Abstract
Background:
FSCN1 and matrix metalloproteinase 14 (MMP14) are both invadopodia-related proteins. We herein elucidate the tumourigenicity of these proteins and identify novel therapeutic agents in esophageal squamous cell carcinoma (ESCC).
Methods:
FSCN1 and MMP14 were evaluated by immunohistochemistry and quantitative PCR, and microRNA (miR)-133a was also evaluated by PCR in surgical ESCC specimens. The roles of FSCN1, MMP14 and miR-133a were established in ESCC cells.
Results:
The expression of FSCN1 or MMP14 was an independent poor prognostic factor according to a multivariate analysis of immunohistochemistry, and their co-expression correlated with the poorest overall survival (OS) out of all the examined factors. Additionally, their mRNAs significantly correlated and both inversely correlated with miR-133a in surgical specimens. Transfection of a miR-133a mimic decreased the mRNA and protein levels of both FSCN1 and MMP14 in ESCC cells. The knockdown of FSCN1 or MMP14 and transfection of a miR-133a mimic inhibited the proliferation and invasion of ESCC cells. Patients with a lower miR-133a expression have a significantly poorer OS than those with a higher expression.
Conclusion:
The combined expression of FSCN1 and MMP14 is associated with a poor prognosis, and miR-133a, which regulates their mRNAs, can serve as a strong tumour suppressor of ESCC.
Keywords: esophageal squamous cell carcinoma, FSCN1, MMP14, invadopodia, anti-oncomir, prognosis
Esophageal squamous cell carcinoma (ESCC) is one of the most malignant cancers of the gastrointestinal tract. Although multimodal therapy has improved the prognosis, the outcome of ESCC is still unsatisfactory. The poor outcomes of ESCC are related to its diagnosis at advanced stages and its propensity for metastasis, even when tumours are superficial (Pennathur et al, 2013).
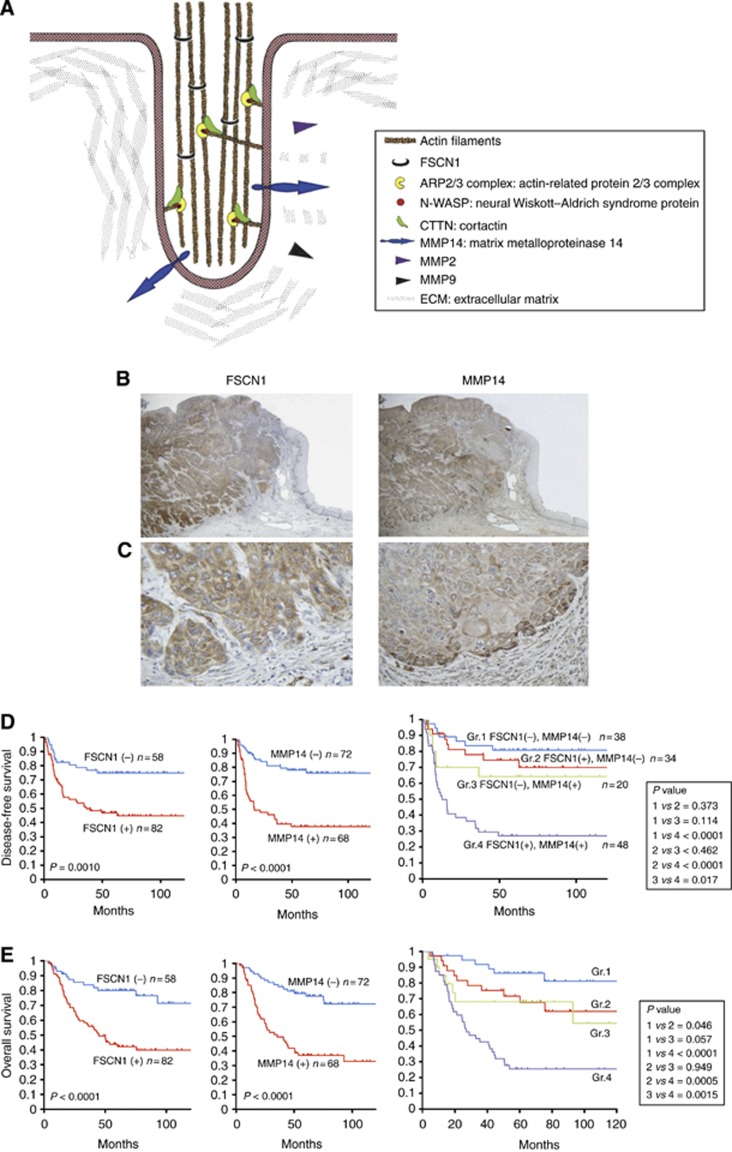
FSCN1 is an evolutionarily conserved actin bundling protein that is localised in microspikes and filopodia underneath the plasma membrane. FSCN1 is widely expressed during embryogenesis in neural and mesenchymal tissues, but in adults, it is largely restricted to specific tissues, including the brain, endothelium and testes (Machesky and Li, 2010). In cancer cells, aberrantly expressed FSCN1 stabilises actin filaments in invasive foot structures termed invadopodia (Figure 1A). Invadopodia promote the degradation of the extracellular matrix (ECM) by coordinating the presentation of matrix metalloprotease 14 (MMP14, also known as membrane type-1 MMP (MT1 MMP)) to the tips of the protruding structure and by increasing the secretion of MMP2 and MMP9 (Murphy and Courtneidge, 2011).
Figure 1.
A schematic drawing of invadopodia and the expression profiles of FSCN1 and matrix metalloproteinase 14 (MMP14) in 140 ESCC samples. (A) FSCN1 bundles actin filaments and contributes to the stabilisation of invadopodia. CTTN (also known as cortactin), also one of the actin-binding proteins, promotes actin branching by the activation of the ARP 2/3 complex in synergy with N-WASP at the edge of the migrating cells. Invadopodia present MMP14 and secret MMP2 and MMP9. (B) A microscopic view of the immunohistochemical analysis of ESCC adjacent to the normal epithelium. The data for FSCN1 (left panel) and MMP14 (right panel) are shown ( × 20); (C) invasive front ( × 400). (D) Kaplan–Meier survival analysis of the disease-free survival in ESCC cases with positive vs negative FSCN1 staining (left panel), MMP14 staining (middle panel) and different FSCN1/MMP14 expression patterns (right panel). (E) Kaplan–Meier survival analysis of the overall survival in ESCC cases with positive vs negative FSCN1 (left panel), MMP14 (middle panel) and FSCN1/MMP14 expression patterns (right panel).
MMPs are zinc-dependent endopeptidases that not only degrade a variety of ECM molecules but also process a number of bioactive molecules. MMPs can degrade collagen, elastin, laminin, fibronectin and proteoglycan and are known to have important roles in cancer growth, migration, invasion, angiogenesis and metastasis (Chambers and Matrisian, 1997). There are two types of MMPs, secreted MMPs and membrane-anchored MMPs. MMP14 is one of the membrane-anchored MMPs, and MMP2 and MMP9 are secreted MMPs. MMP14 has a central role in tumour invasion and not only degrades the ECM itself (Zarrabi et al, 2011) but also promotes the secretion of proMMP2 and proMMP9 by cancer-associated fibroblasts and activates them (Egawa et al, 2006). MMP14 cleaves other functional proteins such as CD44, the integrin v chain, low-density lipoprotein receptor-related protein, interleukin 8 and pro-tumour necrosis factor. Thus the proteolytic activity of MMP14 contributes to the maintenance of the tumour microenvironment and promotes tumour growth and invasion (Egawa et al, 2006). In addition, recent studies have revealed that MMP14 regulates both cell growth and motility by a non-proteolytic mechanism (D'Alessio et al, 2008; Sakamoto and Seiki, 2009).
The effect of FSCN1 on cell invasiveness occurs as a result of its cooperation with MMPs, such as MMP2 and MMP9, in several cancers (Xie et al, 2005; Onodera et al, 2009; Ozerhan et al, 2010; Al-Alwan et al, 2011; Hayashi et al, 2011; Hanker et al, 2013). However, secreted MMPs are not crucial for basement membrane invasion, whereas MMP14 is (Sabeh et al, 2004; Hotary et al, 2006; Li et al, 2008; Poincloux et al, 2009). There are no previous reports investigating the correlation between FSCN1 and MMP14. We herein show the significance of FSCN1 and MMP14 expression in ESCC specimens and demonstrate that these molecules are regulated by one microRNA. Moreover, this microRNA, miR-133a, exerts its tumour-suppressive activities by regulating these molecules.
Materials and methods
Clinical ESCC specimens and ESCC cell lines
Formalin-fixed and paraffin-embedded tissue blocks were obtained from 140 patients, and frozen tissues were obtained from 84 patients. All patients underwent primary surgical treatment and were pathologically proven to have ESCC at the Chiba University Hospital, Chiba, Japan from 1997 to 2011. Written consent for tissue donation was obtained from each patient. The protocol was approved by the Institutional Review Board of Chiba University.
The TE2 human ESCC cell lines were provided by the Cell Resource Center for Biomedical Research Institute of Development, Aging and Cancer, Tohoku University, Sendai, Japan. T.Tn human ESCC cell lines were provided by the Japanese Cancer Research Resources Bank. All cells were cultured in Dulbecco's modified Eagle's medium nutrient mixture (DMEM; Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum at 37 °C in a 5% CO2 atmosphere.
Western blot analysis
Anti-human MMP14 rabbit polyclonal IgG (1 : 250; Millipore, Temecula, CA, USA; Cat. No. AB6004) and anti-human FSCN1 mouse monoclonal immunoglobulin (IG) G (1 : 100; Dako, Tokyo, Japan; code: M3567) were used as primary antibodies. The western blot analysis using MMP14-transfected A431 cells as a positive control, T.Tn control cell and two MMP14-silenced cells revealed that the MMP14 antibody recognised MMP14, and no non-specific bands were observed. In addition, a western blot analysis of T.Tn control cells and two FSCN1-silenced cells was performed and revealed that the FSCN1 antibody recognised FSCN1, and no non-specific bands were observed.
The cells were harvested and lysed with RIPA (Radio-Immunoprecipitation Assay) buffer 72 h after transfection. The western blot analysis was performed on the XV PANTERA Gel 7.5–15% (DRC, Tokyo, Japan). The proteins in the gels were transferred electrophoretically onto nitrocellulose membranes. The membranes were incubated with primary antibodies against FSCN1, MMP14 and β-actin (1 : 1000; Abcam Limited, Cambridge, UK; clone No. mAbcam 8226). The expression of each protein was detected using secondary antibodies conjugated to peroxidase-labelled polymers. Bands were visualised using the Amersham ECL Prime Western blotting reagent (GE Healthcare Life Sciences, Uppsala, Sweden). Densitometry was done using the ImageJ software program (NIH, Bethesda, MD, USA).
Immunohistochemistry
The sections were mechanically deparaffinised and incubated in Target Retrieval Solution (Dako, Carpinteria, CA, USA) at 95 °C for 40 min. After the endogenous peroxidase activity was blocked with methanol containing 3% hydrogen peroxide (Dako) for 30 min, the tissue sections were incubated with FSCN1 antibody or MMP14 antibody at 4 °C overnight. The sections were then incubated in secondary antibody at 37 °C for 60 min. Subsequently, the sections were counterstained with hematoxylin for 1 min. The evaluation of immunohistochemical (IHC) staining was done as described previously (Usui et al, 2012).
Fluorescence microscopy
Cells cultured on coverslips were fixed in 4% paraformaldehyde for 10 min and permeabilised with 0.1% Triton X-100 for 5 min. After blocking with 10% FBS/PBS, the cells were incubated with anti-human FSCN1 antibody at 37 °C for 60 min. Then, cells were incubated with Alexa Fluor 532 gout anti-mouse IgG (Invitrogen, Carlsbad, CA, USA) as secondary antibodies and Acti-stain 488 Fluorescent Phalloidin (Cytoskeleton, Denver, CO, USA) for F-actin at 37 °C for 60 min. ProLong Gold antifade reagent with DAPI (Invitrogen) was used for the counterstaining of DNA and for the antifade reagent.
microRNA target prediction
miRNA targets were predicted by the algorithms of the miRWalk online prediction software program (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/). The miRwalk miRNA target prediction was performed using 3′UTRs incorporating eight other established miRNA-target prediction programs (Diana-microT, miRanda, miRDB, PICTAR, PITAm, RNA22, RNAhybrid, Targetscan) (Dweep et al, 2011).
Target site inhibition assays
The miScript Target Protector for the miR-133a-binding site in the 3′UTR of MMP14 mRNA (target-binding site sequence provided: 5′-CCAGAGGGGGCAGGAGGGGACCAAAAAGGAAAATGAGGAC-3′) and Negative control miScript Target Protector with no homology to any known mammalian gene (Qiagen, Hilden, Germany) was used. The miScript Target Protector is a single-stranded, modified RNA sequence that specially interferes with the interaction of a miRNA with a single target, while leaving the regulation of other targets of the same miRNA unaffected. Mature miRNA molecules (mirVana miRNA hsa-miR-133a mimic; assay ID: MH10413, Applied Biosystems, Foster City, CA, USA) and miScript Target Protector or Negative control miScript Target Protector were co-transfected into ESCC cell lines. After 72 h of transfection, total RNA was isolated.
Transfection of the small interfering RNAs, microRNA mimic and microRNA inhibitor
The small interfering RNA (siRNA) sequences (Stealth RNAi siRNA, Invitrogen) targeting FSCN1 (siFSCN1-1: Cat. No. HSS186009, siFSCN1-2: Cat. No. HSS110044), MMP14 (siMMP14-1: Cat. No. HSS106639, siMMP14-2: Cat. No. HSS106640), the negative control (Negative Control Duplex medium GC Duplex No. 2: Cat. No. 12935-112), the miR-133a mimic (mirVana miRNA Mimic, assay ID MC10413) and the negative control (mirVana miRNA Mimic Negative Control No. 1, Applied Biosystems), the miR-133a inhibitor (mirVana miRNA Inhibitor, assay ID MH10413) and the negative control (mirVana miRNA Inhibitor Negative Control No. 1, Applied Biosystems) were transfected into cell lines by electroporation using the Neon Transfection System (Invitrogen).
Quantitative reverse transcription PCR (qRT–PCR)
Total RNA was extracted from frozen surgical specimens and cell lines using Trizol reagent (Invitrogen) and reverse transcribed into cDNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real-time PCR was done using the SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) with the following primers (PCReady PCR and Sequencing Primers, Operon, Biotechnology, Tokyo, Japan): FSCN1, 5′-CAACGATGGCGCCTACAA-3′ and 5′-CCTTGTTATAGTCGCAGAACTCGAA-3′ and MMP14, 5′-AAGAGGAGAAGAGCAAACAG-3′ and 5′-CGGTAGGCACTGAACTTG-3′. ACTA1 was used as an internal control, and the sequences used were 5′-CCTTCATCGGTATGGAGTC-3′ and 5′-GTTGGCATACAGGTCCTT-3′.
The miR-133a expression levels (assay ID: 002246) were analysed by TaqMan quantitative real-time PCR (TaqMan MicroRNA Assay, Applied Biosystems) and normalised to the RNU6B level (assay ID: 001093). All reactions were performed in triplicate.
Cell proliferation, migration and invasion assays in ESCC cell lines
After transfection of siFSCN1, siMMP14 or the miR-133a mimic, cells were plated in 96-well plates at 5 × 103 cells per well in 100 μl of medium containing 10% FBS. After 120 h, the cell proliferation was assessed with the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). Triplicate wells were measured for cell viability in each treatment group. Cell motility was determined using a micropore chamber assay, and the cell invasion was determined using a Matrigel-coated (BD Biosciences, Bedford, MA, USA) and collagen I-coated micropore membrane filter with 8 μm pores. The collagen I-coated chambers were prepared as described previously (Wang et al, 2010).
In brief, 5 × 104 cells were seeded onto the top chamber of a 24-well micropore polycarbonate membrane filter with 8 μm pores (BD Biosciences), and the bottom chamber was filled with DMEM containing 10% FBS as a chemoattractant. After 24 h of incubation, the membranes were fixed and stained by the Diff-Quik reagent (International Reagents, Kobe, Japan), and the cells on the upper surface were carefully removed with a cotton swab. Cell migration/invasion was quantified by counting the average number of migrated/invaded cells in four random high-powered fields per filter.
Statistical analyses
The categorical variables of the 140 clinical samples examined by IHC were compared by the chi-square test. The survival analysis using the Kaplan–Meier method was performed by the log-rank test. The relationship between two variables and numerical values obtained by real-time RT–PCR were analysed using t-tests. The relationships among miR-133a expression, FSCN1 expression and MMP14 expression were analysed using the Spearman's rank correlation. Statistical significance was defined as P<0.05. All statistical analyses were performed using the JMP statistical package (SAS Institute, Cary, NC, USA).
Results
Expression of two invadopodia-related proteins, FSCN1 and MMP14, in clinical ESCC specimens
Neither FSCN1 nor MMP14 was detected in the normal esophageal epithelium by IHC staining. Positive staining for both FSCN1 and MMP14 was apparent in cancer cells, especially in the cytoplasm, and strong staining was observed at the invasive front of tumours (Figure 1B and C).
Relationship between FSCN1 or MMP14 expression and the clinicopathological features
Among the 140 ESCC samples, 82 (58.6%) were positive for FSCN1 and 68 (48.6%) were positive for MMP14. Patients with positive FSCN1 showed significantly higher T stages (P<0.0005), higher clinical stages (P<0.0005) and a higher rate of recurrence (P<0.005 by the chi-square test), and patients with positive MMP14 tumours showed significantly higher T stages (P<0.05), higher N stages (P<0.005), higher clinical stages (P<0.001), a higher level of lymphatic invasion (P<0.05), a higher level of vascular invasion (P<0.05) and higher rates of recurrence (P<0.0001) (Table 1). Both the FSCN1-positive and MMP14-positive patients had a significantly shorter disease-free survival rate (DFS) (P=0.0010 and P<0.0001, respectively) and OS (P<0.0001 and P<0.0001, respectively) (Figure 1D and E). The 5-year survival rates were 79.9% vs 43.8% for those with no FSCN1 staining and positive FSCN1 staining, respectively. The 5-year survival rates were 79.4% and 36.9% for those without and with MMP14 expression, respectively.
Table 1. Relationship between the FSCN1 and MMP14 expression and clinicopathological features.
|
FSCN1 expression |
MMP14 expression |
|||||
|---|---|---|---|---|---|---|
| − (n=58) | + (n=82) | P-value | − (n=72) | + (n=68) | P-value | |
|
Age, years | ||||||
| <65 (n=81) | 37 | 44 | 42 | 39 | ||
| ⩾65 (n=59) |
21 |
38 |
0.23 |
30 |
29 |
0.91 |
|
Gender | ||||||
| Male (n=121) | 50 | 7 | 63 | 57 | ||
| Female (n=20) |
8 |
12 |
0.89 |
9 |
11 |
0.70 |
|
Tumour depth | ||||||
| Tis, T1 (n=60) | 36 | 24 | 38 | 22 | ||
| T2, T3, T4 (n=80) |
22 |
58 |
0.00011 |
34 |
46 |
0.015 |
|
Regional lymph nodes | ||||||
| N0 (n=66) | 33 | 33 | 43 | 23 | ||
| N1 (n=74) |
25 |
49 |
0.052 |
29 |
45 |
0.0022 |
|
Stage | ||||||
| stage0, I (n=35) | 24 | 11 | 27 | 8 | ||
| stageII, III, IV (n=105) |
34 |
71 |
0.00017 |
45 |
60 |
0.00090 |
|
Lymphatic invasion | ||||||
| ly0 (n=51) | 25 | 26 | 33 | 18 | ||
| ly1, 2, 3 (n=89) |
33 |
56 |
0.17 |
39 |
50 |
0.017 |
|
Vacular invasion | ||||||
| v0 (n=57) | 28 | 29 | 36 | 21 | ||
| v1, 2, 3 (n=83) |
30 |
53 |
0.13 |
36 |
47 |
0.021 |
|
Histology | ||||||
| Well, mod (n=111) | 45 | 66 | 59 | 52 | ||
| Poor (n=29) |
13 |
16 |
0.68 |
13 |
16 |
0.42 |
|
Recurrence | ||||||
| Recurrence (n=56) | 14 | 42 | 16 | 40 | ||
| No recurence (n=84) | 44 | 40 | 0.0013 | 56 | 28 | 0.000010 |
Abbreviation: MMP14=matrix metalloproteinase 14.
Relationship between FSCN1 and MMP14 expression and the survival time
We classified patients into four groups: group 1 included those who were both FSCN1 and MMP14 negative (n=38, 27.1%), group 2 included those who were FSCN1 positive and MMP14 negative (n=34, 24.3%), group 3 included those who were MMP14 positive and FSCN1 negative (n=20, 14.3%) and group 4 included patients who were positive for both FSCN1 and MMP14 (n=48, 34.3%). Out of the 68 cases with positive MMP14 expression, 48 cases (70.6%) also showed positive FSCN1 expression. However, 34 out of the 72 cases with negative MMP14 staining (47.2%) showed a positive FSCN1 expression. MMP14 expression was significantly correlated with the FSCN1 expression (P=0.0050). A survival analysis showed significant differences in the DFS and OS among the four groups of patients, with the fourth group having a significantly lower survival rate compared with the other groups (Figure 1D and E). The 5-year survival rates of each group were 86.2, 71.7, 69.1 and 25.4% for groups 1, 2, 3 and 4, respectively.
A multivariable Cox proportional hazards analysis was performed to identify the independent factors associated with the OS. Positive FSCN1 staining, positive MMP14 staining, the T stage and the N stage were identified as significant independent prognostic factors (Table 2).
Table 2. Cox multivariate analysis results.
| Terms | Risk ratio (95% CI) | P-value |
|---|---|---|
| Age >65 years only |
0.682 (0.391–1.160) |
0.16 |
| Male sex |
1.564 (0.733–3.836) |
0.28 |
| T2,T3,T4 |
2.315 (1.065–5.626) |
0.045 |
| N1 |
2.088 (1.094–4.230) |
0.032 |
| Stage II,III,IV |
0.535 (0.151–1.882) |
0.33 |
| Lymphatic invasion |
1.230 (0.673–2.351) |
0.51 |
| Vascular invasion |
1.451 (0.808–2.720) |
0.23 |
| Poor tumour differentiation |
1.104 (0.567–2.002) |
0.76 |
| FSCN1-positive staining |
2.348 (1.200–4.862) |
0.016 |
| MMP14-positive staining | 2.458 (1.364–4.595) | 0.0035 |
Abbreviations: CI=confidence interval; MMP14=matrix metalloproteinase 14.
Effects of transfection of siRNAs targeting FSCN1 mRNA and MMP14 mRNA on ESCC cell lines
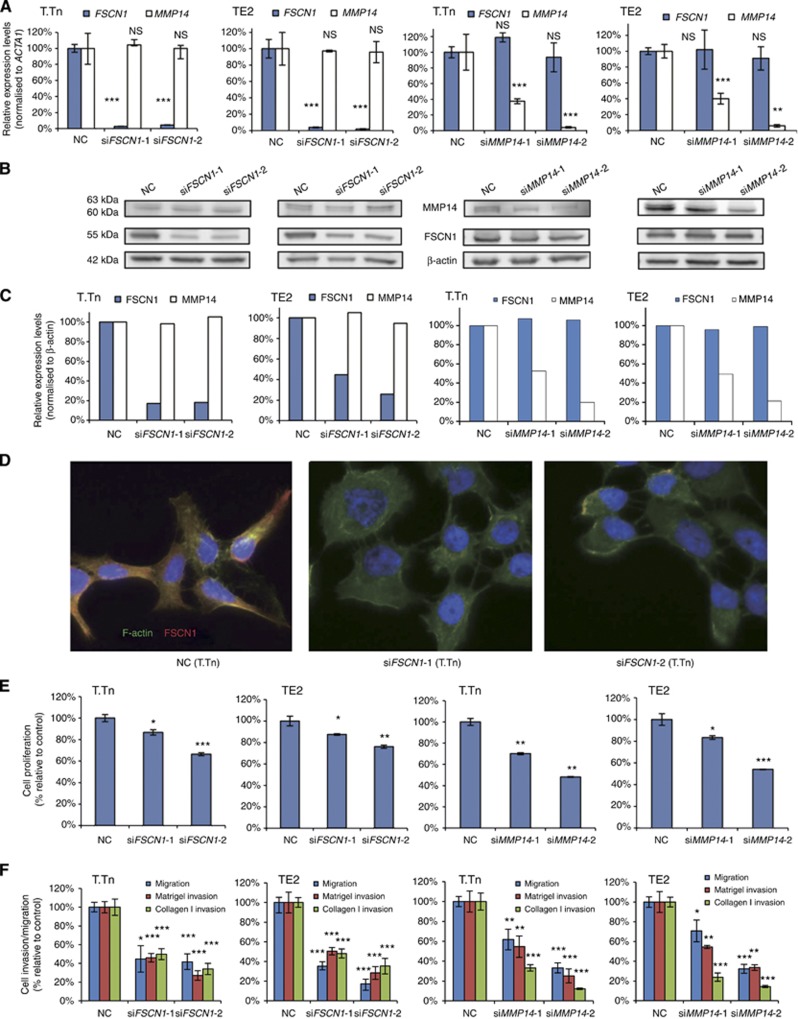
The introduction of each of the four siRNA sequences (siFSCN1-1, siFSCN1-2, siMMP14-1 and siMMP14-2) into T.Tn and TE2 cells led to a significant decrease of the target mRNA and a remarkable decrease in the amount of the target protein. However, the knockdown of FSCN1 did not lead to a decrease in MMP14 mRNA or protein in the two cell lines. Similarly, the knockdown of MMP14 did not affect the expression of FSCN1 mRNA or protein (Figure 2A–C).
Figure 2.
The effects of FSCN1 and MMP14 knockdown on the mRNA levels, protein levels, morphological changes, proliferation and migration/invasion activities in ESCC cell lines 72 h after the transfection. Two siFSCN1, siFSCN1-1 and siFSCN1-2, and two siMMP14, siMMP14-1 and siMMP14-2, were transfected into ESCC cell lines (T.Tn and TE2 cells). (A) Results of the evaluation of the mRNA expression levels by real-time RT–PCR. (B and C) The results of an evaluation of the protein expression levels determined by a western blot analysis. The MMP14 protein was seen as two bands, 63 kDa for the latent form and 60 kDa for the active form. (D) Morphological changes of T.Tn cells. The cells were stained with an anti-FSCN1 antibody (red), phalloidin (green) and DAPI (blue). (E) Results of the proliferation assay. (F) Results of the migration/Matrigel invasion/collagen I invasion assay. Abbreviations: NC, negative control, NS, not significant. *P<0.05, **P<0.01, ***P<0.005. The full colour version of this figure is available at British Journal of Cancer online.
Immunofluorescent staining of T.Tn and TE2 cells demonstrated that siFSCN1 cells lost their normal spindle shape as seen in negative controls and also exhibited a more rounded morphology (Figure 2D, the data for TE2 cells not shown). However, the siMMP14-transfected cells did not show morphological changes (data not shown).
In the proliferation assays, transfection of each of the four siRNAs led to a significant decrease in cell growth 120 h after transfection (Figure 2E).
In the migration assay, the Matrigel invasion assay and the collagen I invasion assay, the rates of penetration of cells through the membrane were significantly decreased by the transfection of the each of the siRNAs. There was no significant difference between the number of siFSCN1-transfected cells that penetrated the Matrigel-coated membrane and that of the cells that penetrated the collagen I-coated membrane. However, the rate of siMMP14-transfected cells that penetrated the collagen I membrane was significantly lower than that of the cells that penetrated the Matrigel-coated membrane (33.3% vs 54.6% of the negative control (siMMP14-1, P=0.0395) and 12.3% vs 25.2% (siMMP14-2, P=0.0362) in T.Tn cells and 33.6% vs 54.4% (siMMP14-1, P=0.0027) and 14.3% vs 23.9% (siMMP14-2, P=0.0026) in TE2 cells (Figure 2F)).
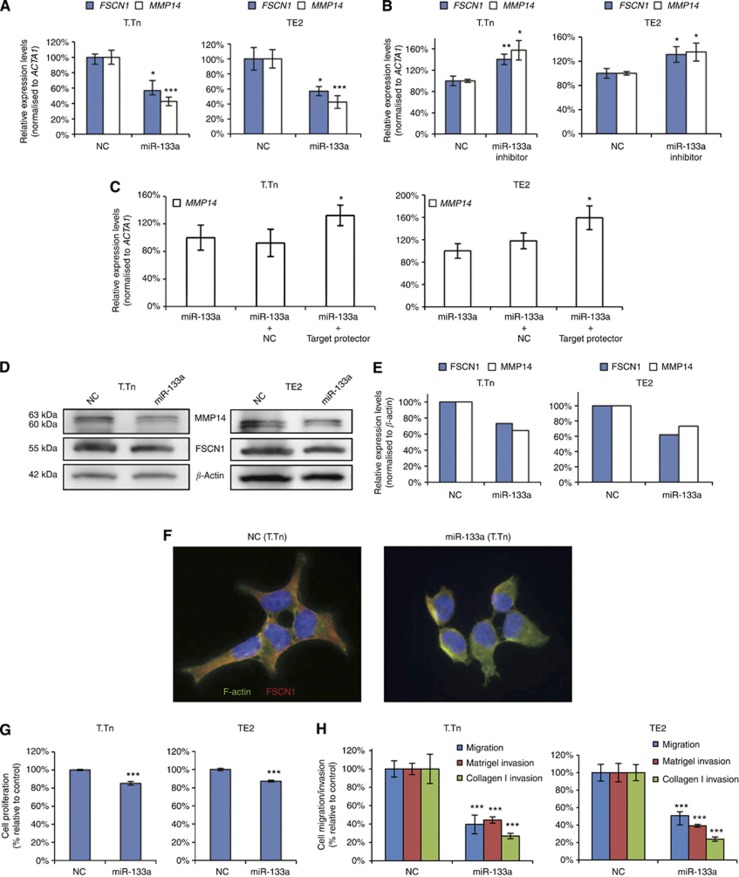
Effects of miR-133a mimic, miR-133a inhibitor and Target Protector on ESCC cell lines
We previously reported that miR-133a regulates FSCN1 (Kano et al, 2010). A bioinformatic analysis performed using the ‘miRwalk' method identified one conserved sequence site for miR-133a in the 3′ UTR of MMP14.
The introduction of a miR-133a mimic into T.Tn and TE2 cells led to significant decreases in both the FSCN1 and MMP14 mRNA levels (56.8% and 42.6% of the negative control in T.Tn cells and 57.0% and 42.5% in the TE2 cells; Figure 3A). On the other hand, the introduction of a miR-133a inhibitor into the two cell lines led to significant increases in both the FSCN1 mRNA and MMP14 mRNA levels (140.4% and 157.6% of the negative control in T.Tn cells and 131.0% and 135.0% in TE2 cells; Figure 3B).
Figure 3.
The effects of the miR-133a mimic, miR-133a inhibitor and Target Protector transfection on the mRNA and protein levels, and the effects of miR-133a mimic transfection on the proliferation and migration/invasion activities in ESCC cell lines 72 h after transfection. (A) The effects of miR-133a mimic transfection on the mRNA levels as evaluated by real-time RT–PCR. (B) The effects on the mRNA levels following miR-133a inhibitor transfection, as evaluated by real-time RT–PCR. (C) The effects of the co-transfection of the miR-133a mimic and Target Protector on the mRNA levels as evaluated by real-time RT–PCR. (D and E) The effects of the transfection of the miR-133a mimic on the protein levels as evaluated by a western blot analysis. (F) Morphological changes of T.Tn cells. (G) Results of the proliferation assay. (H) Results of the migration/Matrigel invasion/collagen I invasion assay. Abbreviation: NC=negative control. *P<0.05, **P<0.01, ***P<0.005.
Then, target site inhibition assays were performed to investigate the interaction between miR-133a and MMP14 mRNA. The two cell lines were co-transfected with the miR-133a mimic and miScript Target Protector designed to interfere with the interaction of miR-133a with MMP14 mRNA by protecting the miR-133a-binding site of the MMP14 mRNA. The results showed a significantly higher expression of MMP14 mRNA than that in the cells transfected with the miR-133a and negative control miScript Target Protector (143.4% of the negative control in T.Tn cells and 135.0% in TE2 cells; Figure 3C). These results suggested that miR-133a directly regulates MMP14 mRNA. The introduction of a miR-133a mimic into T.Tn and TE2 cells led to a decrease in the FSCN1 and MMP14 protein levels (Figure 3D and E).
Immunofluorescent staining of the cells demonstrated that miR-133a-transfected cells lost their normal spindle shape as seen in negative controls and exhibited a more rounded morphology (Figure 3F, the data for TE2 cells not shown).
In the proliferation assay, there was a small but significant decrease in cell growth (85.3% (P=0.0027) of negative control in T.Tn and 87.2% (P<0.001) in TE2 cells) (Figure 3G). There were also significant decreases in the migration, the Matrigel invasion and the collagen I invasion. As was observed in the siMMP14-transfected cells, the rate of miR-133a-transfected cells that penetrated the collagen I-coated membrane was significantly lower than that of the cells that penetrated the Matrigel-coated membrane (27.9% vs 44.2% of the negative control (P=0.00390) in T.Tn cells and 24.0% vs 39.2% (P=0.0342) in TE2 cells (Figure 3H)).
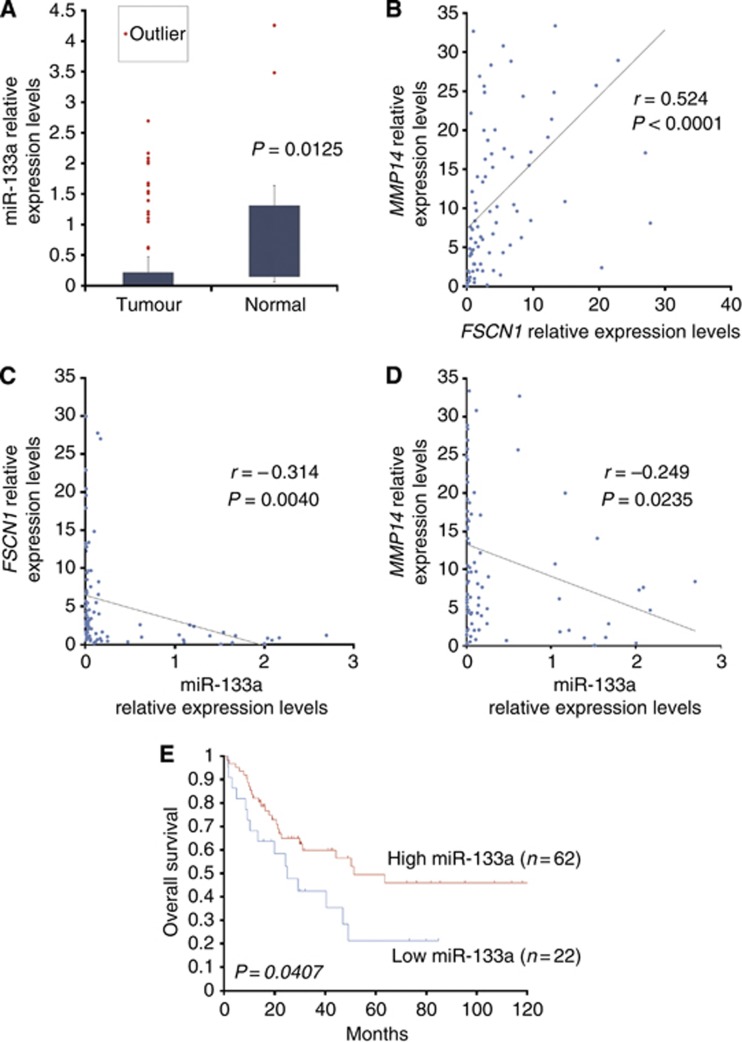
Relationships between miR-133a, FSCN1 and MMP14 mRNA expression in 84 ESCC clinical specimens
Total RNA was isolated from 84 matched non-cancerous esophageal epithelia and ESCC tissues from which the miR-133a, FSCN1 and MMP14 levels were determined by real-time PCR. In all the 84 matched normal and tumour specimens, the expression levels of miR-133a were significantly lower in the tumour tissues than in the normal tissues (P=0.0125; Figure 4A). The Spearman's rank correlation test revealed a significant correlation between the expression of FSCN1 and MMP14 (r=0.524, P<0.0001; Figure 4B) and an inverse correlation between miR-133a and FSCN1 (r=−0.314, P=0.0040, Figure 4C) and between miR-133a and MMP14 (r=−0.249, P=0.0235, Figure 4D).
Figure 4.
The expression profiles of FSCN1 and MMP14 in 84 ESCC samples. (A) The expression levels of miR-133a in tumour tissues and normal tissues. (B) Correlation between FSCN1 and MMP14. (C) Correlation between miR-133a and FSCN1. (D) Correlation between miR-133a and MMP14. (E) Results of the Kaplan–Meier survival analysis of the OS in the high miR-133a group vs the low miR-133a group.
We divided the patients into two groups, high and low miR-133a groups, based on the relative miR-133a expression levels normalised to that of RNU6B. The cutoff value was set at 0.012. A survival analysis using the Kaplan–Meier method and the log-rank test showed a significant decrease in the OS in the low miR-133a group (P=0.0407, Figure 4E), and the 5-year survival rate was 21.2% (vs 49.4% in the high miR-133a group).
Discussion
These findings showed that FSCN1 and MMP14, which are both located in the invadopodia of cancer cells, contribute to tumour growth, migration and invasion, and the overexpression of each of them is an independent poor prognostic factor for ESCC. The co-expression of these two molecules correlated with the poorest 5-year survival out of all of the factors examined. miR-133a also regulates both of these invadopodia-related genes and exerts a tumour-suppressive activity in ESCC.
Recently, Chen et al (2010) reported that migrastatin analogues exert their tumour-suppressive activity by inhibiting FSCN1. Migrastatin is a natural product secreted by Streptomyces (Nakae et al, 2000), and synthetic migrastatin analogues are potent inhibitors of tumour cell migration, invasion and tumour metastasis (Takemoto et al, 2001; Lecomte et al, 2011).
Xie et al (2005) reported that the effect of FSCN1 on tumour cell invasion depended on the activation of MMP2 and MMP9. Onodera et al (2009) showed that tumour necrosis factor-α (TNF-α) induced the overproduction of FSCN1, which in turn upregulated MMP9 expression in two cholangiocarcinoma cell lines. Al-Alwan et al (2011) reported that FSCN1 enhanced the urokinase-type plasminogen activator secretion after TNF-α activation via the enhancement of NF-kappa B activity, which in turn can activate MMP2 and MMP9. However, Sabeh and other researchers reported that secreted MMPs are not crucial for basement membrane invasion, whereas MMP14 is (Sabeh et al, 2004; Hotary et al, 2006; Li et al, 2008; Poincloux et al, 2009). Additionally, Onodera et al (2009) also reported that FSCN1 siRNA did not influence the expression level of MMP9 under the normal culture conditions in cholangiocarcinoma cell lines, and similarly, our present study shows that FSCN1 knockdown does not affect the expression level of MMP14.
In the siFSCN1-transfected cells, the inhibition of Matrigel invasion and collagen I invasion paralleled that of the migration. In contrast, in the siMMP14- or miR-133a-transfected cells the inhibition of collagen I invasion was stronger than that of Matrigel invasion, which still paralleled that of migration. These results are consistent with a previous report that collagen I, but not Matrigel matrices, provide an MMP-dependent barrier function for the basement membrane (Sodek et al, 2008) and reflect our finding that FSCN1 knockdown does not affect the expression level of MMP14. In addition, although some studies have reported that inhibition of MMP14 did not affect the 2D proliferation, D'Alessio et al (2008) reported that MMP14 contributes to cell migration and proliferation in vitro (in the absence of ECM) through a proteolysis-independent mechanism. The finding of our study that the introduction of siMMP14 led to a decrease of cell growth and migration is consistent with this previous report.
Accordingly, these results suggest that tumour-suppressive effect of FSCN1 inhibitors, such as migrastatin analogues, may be limited.
Although inhibitors of MMPs, especially of MMP14, seem to be a strong candidate tumour suppressor, there are currently no synthetic or biological MMP inhibitors in clinical trials for cancer (Devy and Dransfield, 2011). MMP-based intervention strategies using compounds containing zinc-chelating groups, such as hydroxamates, have met with limited clinical success and had severe side effects. These are assumed to be due to the poor selectivity of these broad-spectrum compounds (Devy et al, 2009). The catalytic domains of all MMPs share high amino-acid similarity, and their active sites are extensively conserved, thus differentiating between MMPs with small molecule substrates or inhibitors is extremely difficult (Zarrabi et al, 2011).
Devy et al (2009) reported that antibody-based biotherapeutic agents may fulfill the selectivity, and they identified a highly selective antibody-based inhibitor of MMP14 (DX-2400). However, MMP14 is necessary for normal physiological processes, such as wound healing, the maintenance of bone (Holmbeck et al, 2005) and normal angiogenesis, so even selective inhibition of MMP14 might cause severe side effects.
We demonstrated that the prognosis of patients with both FSCN1- and MMP14-positive IHC staining was significantly worse than that of patients whose tumours were positive for only one of the proteins.
Because a FSCN1 inhibitor might not exert sufficient tumour-suppressive activity, and a MMP14 inhibitor might cause adverse effects, an upstream regulator of both FSCN1 and MMP14, not an inhibitor of either of them, could serve as an ideal tumour-suppressive agent. We have previously reported that miR-133a regulates FSCN1 mRNA (Kano et al, 2010). We found that miR-133a also regulates MMP14 mRNA, and demonstrated an inverse correlation between miR-133a and FSCN1 and between miR-133a and MMP14, and also demonstrated miR-133a to have tumour-suppressive effects. MicroRNAs are endogenous small (⩽22 nucleotide) ‘non-messenger' RNAs that target mRNAs for cleavage or translational repression (Ambros, 2004; Bartel, 2004). In animals, miRNAs are imprecisely complementary to their mRNA targets. The impreciseness makes it possible for one miRNA to target several mRNAs. A single mRNA can also be targeted by several miRNAs (Gomes et al, 2013). Many recent experimental and clinical studies have shown that the aberrant expression of miRNAs is associated with tumour aggressiveness. For example, it has been shown that some miRNAs can function as tumour promoters (oncomirs) and other miRNAs function as tumour suppressors (anti-oncomirs) (Zhang et al, 2007; Ghelani et al, 2012).
Promoting the activity of anti-oncomirs using the delivery of miRNA mimics could be a novel therapeutic option for cancer (Ghelani et al, 2012). As most normal cells already express the anti-oncomirs, the delivery of miRNA mimics to the cells is unlikely to induce adverse effects, as the cellular pathways affected by the mimic have already been activated or inactivated by the endogenous miRNA. However, it is not easy to deliver these agents successfully, because it is difficult for them to penetrate the cellular lipid bilayer membrane as a result of their negative charge and because they are easily degraded by RNase in vivo. Recently, numerous trials targeting miRNA have been conducted by researchers and biopharmaceutical companies using cationic lipids, polymers, nanoparticles and so on as delivery reagents for more stable, safer and more effective drug delivery systems (Uchino et al, 2013), and some have yielded good results without major adverse effects.
In conclusion, the combination of FSCN1 and MMP14 is a poor prognostic factor, and miR-133a, which directly regulates both FSCN1 and MMP14, can serve as a strong tumour suppressor of ESCC.
Acknowledgments
We thank Dr Motoharu Seiki and Dr Naohiko Koshikawa (The Institute of Medical Science, the University of Tokyo, Tokyo, Japan) for the generous gift of the MMP14-transfected A431 cells. This study was financially supported by a Grant-Aid for Scientific Research (JSPS KAKENHI Grant no. 25861175) from the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Al-Alwan M, Olabi S, Ghebeh H, Barhoush E, Tulbah A, Al-Tweigeri T, Ajarim D, Adra C. Fascin is a key regulator of breast cancer invasion that acts via the modification of metastasis-associated molecules. PloS One. 2011;6 (11:e27339. doi: 10.1371/journal.pone.0027339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431 (7006:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116 (2:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Chambers AF, Matrisian LM. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89 (17:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- Chen L, Yang S, Jakoncic J, Zhang JJ, Huang XY. Migrastatin analogues target fascin to block tumour metastasis. Nature. 2010;464 (7291:1062–1066. doi: 10.1038/nature08978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio S, Ferrari G, Cinnante K, Scheerer W, Galloway AC, Roses DF, Rozanov DV, Remacle AG, Oh ES, Shiryaev SA, Strongin AY, Pintucci G, Mignatti P. Tissue inhibitor of metalloproteinases-2 binding to membrane-type 1 matrix metalloproteinase induces MAPK activation and cell growth by a non-proteolytic mechanism. J Biol Chem. 2008;283 (1:87–99. doi: 10.1074/jbc.M705492200. [DOI] [PubMed] [Google Scholar]
- Devy L, Dransfield DT. New strategies for the next generation of matrix-metalloproteinase inhibitors: selectively targeting membrane-anchored MMPs with therapeutic antibodies. Biochem Res Int. 2011;2011:191670. doi: 10.1155/2011/191670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devy L, Huang L, Naa L, Yanamandra N, Pieters H, Frans N, Chang E, Tao Q, Vanhove M, Lejeune A, van Gool R, Sexton DJ, Kuang G, Rank D, Hogan S, Pazmany C, Ma YL, Schoonbroodt S, Nixon AE, Ladner RC, Hoet R, Henderikx P, Tenhoor C, Rabbani SA, Valentino ML, Wood CR, Dransfield DT. Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer Res. 2009;69 (4:1517–1526. doi: 10.1158/0008-5472.CAN-08-3255. [DOI] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44 (5:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Egawa N, Koshikawa N, Tomari T, Nabeshima K, Isobe T, Seiki M. Membrane type 1 matrix metalloproteinase (MT1-MMP/MMP-14) cleaves and releases a 22-kDa extracellular matrix metalloproteinase inducer (EMMPRIN) fragment from tumor cells. J Biol Chem. 2006;281 (49:37576–37585. doi: 10.1074/jbc.M606993200. [DOI] [PubMed] [Google Scholar]
- Ghelani HS, Rachchh MA, Gokani RH. MicroRNAs as newer therapeutic targets: a big hope from a tiny player. J Pharmacol Pharmacother. 2012;3 (3:217–227. doi: 10.4103/0976-500X.99416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CP, Cho JH, Hood L, Franco OL, Pereira RW, Wang K. A review of computational tools in microRNA discovery. Front Genet. 2013;4:81. doi: 10.3389/fgene.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanker LC, Karn T, Holtrich U, Graeser M, Becker S, Reinhard J, Ruckhaberle E, Gevensleben H, Rody A. Prognostic impact of fascin-1 (FSCN1) in epithelial ovarian cancer. Anticancer Res. 2013;33 (2:371–377. [PubMed] [Google Scholar]
- Hayashi Y, Osanai M, Lee GH. Fascin-1 expression correlates with repression of E-cadherin expression in hepatocellular carcinoma cells and augments their invasiveness in combination with matrix metalloproteinases. Cancer Sci. 2011;102 (6:1228–1235. doi: 10.1111/j.1349-7006.2011.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Pidoux I, Inoue S, Billinghurst RC, Wu W, Chrysovergis K, Yamada S, Birkedal-Hansen H, Poole AR. The metalloproteinase MT1-MMP is required for normal development and maintenance of osteocyte processes in bone. J Cell Sci. 2005;118 (Pt 1:147–156. doi: 10.1242/jcs.01581. [DOI] [PubMed] [Google Scholar]
- Hotary K, Li XY, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20 (19:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer J. 2010;127 (12:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- Lecomte N, Njardarson JT, Nagorny P, Yang G, Downey R, Ouerfelli O, Moore MA, Danishefsky SJ. Emergence of potent inhibitors of metastasis in lung cancer via syntheses based on migrastatin. Proc Natl Acad Sci USA. 2011;108 (37:15074–15078. doi: 10.1073/pnas.1015247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Ota I, Yana I, Sabeh F, Weiss SJ. Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol Biol Cell. 2008;19 (8:3221–3233. doi: 10.1091/mbc.E08-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Li A. Fascin: invasive filopodia promoting metastasis. Communicative Integr Biol. 2010;3 (3:263–270. doi: 10.4161/cib.3.3.11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DA, Courtneidge SA. The 'ins' and 'outs' of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12 (7:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae K, Yoshimoto Y, Sawa T, Homma Y, Hamada M, Takeuchi T, Imoto M. Migrastatin, a new inhibitor of tumor cell migration from Streptomyces sp. MK929-43F1. Taxonomy, fermentation, isolation and biological activities. J Antibiot. 2000;53 (10:1130–1136. doi: 10.7164/antibiotics.53.1130. [DOI] [PubMed] [Google Scholar]
- Onodera M, Zen Y, Harada K, Sato Y, Ikeda H, Itatsu K, Sato H, Ohta T, Asaka M, Nakanuma Y. Fascin is involved in tumor necrosis factor-alpha-dependent production of MMP9 in cholangiocarcinoma. Lab Invest. 2009;89 (11:1261–1274. doi: 10.1038/labinvest.2009.89. [DOI] [PubMed] [Google Scholar]
- Ozerhan IH, Ersoz N, Onguru O, Ozturk M, Kurt B, Cetiner S. Fascin expression in colorectal carcinomas. Clinics (Sao Paulo) 2010;65 (2:157–164. doi: 10.1590/S1807-59322010000200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381 (9864:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- Poincloux R, Lizarraga F, Chavrier P. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J Cell Sci. 2009;122 (Pt 17:3015–3024. doi: 10.1242/jcs.034561. [DOI] [PubMed] [Google Scholar]
- Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, Allen E, Chung D, Weiss SJ. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167 (4:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Seiki M. Cytoplasmic tail of MT1-MMP regulates macrophage motility independently from its protease activity. Genes Cells. 2009;14 (5:617–626. doi: 10.1111/j.1365-2443.2009.01293.x. [DOI] [PubMed] [Google Scholar]
- Sodek KL, Brown TJ, Ringuette MJ. Collagen I but not Matrigel matrices provide an MMP-dependent barrier to ovarian cancer cell penetration. BMC Cancer. 2008;8:223. doi: 10.1186/1471-2407-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto Y, Nakae K, Kawatani M, Takahashi Y, Naganawa H, Imoto M. Migrastatin, a novel 14-membered ring macrolide, inhibits anchorage-independent growth of human small cell lung carcinoma Ms-1 cells. J Antibiot. 2001;54 (12:1104–1107. doi: 10.7164/antibiotics.54.1104. [DOI] [PubMed] [Google Scholar]
- Uchino K, Ochiya T, Takeshita F. RNAi therapeutics and applications of microRNAs in cancer treatment. Jpn J Clin Oncol. 2013;43 (6:596–607. doi: 10.1093/jjco/hyt052. [DOI] [PubMed] [Google Scholar]
- Usui A, Hoshino I, Akutsu Y, Sakata H, Nishimori T, Murakami K, Kano M, Shuto K, Matsubara H. The molecular role of Fra-1 and its prognostic significance in human esophageal squamous cell carcinoma. Cancer. 2012;118 (13:3387–3396. doi: 10.1002/cncr.26652. [DOI] [PubMed] [Google Scholar]
- Wang FQ, Ariztia EV, Boyd LR, Horton FR, Smicun Y, Hetherington JA, Smith PJ, Fishman DA. Lysophosphatidic acid (LPA) effects on endometrial carcinoma in vitro proliferation, invasion, and matrix metalloproteinase activity. Gynecologic Oncol. 2010;117 (1:88–95. doi: 10.1016/j.ygyno.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Xie JJ, Xu LY, Zhang HH, Cai WJ, Mai RQ, Xie YM, Yang ZM, Niu YD, Shen ZY, Li EM. Role of fascin in the proliferation and invasiveness of esophageal carcinoma cells. Biochem Biophys Res Commun. 2005;337 (1:355–362. doi: 10.1016/j.bbrc.2005.09.055. [DOI] [PubMed] [Google Scholar]
- Zarrabi K, Dufour A, Li J, Kuscu C, Pulkoski-Gross A, Zhi J, Hu Y, Sampson NS, Zucker S, Cao J. Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer cell migration. J Biol Chem. 2011;286 (38:33167–33177. doi: 10.1074/jbc.M111.256644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302 (1:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]