google Scholar]
- 13.Yavar R, Abedin S, Reza AM, Ali OM, Sina R, Mehdi M, Reza YE, Fatemeh M, Babak F. Phlebotomus papatasi and Meriones libycus as the vector and reservoir host of cutaneous leishmaniasis in Qomrood district, Qom province, and central Iran. Asian Pac J Trop Med. 2011;4:97–100. doi: 10.1016/S1995-7645(11)60045-X. [DOI] [PubMed] [Google Scholar]
- 14.Shahbazi F, Shahabi S, Kazemi B, Mohebali M, Abadi AR, Zare Z. Evaluation of PCR assay in diagnosis and identification of cutaneous leishmaniasis: a comparison with the parasitological methods. Parasitol Res. 2008;103:1159–1162. doi: 10.1007/s00436-008-1111-4. [DOI] [PubMed] [Google Scholar]
- 15.Vaeznia H, Dalimi A, Sadraei J, Pirstani M. Determination of Leishmania species causing cutaneous leishmaniasis in Mashhad by PCR-RFLP method. Archives of Razi Institute. 2009;64:39–44. [Google Scholar]
- 16.Razmjou S, Hejazya H, Motazedianb MH, Baghaeia M, Emamyc M, Kalantaryb M. A new focus of zoonotic cutaneous leishmaniasis in Shiraz, Iran. Trop Med Hyg. 2009;103:727–730. doi: 10.1016/j.trstmh.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Maraghi S, Samarbaf Zadeh A, Sarlak AA, Ghasemian , Vazirianzadeh B. Identification of cutaneous leishmaniasis agents by Nested Polymerase Chain Reaction (Nested-PCR) in Shush city, Khuzestan province, Iran. Iranian J Parasitol. 2007;2:13–15. [Google Scholar]
- 18.Kheirandish F, Chegeni Sharafi A, Kazemi B, Bandehpour M, Tarahi MJ, Khamesipour A. First molecular identification of Leishmania species in a new endemic area of cutaneous leishmaniasis in Lorestan, Iran. Asian Pac J Trop Biomed. 2013;6(9):713–717. doi: 10.1016/S1995-7645(13)60124-8. [DOI] [PubMed] [Google Scholar]
- 19.Chegeni Sharafi A, Amani H, Kayedi MH, Yarahahmadi A, Saki M, Mehrdad M, Nasiri E. Epidemiological Survey of Cutaneous Leishmaniasis in Lorestan Province(Iran) and Introduction of Disease Transmission in New Local Areas. J IUMS. 2011;19(1):54–60. In Persian. [Google Scholar]
- 20.Gomes AH, Ferreira IM, Lima ML, Cunha EA, Garcia AS, Araujo MF, Pereira-Chioccola VL. PCR identification of Leishmania in diagnosis and control of canine leishmaniasis. Vet Parasitol. 2007;144(3-4):234–241. doi: 10.1016/j.vetpar.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Alimoradi S, Hajjaran H, Mohebali M, Mansouri F. Molecular identification of Leishmania species isolated human cutaneous leishmaniasis by RAPD-PCR. Iranian J Publ Health. 2009;38(2):44–50. [Google Scholar]
- 22.Schonian G, Nasereddin A, Dinse N. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diag Microbiol Infect Dis. 2003;47(1):349–58. doi: 10.1016/s0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 23.Eroglu F, Koltas IS, Genc A. Identification of Causative Species in Cutaneous Leishmaniasis Patients Using PCR-RFLP. J Bacteriol Parasitol. 2011;2:113. doi: 10.4172/2155-9597.1000113. [DOI] [Google Scholar]
- 24.Marfurt J, Nasereddin A, Niederwieser I, Jaffe CL, Beck HP, Felger I. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the Miniexon sequence and subsequent Restriction Fragment Length Polymorphism analysis. J Clin Microbiol. 2003;41(7):3147–53. doi: 10.1128/JCM.41.7.3147-3153.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dweik A, Schonian G, Mosleh IM, Karanis P. Evaluation of PCR_RFLP (based on ITS-1 and HeaIII) for the detection of Leishmania species, using Greek canine isolates and Jordanian clinical material. Ann Trop Med Parasitol. 2007;101(5):399–407. doi: 10.1179/136485907X176436. [DOI] [PubMed] [Google Scholar]
- 26.Martínez LP, Rebollo JA, Luna AL, Cochero S, Bejarano EE. Molecular identification of the parasites causing cutaneous leishmaniasis on the Caribbean coast of Colombia. Parasitol Res. 2010;106(3):647–652. doi: 10.1007/s00436-009-1712-6. [DOI] [PubMed] [Google Scholar]
- 27.Motazedian H, Karamian M, Noyes HA, Ardehali S. DNA extraction and amplification of Leishmania from archived, Giemsa-stained slides, for the diagnosis of cutaneous leishmaniasis by PCR. Ann Trop Med Parasitol. 2002;96(1):31–34. doi: 10.1179/000349802125000484. [DOI] [PubMed] [Google Scholar]
- 28.Kazemi-Rad E, Mohebali M, Hajaran H, Rezeai S, Memishi S. Diagnosis and characterization of Leishmania species in Giemsa-stained slides by PCR-RFLP. Iranian J Publ Health. 2008;37(1):454–60. [Google Scholar]
- 29.Mahdy MAK, Al-Mekhlafi HM, Al-Mekhlafi AM, Lim YAL, Bin Shuaib NOM, Azazy AA, Mahmud R. Molecular Characterization of Leishmania Species Isolated from Cutaneous Leishmaniasis in Yemen. PLoS ONE. 2010;5:e12879. doi: 10.1371/journal.pone.0012879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volpini AC, Marques MJ, Lopes dos Santos S, Machado-Coelho GL, Mayrink W, Romanha AJ. Leishmania identification by PCR of Giemsa-stained lesion imprint slides stored for up to 36 years. Clin Microbiol Infec. 2006;12:815–818. doi: 10.1111/j.1469-0691.2006.01422.x. [DOI] [PubMed] [Google Scholar]
- 31.Khademvatan S, Neisi N, Maraghi S, Saki J. Diagnosis and identification of Leishmania spp. from Giemsa-stained slides, by real-time PCR and melting curve analysis in south-west of Iran. Ann Trop Med Parasitol. 2011;105:559–65. doi: 10.1179/2047773211Y.0000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yvone Maia Brustoloni, Rosimar Batista Lima, Rivaldo Venâncio da Cunha, Maria Elizabeth Dorval, Elisa Teruya Oshiro, Ana Lúcia Lyrio de Oliveira, Claude Pirmez. Sensitivity and specificity of polymerase chain reaction in Giemsa-stained slides for diagnosis of visceral leishmaniasis in children. Mem Inst Oswaldo Cruz, Rio de Janeiro. 2007;102:497–500. doi: 10.1590/s0074-02762007005000036. [DOI] [PubMed] [Google Scholar]
- 33.Al-Jawabreh A, Schoenian G, Hamarsheh O, Presber W. Clinical diagnosis of cutaneous leishmaniasis: A comparison study between standardized graded direct microscopy and ITS1-PCR of Giemsa-stained smears. Acta Trop. 2006;99:55–61. doi: 10.1016/j.actatropica.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Alavinia SM, Arzamani K, Reihani MH, Jafari J. Some Epidemiological Aspects of Cutaneous Leishmaniasis in Northern Khorasan Province, Iran. Iranian J Arthropod-Borne Dis. 2009;3(2) [PMC free article] [PubMed] [Google Scholar]
- 35.Rassi Y, Javadian E, Amin M, Rafizedeh S, vatandoost H, Motazedian H. Meriones libycus is the main reservoir of zoonotic cutaneous leishmaniasis in south of Islamic Republic of Iran. East Mediterr Health J. 2006;12(3-4):474–7. [PubMed] [Google Scholar]
- 36.Rassi Y, Sofizadeh A, Abai MR, Oshaghi MA, Rafizadeh S, Mohebail M, et al. Molecular detection of L. major in the vectores and reservoir hosts of cutaneous leishmaniasis in Kalaleh district, Golestan Province, Iran. Iranian J Arthropod-Borne Dis. 2008;2(2):21–27. [Google Scholar]
- 37.Hajjaran H, Mohebali M, Alimoradi S, Abai MR, Edrissian GhH. Isolation and characterization of pathogenic Leishmania turanica from Nesokia indica (Rodentia, Muridea) by PCR-RFLP and ITSI sequencing in Iran. Trans R Soc Trop Med Hyg. 2009;103(11):1177–9. doi: 10.1016/j.trstmh.2008.08.016. [DOI] [PubMed] [Google Scholar]