Abstract
Background:
The majority of patients diagnosed with advanced epithelial ovarian carcinoma (EOC) relapse with resistant disease, and there are no biomarkers that possess clinical utility to identify or monitor these patients. This study aimed to identify secreted proteins (‘secretome') collected from human EOC cell lines that differ in their inherent platinum sensitivity.
Methods:
Secreted proteins collected from conditioned medium from ovarian cancer cell lines that vary in their sensitivity to cisplatin were digested with trypsin and analysed by liquid chromatography-tandem mass spectrometry for peptide identification.
Results:
Of the 1688 proteins identified, 16 possessed significant differential abundances (P<0.05) between the platinum-resistant and -sensitive cell lines. A number of these were verified by immunoblot, including COL11A1, which was also found to be associated with worse progression-free survival (PFS; N=723) and overall survival (OS; N=1183) as assessed from publicly available transcript expression data from ovarian cancer tumour specimens.
Conclusion:
Secretome proteomics of EOC cells resulted in the identification of a novel candidate biomarker, COL11A1. The expression level of COL11A1 correlates to worse PFS and OS, and is predicted to reside in peripheral circulation making this an attractive candidate for validation in sera as a biomarker of cisplatin resistance and poor outcome.
Keywords: secretome, proteomics, cisplatin resistance, ovarian cancer, biomarker
Epithelial ovarian carcinoma (EOC) is one of the top 10 most frequently occurring cancers in women with an estimated 22 240 new cases in the United States in 2013, 14 030 of whom are predicted to succumb to their disease (Siegel et al, 2013). Epithelial ovarian carcinoma is the most lethal gynaecological malignancy largely attributable to its typical late stage (stage III or IV) diagnosis (Heintz et al, 2006). Advanced-stage EOCs are characterised by extensive tumour heterogeneity, genomic instability and defects in p53 signalling and homologous recombination (The Cancer Genome Atlas Research Network, 2011). Current front-line therapies include cytoreductive surgery followed by adjuvant platinum- and taxane-based chemotherapy. Despite the high initial response to primary therapy, a high proportion of patients relapse with platinum-resistant disease, which represents a major hurdle in successful management of ovarian cancer patients.
Platinum-based drugs are among the most active and commonly used chemotherapy agents for solid tumors including ovarian cancer, lung cancer and colorectal cancer. Platinum drugs, such as cisplatin and carboplatin, are alkylating-like agents that adduct DNA resulting in intra- and inter-strand crosslinks that accumulate to produce overwhelming levels of DNA damage, and ultimately apoptosis. The mechanisms underlying platinum resistance are manifold and poorly understood, but likely involve reduced uptake of and increased inactivation of the drug, increased DNA repair capacity and modulation of apoptosis evasion mechanisms (Siddik, 2003; Galluzzi et al, 2012). Identification and clinical validation of surrogate biomarkers of platinum resistance would represent a major advance in ovarian cancer patient care; however, no such biomarkers have yet been identified.
Unfortunately, serum-based proteomic biomarker discovery efforts have been met with serious limitations due to the high dynamic range of serum proteins (Anderson et al, 2004; Hanash et al, 2008). These limitations prompted discovery strategies from alternative sample sources, such as conditioned media or proximal fluids (Faca et al, 2008; Gortzak-Uzan et al, 2008; Gunawardana et al, 2009; Kuk et al, 2009). This rationale is supported by the frequent observation that secreted and/or shed proteins from the tumour microenvironment, such as CA-125, are often present at higher relative concentrations in proximity to the tumour site than in peripheral circulation (Sedlaczek et al, 2002). Therefore, characterisation of secreted proteins from tumour cells may provide an ideal source to identify candidate protein biomarkers for validation in serological samples.
In this study, conditioned media was harvested from three papillary serous ovarian cancer cell lines (CaOV3, OVCAR3 and OV90) along with that from an undifferentiated EOC cell line A2780 and its isogenic, yet cisplatin-resistant daughter cell line A2780-CP20. Proteins harvested from the conditioned media from these cell lines were digested and analysed by liquid chromatography-tandem mass spectrometry (LC-MS/MS), and the relative abundances of the identified proteins were determined by spectral counting, ultimately resulting in 16 proteins that possessed significant differential abundances (Z-statistic test, P<0.05) between the platinum-resistant and -sensitive cell lines. Proteins that were elevated in the conditioned medium from platinum-resistant EOC cells were further evaluated for potential clinical utility by determining the relationship between the level of candidate biomarkers and measures of clinical outcome including progression-free survival (PFS) and overall survival (OS) in ovarian cancer patients using publicly available transcript expression data with corresponding clinical information (Ganzfried et al, 2013). Of the candidates identified from the secretome analysis, higher than median expression of collagen, type XI, alpha 1 chain (COL11A1) was associated with worse PFS and OS.
Materials and Methods
Cell lines and tissue culture conditions
The human EOC cell lines A2780 and A2780-CP20 were purchased from the European Collection of Cell Cultures (Salisbury, UK). A2780 and A2780-CP20 cells were maintained in RPMI-1640 media supplemented with 10% foetal bovine serum albumin (FBS; ATCC, Manassas, VA, USA). The human epithelial OVCAR3 cell line, established from malignant ascites of a patient with progressive ovarian adenocarcinoma and resistant to clinically relevant concentrations of adriamycin, melphalan and cisplatin, was obtained from ATCC. OVCAR3 was maintained in RPMI-1640 media with 20% FBS and 0.01 mg ml−1 human insulin (Sigma-Aldrich, St Louis, MO, USA). CaOV3 and OV90 were also obtained from ATCC and were cultured in DMEM media containing 10% FBS. All cells were cultured in monolayer using 75 cm2 tissue culture flasks and maintained at 37 °C supplemented with 5% CO2. The identities of all cell lines used were verified by short tandem-repeat testing and were confirmed to be mycoplasma-free (Universal Mycoplasma Detection Kit; ATCC, Manassas, VA, USA).
Cell proliferation assay
Cells were trypsinised with 0.25% trypsin-EDTA (ATCC) and counted with 0.4% trypan blue using a TC10 automated cell counter (Bio-Rad, Hercules, CA, USA). Cells were seeded in 96-well plates with 5000–10 000 cells per well. Once cells were attached (∼24 h post seeding), media was removed and replaced with fresh media containing cisplatin (0, 1.56, 3.13, 6.25, 12.5, 25, 50 and 100 μM). An MTS cell proliferation assay was performed 72 h post treatment using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) according to the manufacturer's instructions. Briefly, 20 μl of the CellTiter 96 Aqueous One Solution reagent was added to each well and incubated at 37 °C for 2 h. Absorbance at 490 nm was measured using the xMark microplate spectrophotometer (Bio-Rad), and the CompuSyn software (ver 3.0.1; ComboSyn, Inc., Paramus, NJ, USA) was used to obtain the half maximal inhibitory (IC50) values for each cell line.
Secretome collection and sample preparation
Ovarian cancer cells were cultured in conditions as appropriate until they were ∼70% confluent (15 × 106 cells per 75 cm2) and washed three times with 10 ml of serum- and phenol red-free RPMI-1640 media to remove residual FBS. To collect the secretome, cells were incubated with 10 ml serum- and phenol red-free RPMI-1640 media for 24 h at 37 °C supplemented with 5% CO2. For each cell secretome sample, cultured media (secretome) was pooled from three 75 cm2 tissue culture flasks and centrifuged at 500 g at 4 °C to remove any floating debris. A protease and phosphatase inhibitor cocktail (Halt, Thermo Fisher Scientific Inc., Rockford, IL, USA) and 0.1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich) were added to the secretome samples. The secretome samples were buffer-exchanged with 25 mM ammonium bicarbonate (AMB; Thermo Fisher Scientific Inc., Fair Lawn, NY, USA) and concentrated to 500 μl using 3 K MWCO Ultra Centrifugal Filters (Amicon Millipore, Billerica, MA, USA). Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific Inc., San Jose, CA, USA). Secretome samples were maintained at −80 °C before sample preparation for proteomic analysis or immunoblotting.
Ten micrograms of proteins harvested from conditioned medium from A2780 and A2780-CP20 cells were electrophoresed briefly into the stacking portion of a one-dimensional gel (4–15% Mini-PROTEAN TGX gel, Bio-Rad). The gel was stained for 10 min with Coomassie (SimplyBlue SafeStain; Invitrogen, Carlsbad, CA, USA) and the stained gel bands containing the entire ‘secreted proteome' were excised and destained in 50% acetonitrile (ACN; Sigma-Aldrich) and 25 mM AMB at ambient temperature on a vortex platform for 1 h. Gel bands were dehydrated in 100% ACN and incubated in 25 mM AMB, 10 mM dithiothreitol (Sigma-Aldrich) at 56 °C for 30 min to reduce disulphide bonds of reactive cysteine residues, followed by alkylation with 25 mM AMB and 45 mM iodoacetamide (Sigma-Aldrich) in darkness for 45 min at ambient temperature. Gel slices were dehydrated in 100% ACN and rehydrated with 20 ng μl−1 sequencing-grade modified trypsin (Promega), and 25 mM AMB on ice for 30 min. Excess trypsin was removed, gel bands were washed with 500 μl of 25 mM AMB and in-gel digestion was performed at 37 °C for 16 h. Tryptic peptides were extracted with 70% ACN and 5% formic acid (Sigma-Aldrich), lyophilised by vacuum centrifugation and resuspended in 25 mM AMB. Samples were centrifuged at 16 000 g to remove particulates, and supernatant was stored at −80 °C.
Liquid chromatography-tandem mass spectrometry
Seven replicate injections (1 μg each) of each secretome digest were analysed by nanoflow LC (Easy-nLC; Thermo Fisher Scientific Inc., San Jose, CA, USA) coupled online with an LTQ-Orbitrap Velos MS (Thermo Fisher Scientific Inc., San Jose, CA, USA). Following system equilibration, each sample was loaded onto a 2-cm reversed-phase (C18) vented pre-column (Thermo Fisher Scientific Inc., San Jose, CA, USA) at 3 μl min−1 for 6 min with mobile phase A (0.1% formic acid in water). Peptides were resolved on a 100 μm inner diameter × 360 μm outer diameter × 200-mm long capillary column (Polymicro Technologies, Phoenix, AZ, USA) slurry packed in house with 5 μm diameter, 100 Å pore size reversed phase (Magic C18 AQ, Bruker Michrom Bioresources, Auburn, CA, USA) at a constant flow rate of 200 nl min−1 by development of a linear gradient of 0.33% mobile phase B (0.1% formic acid in ACN) per min for 120 min. The column was washed for 15 min at 95% mobile phase B and then equilibrated to 100% mobile phase A before the next sample injection. The LTQ-Orbitrap Velos MS was configured to collect high resolution (R=60 000 at m/z 400) broadband mass spectra (m/z 375–1800) using the lock mass feature for the polydimethylcyclosiloxane ion generated in the electrospray process (m/z 445.12002) from which the 20 most abundant peptide molecular ions dynamically determined from the MS scan were selected for tandem MS. MS conditions were set as follows: electrospray voltage, 1.9 kV; no sheath and auxiliary gas flow; capillary temperature, 220 °C; and S-Lens RF level, 69%. The ion selection threshold for the Orbitrap (MS) was set at 1.0 × 106 with a maximum ion accumulation time of 500 ms. Tandem MS were collected in the high-pressure linear ion trap (MS/MS) with the following settings: ion threshold, 5000; minimum intensity, 3000; maximum ion accumulation time, 25 ms; and activation time, 10 ms. Dynamic exclusion (60 s) was used to minimise redundant selection of peptides for MS/MS.
Peptide identification and spectral counting
Tandem mass spectra were searched against the UniProt human protein database (downloaded on 25/10/2012, 68 109 sequences) from the European Bioinformatics Institute (http://www.ebi.ac.u/integr8/) using the Mascot Daemon (Matrix Science Inc., Boston, MA, USA). The data were searched with a precursor mass tolerance of 10 p.p.m. and a fragment ion tolerance of 0.6 Da. Cysteine carbamidomethylation (m/z 57.021464) was set as a fixed modification and methionine oxidation (m/z 15.99492) was set as a dynamic modification, and a maximum of two missed tryptic cleavages were allowed. Identified peptides were filtered using an ion score cutoff of 33 resulting in a false peptide discovery rate of <1% for all peptides identified (determined from an automatic decoy database search). Peptides whose sequence mapped to multiple protein isoforms were grouped as per the principle of parsimony (Marengo et al, 2010). Total peptide spectral matches (spectral counts) were derived by summing the results for each UniProt accession from each of the seven replicate LC-MS/MS analyses for each secretome sample, and normalisation was performed using the lowest total peptide spectral matches value across all samples in the comparison group. A value of 0.5 was added to each spectral count value before log2 transformation to enable ratio values to be calculated for proteins identified in one cell line but not another (Mcdonald, 2009).
Statistical analyses for proteomics data
Statistical analyses for the enrichment of specific proteins among differentially expressed proteins were performed using a Z-statistic test on the log2-transformed spectral count ratios. The Z-statistic test was used to calculate P-values with the minimum significance level set to P<0.05. Primary accessions, with isoform designations removed, were used when comparing EOC secretome protein identifications to other published data sets. Secretome proteins identified from the cisplatin-resistant A2780-CP20 cell line were also compared with those identified from cisplatin-sensitive A2780, CaOV3 and OVCAR3 cell lines to derive mutually significant differentially abundant proteins between the sensitive and resistant phenotypes.
Immunoblot analyses
Thirty micrograms of secretome were resolved by 4–15% mini-PROTEIN TGX gels (Bio-Rad) and transferred to PVDF membranes using Trans-Blot Turbo Transfer System (Bio-Rad) according to the manufacturer's protocol. Membranes were blocked for 1 h at ambient temperature in 5% blocking grade non-fat dry milk (Bio-Rad), 0.1% Tween-20, and 1 × tris-buffered saline. Membranes were incubated with primary antibody overnight at 4 °C with gentle rocking, washed three times for 15 min in 0.1% Tween-20, 1 × tris-buffered saline and incubated with secondary antibody for 1 h at ambient temperature with gentle rocking. Primary antibodies used were as follows: anti-matrix-degrading metalloproteinases (MMP)1 (1 : 1000, clone H00004312-B01P; Abnova, Taipei, Taiwan), anti-MMP10 (1 : 1000, clone H00004319-B01P; Abnova), anti-CTH (1 : 1000, clone HPA023300; Sigma-Aldrich) and anti-Col11A1 (1 : 500, clone NBP1–55803; Novus Biologicals, Littleton, CO, USA). The secondary antibodies were goat anti-rabbit IgG HRP-linked (1 : 2000, clone 7074S, Cell Signaling, Beverly, MA, USA) and goat anti-mouse IgG (H+L)-HRP (1 : 2000, clone 31430, Thermo Fisher Scientific Inc., San Jose, CA, USA). Membranes were washed three times for 15 min in 0.1% Tween-20, 1 × tris-buffered saline and incubated with SuperSignal West Pico or Dura Chemiluminescent Substrate (Thermo Scientific, San Jose, CA, USA) for 5 min. Images were acquired using a ChemiDoc XRS+ system (Bio-Rad). Membranes were stained with Ponceau S (Sigma-Aldrich) to verify loading equivalency.
Statistical analysis of public gene expression and patient outcome data
CuratedOvarianData, a manually curated data collection by Ganzfried et al (2013), were downloaded on 23 April 2013 from the web site (http://bcb.dfci.harvard.edu/ovariancancer) using the statistical computing language ‘R' with the Bioconductor package. It included 23 data sets from 11 platforms and provided curated gene expression and clinical data for a total of 2970 samples. In this collection, there were 15 data sets from Affymetrix platforms. From a total of 2970 cases with public gene expression data, 1183 cases meet the eligibility criteria for this candidate gene analysis. Log2-transformed gene expression data must be available from frozen primary tumors using one of three platforms (Affymetrix HG-U133A, Affymetrix HG-U133 Plus 2.0 and Affymetrix HG-HT U133A) from women with a diagnosis of invasive serous adenocarcinoma of the ovary and survival data. Data from cell lines, normal human surface epithelium, breast cancer metastasis, unknown samples, borderline tumors, non-serous ovarian cancers and patients who died within 2 weeks of diagnosis were excluded from the analysis. Data from Agilent platforms or from formalin-fixed and paraffin-embedded tumors were also excluded. Disease site, cell type, tumour stage and survival data including survival status and survival time in months from diagnosis were available on all of the eligible patients (N=1 183). PFS data was available in a subset (N=723). A total of 22 277 probe sets (representing 13 435 unique genes) had expression data across the eligible 1183 samples in data sets from six sources (GSE14764, GSE18520, GSE26712, GSE9891, PMID17290060 and TCGA) (Dressman et al, 2007; Bonome et al, 2008; Tothill et al, 2008; Denkert et al, 2009; Mok et al, 2009; The Cancer Genome Atlas Research Network, 2011). The combat method was implemented in the Surrogate Variable Analysis Bioconductor package (R package version 3.6.0; www.bioconductor.org). SVA was used to combine log2-transformed gene expression data from the six data sets to adjust for the between-experiments batch effects.
Statistical analysis of the clinical relevance of the candidate biomarkers was performed using R and SPSS v. 20 (IBM Corp., Pittsburgh, PA, USA). Cox regression analysis was performed to evaluate the relationship between expression of candidate genes of interest and PFS or OS with unadjusted hazard ratios (HRs) and 95% confidence intervals (CIs) estimated for each unit increase in average gene expression for each of the candidate biomarkers. Cox regression modelling was also performed with expression levels categorised at the median without and with adjustments for tumour stage. Kaplan–Meier method was used to estimate PFS and OS probabilities for average gene expression data categorised at the median as low (⩽ median) vs high (> median). Differences in PFS and OS distributions between groups were assessed using the log-rank test. The Mantel–Haenszel common odds ratio and 95% CI were estimated to examine the relationship between candidate genes categorised at the median and tumour stage. The association between categorised expression of an individual gene and stage was examined using Fisher's exact test. All tests were two-sided.
Results
Secretome collection
The half maximal inhibitory (IC50) values for each EOC cell line were determined before the collection and secretome proteomic analysis as follows (listed from the most platinum sensitive to most platinum resistant): OVCAR3, IC50=3.9 μM; A2780 IC50=4.2 μM; CaOV3, IC50=5.0 μM; OV90, IC50=11.2 μM; and A2780-CP20 IC50=23.3 μM. Secretome samples were harvested at 24 h after incubation in serum-free media when all cell lines displayed healthy morphologies and none of the cultures had evidence of free, floating or dead cells. Thirty millilitres of conditioned medium yielded ∼500 μg of total protein for each cell line.
Protein identification from ovarian carcinoma cell line secretomes
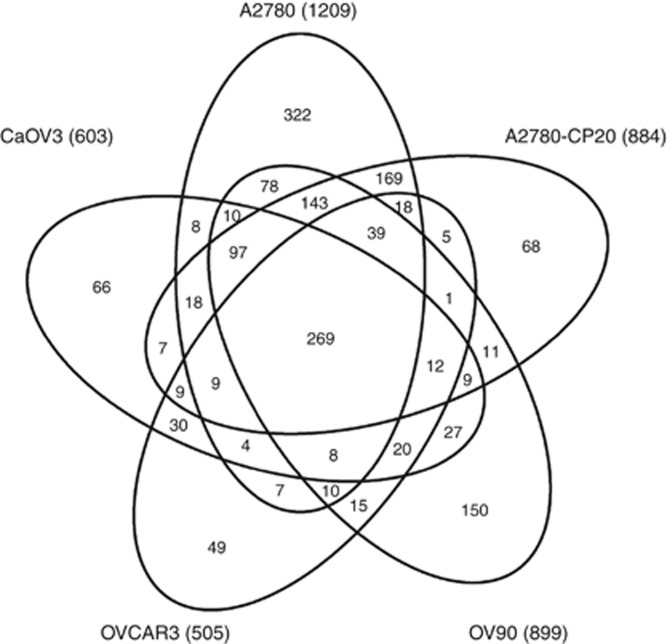
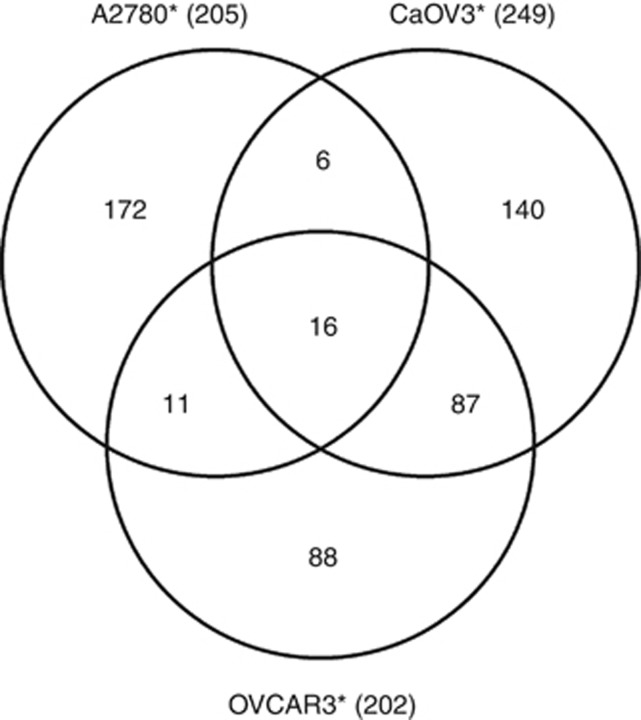
The LC-MS/MS analyses yielded a total of 160 411 peptide identifications representing 1983 aggregate proteins from the five EOC cell line (A2780, A2780-CP20, CaOV3, OVCAR3 and OV90) secretome samples (Supplementary Tables 1 and 4). The number of peptides and proteins identified from each cell line is shown in Table 1. There were a total of 1688 proteins identified with two or more peptide sequences in any of the five EOC secretome samples, among which 269 were identified in common across the secretome samples from these five cell lines (Figure 1). The spectral count values were used to identify statistically significant alterations in protein abundances from the secretome of the cisplatin-sensitive A2780 cell line compared with the isogenic cisplatin-resistant daughter cell line (A2780-CP20), where 205 proteins were found to be differentially abundant (P<0.05, Supplementary Table 2, Supporting Information), of which 67 proteins were elevated in the secretome from A2780-CP20 as compared with A2780. The proteins identified in the secretome from A2780-CP20 were also compared with those from OVCAR3 and CaOV3 (both having similar cisplatin IC50 values as the A2780 cells). Sixteen proteins were identified as differentially abundant in platinum-sensitive cells (A2780, CaOV3 and OVCAR3) compared with platinum-resistant cells (A2780-CP20) (Figure 2, Table 2 and Supplementary Table 3).
Table 1. Numbers of peptides and proteins identified from the EOC secretome.
| Secretome sample | Total peptide sequences identified | Unique peptide sequences identified | Total protein accessions identified | Protein accessions identified with⩾two unique peptides |
|---|---|---|---|---|
| A2780 |
46689 |
6802 |
1396 |
1209 |
| A2780-CP20 |
32840 |
4738 |
1046 |
884 |
| CaOV3 |
20643 |
2955 |
712 |
603 |
| OV90 |
38558 |
5654 |
1060 |
899 |
| OVCAR3 | 21681 | 2564 | 607 | 505 |
Abbreviation: EOC=epithelial ovarian carcinoma.
Figure 1.
Venn diagram of 1688 secretome proteins identified with two or more spectral counts from five EOC cell lines.
Figure 2.
Venn diagram of significant differential proteins between cisplatin-resistant A2780-CP20 and cisplatin-sensitive A2780, OVCAR3 and CaOV3, where 16 mutually differential proteins were identified. *Significant differential proteins compared with A2780-CP20.
Table 2. Sixteen secretome proteins identified with significant differential abundances (P<0.05) between cisplatin-sensitive (A2780) and cisplatin-resistant A2780-CP20) cells compared with publicly available proteomic data sets and secreted/plasma databases.
| |
|
Secretome analysis |
|
|
Hoskins et al (2011) |
Faca et al (2008) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Differentially abundant proteins in cisplatin-resistant (A2780-CP20) relative to cisplatin-sensitive (A2780) ovarian cancer cells | Uniprot accession | OVCAR3 | A2780 | CaOV3 | OV90 | A2780-CP20 | Human plasma proteomeb | Human secreted proteome databasec | TIF | Ascites | OVCAR3 | CaOV3 | Ascites |
|
Increased | |||||||||||||
| 3-ketoacyl-CoA thiolase (ACAA2) | P42765 | 0a | 0 | 0 | 0 | 22 | X | 0 | 0 | 0 | 0 | 15 | |
| Cerebellin-2 (CBLN2) | Q8IUK8 | 0 | 8 | 0 | 0 | 58 | 0 | 0 | 0 | 0 | 0 | ||
| Collagen alpha-1(II) chain (COL2A1) | P02458 | 0 | 0 | 0 | 0 | 35 | X | 0 | 0 | 1 | 5 | 0 | |
| Collagen alpha-1(XI) chain (COL11A1) | P12107 | 0 | 0 | 0 | 0 | 212 | X | 0 | 0 | 0 | 0 | 0 | |
| Cystathionine gamma-lyase (CTH) | P32929 | 0 | 3 | 0 | 7 | 40 | 0 | 0 | 0 | 1 | 0 | ||
| Inhibin beta A chain (INHBA) | P08476 | 0 | 0 | 0 | 0 | 64 | 0 | 0 | 0 | 0 | 6 | ||
| Interstitial collagenase (MMP1) | P03956 | 0 | 5 | 0 | 48 | 189 | X | 0 | 0 | 0 | 0 | 2 | |
| Stromelysin-1 (MMP3) | P08254 | 0 | 60 | 0 | 0 | 944 | X | 0 | 0 | 0 | 0 | 0 | |
| Stromelysin-2 (MMP10) | P09238 | 0 | 109 | 0 | 16 | 740 | X | 0 | 0 | 0 | 0 | 0 | |
| Pappalysin-1 (PAPPA) | Q13219 | 0 | 0 | 0 | 0 | 23 | 0 | 0 | 0 | 0 | 0 | ||
| Protocadherin-10 (PCDH10) | Q9P2E7 | 0 | 0 | 0 | 0 | 31 | 0 | 0 | 0 | 0 | 0 | ||
| Semaphorin-3D (SEMA3D) | O95025 | 0 | 0 | 0 | 0 | 63 | 0 | 0 | 0 | 0 | 0 | ||
| Torsin-1B (TOR1B) |
O14657 |
0 |
2 |
0 |
2 |
19 |
|
X |
0 |
0 |
0 |
0 |
0 |
|
Decreased | |||||||||||||
| Agrin (AGRN) | O00468 | 465 | 24 | 108 | 304 | 1 | X | X | 15 | 2 | 277 | 60 | 109 |
| D-3-phosphoglycerate dehydrogenase (PHGDH) | O43175 | 39 | 144 | 12 | 76 | 0 | X | 58 | 0 | 50 | 8 | 11 | |
| Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) | P09936 | 17 | 35 | 36 | 0 | 0 | X | 38 | 0 | 14 | 27 | 2 | |
Abbreviation: TIF=tissue interstitial fluid.
Spectral count.
Evidence of biological and translational potential
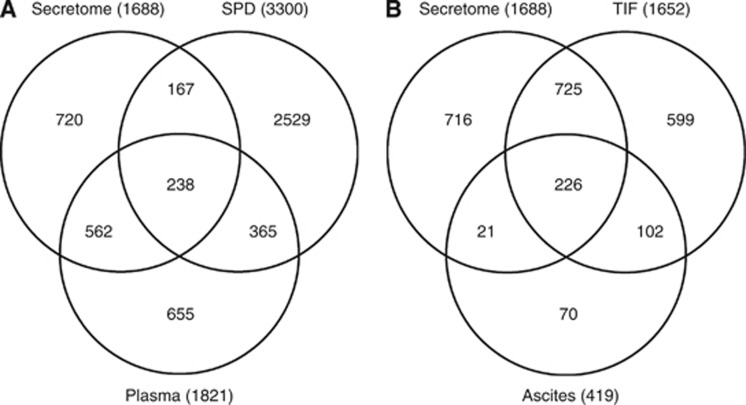
Public proteomic and gene expression data were used to further interrogate the candidate biomarkers from our secretome analyses to determine whether the secretome from human ovarian cancer cell lines represents a rich source for candidate secreted or shed proteins with biological and translational potential as candidate circulating biomarkers. The in vitro EOC secretome data were compared with the human plasma proteome (Farrah et al, 2011) and the human-derived protein entries in the secreted protein database (Figure 3A and Table 2) (Chen et al, 2005). Overall ∼60% of the secretome proteins identified in this investigation were also identified in the human plasma proteome and/or were annotated in the secreted protein database, strongly suggesting that the secretome from in vitro cultures are viable and valuable sources of candidate circulating biomarkers in humans. Of the 16 most differential proteins in our secretome analysis of cisplatin-sensitive compared with cisplatin-resistant cells, four were also found in the human plasma proteome (ACAA2, Agrin (AGRN), D-3-phosphoglycerate dehydrogenase (PHGDH) and ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1)) and eight were also detected in the secreted protein database (COL2A1, COL11A1, MMP1, MMP3, MMP10, TOR1B and AGRN). AGRN, an extracellular matrix (ECM) protein that was observed to be lower in abundance in the cisplatin-resistant A2780-CP20 secretome, was present in both the human plasma proteome and the secreted proteome databases (Table 2). Matrix-degrading metalloproteinases have important roles in ECM remodelling and tumour microenvironment dynamics (Sole et al, 2004). Of the three MMPs elevated in the platinum-resistant A2780-CP20, MMP3 was also detected in the human secreted proteome database and has been shown to degrade AGRN.
Figure 3.
Comparison of proteins identified from the current ovarian cancer cell secretome analyses to publicly available databases/data sets, including (A) the secreted protein database (SPD) and the Human Proteome Organization (HUPO) plasma proteome; and (B) TIF and ascites from ovarian cancer patients.
We then compared our EOC secretome results with previously published data from proximal fluid studies, including those derived from proteomic analyses of ascites and tissue interstitial fluid (TIF) collected from papillary serous EOC patients (Figure 3B and Table 2) (Hoskins et al, 2011). Ascites is fluid in the peritoneal cavity that commonly accumulates in ovarian cancer patients and represents a complex milieu of detached ovarian cancer cells and soluble protein (Gortzak-Uzan et al, 2008; Kuk et al, 2009; Hoskins et al, 2011), whereas TIF is harvested by ex vivo incubation of finely diced solid tumors in buffered solutions (Celis et al, 2005; Teng et al, 2010; Teng et al, 2011). Approximately 60% of the secretome proteins identified from EOC cells in this study have been previously reported from proteomic studies of ascites and/or ex vivo TIF. When comparing the 16 differentially abundant secretome proteins from platinum-sensitive vs platinum-resistant EOC cells to those observed in TIF and ascites, AGRN was observed in both ascites and TIF, whereas UCHL1 and PHGDH were observed only in TIF. UCHL1 and PHDGH were more abundant in the secretome from platinum-sensitive EOC cells. UCHL1, a deubiquitinating enzyme, has been suggested to be a tumour suppressor in multiple cancers, including ovarian cancer (Okochi-Takada et al, 2006; Yu et al, 2008; Seliger et al, 2009), that is silenced by promoter methylation (Okochi-Takada et al, 2006; Yu et al, 2008; Seliger et al, 2009). In addition, UCHL1, which induces G0/G1 cell cycle arrest and apoptosis by stabilising p53, was shown to be expressed in normal breast cell lines and tissues but downregulated or silenced in breast cancer cell lines and tissues (Xiang et al, 2012). PHDGH, amplified in melanoma and breast cancer, was demonstrated to contribute to carcinogenesis by metabolic transformation specifically through diversion of glycolytic flux (Locasale et al, 2011). Liu et al (2013) showed that PHGDH was over expressed in aggressive glial brain tumors and provided evidence of a non-metabolic role in promoting tumorigenesis. Our results showed a decrease of PHDGH in the cisplatin-resistant ovarian cancer cells compared with the sensitive cells.
Finally, we compared our in vitro secretome results in the cisplatin-sensitive CaOV3 and OVCAR3 ovarian cancer cells with those published by Faca et al (2008) in the same cell lines and found an overlap of 61% and 70% of the proteins, respectively . When we focused on the 16 candidate biomarkers, AGRN, PHGDH and UCHL1 were the most abundant proteins observed in secretome samples from CaOV3 and OVCAR3 in our study and that reported by Faca et al (2008) (Table 2). In addition, both studies provide confirmation that the cisplatin-sensitive CaOV3 and OVCAR3 did not secrete or shed detectable levels of 11 biomarkers and only low levels of two others (Faca et al, 2008). Of note, COLL11A1 was only identified in this study from the secretome of the cisplatin-resistant cell line, A2780-CP20. Faca et al (2008) also studied the secretome harvested from ovarian cancer ascites cells and detected three candidate biomarkers associated with cisplatin sensitivity (ACAA1, INHBA and MMP1) and three associated with cisplatin resistance (AGRN, PHGDH and UCHL1), suggesting that the patient may had a heterogeneous cancer made up of clonally distinct populations of sensitive and resistant tumour cells (Table 2).
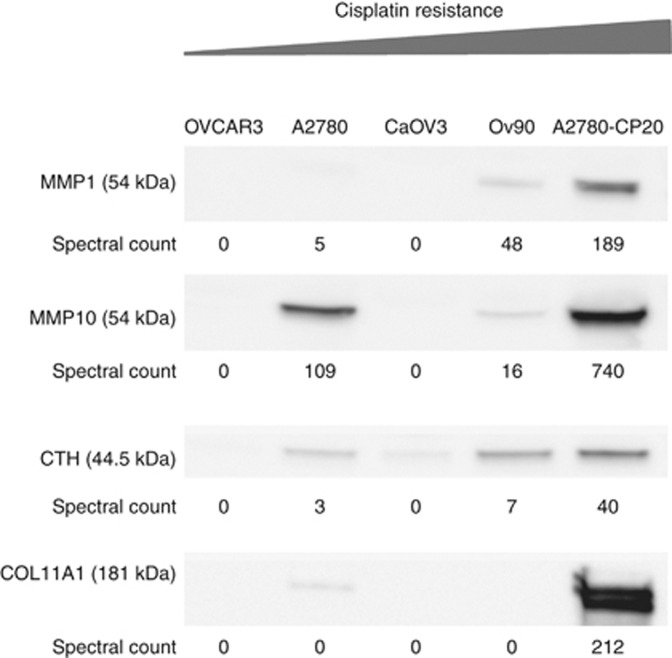
Immunoblot verification
From the panel of 16 differentially abundant secretome proteins from platinum-sensitive compared with platinum-resistant EOC cells (Table 2), MMP1, MMP10, CTH and COL11A1 were selected for verification by immunoblot in independent biological replicate samples based on known mechanisms of action and relevance in cancer. These secretome immunoblots unequivocally verify the spectral count-based proteomic findings from all five EOC cell lines (Figure 4).
Figure 4.
Immunoblotting verification of selected target secretome proteins. MMP1, MMP10, CTH and COL11A1 were selected for validation by western blotting in EOC secretome.
Evidence of clinical relevance
Public gene expression data was used to further interrogate the four candidate biomarkers from our in vitro secretome analysis that passed immunoblot verification to evaluate the clinical relevance of some of the candidate biomarkers. Table 3 provides the clinical characteristics for the eligible 1183 women with invasive serous ovarian cancer, batch corrected Affymetrix gene expression data from primary tumors with PFS (N=723) and survival data (N=1183) from six sources (Dressman et al, 2007; Bonome et al, 2008; Tothill et al, 2008; Denkert et al, 2009; Mok et al, 2009; The Cancer Genome Atlas Research Network, 2011). Cox regression analysis was initially performed to evaluate the relationship between expression of the four candidate biomarkers of interest (MMP1, MMP10, CTH and COL11A1) and risk of disease progression and death for each unit increase in average gene expression across a probe set (Table 4). Among the candidates evaluated, COL11A1 was strongly associated with both PFS and OS. Higher levels of COL11A1 expression were associated with an increased risk of disease progression (unadjusted HR=1.078, 95% CI=1.035–1.124, P=0.0003) and death (unadjusted HR=1.070, 95% CI=1.035–1.106, P=7 × 10–5). The associations between higher expression of COL11A1 and worse PFS (adjusted HR=1.053, 95% CI=1.010–1.097, P=0.015) and OS (adjusted HR=1.059, 95% CI=1.024–1.094, P=0.001) held after adjusting for tumour stage.
Table 3. Clinical characteristics for invasive serous ovarian cancer patients with progression-free survival and/or overall survival data.
|
Progression-free survival data |
Overall survival data |
|||
|---|---|---|---|---|
| Cases | Percent | Cases | Percent | |
|
Age at diagnosis (years) | ||||
| <50 | 139 | 19.2 | 185 | 15.6 |
| 50–59 | 249 | 34.4 | 292 | 24.7 |
| 60–69 | 173 | 23.9 | 242 | 20.5 |
| 70–79 | 138 | 19.1 | 190 | 16.1 |
| 80+ | 24 | 3.3 | 34 | 2.9 |
| Missing |
|
|
240 |
20.3 |
|
Site | ||||
| Ovarian |
723 |
100 |
1183 |
100 |
|
Stage | ||||
| I | 31 | 4.3 | 34 | 2.9 |
| II | 37 | 5.1 | 41 | 3.5 |
| III | 564 | 78.0 | 951 | 80.4 |
| IV | 91 | 12.6 | 154 | 13.0 |
| Advanced (III or IV) |
|
|
3 |
0.3 |
|
Cell type | ||||
| Serous adenocarcinoma |
723 |
100 |
1183 |
100 |
|
Grade | ||||
| 1 | 20 | 2.8 | 26 | 2.2 |
| 2 | 134 | 18.5 | 219 | 18.5 |
| 3 | 556 | 76.9 | 921 | 77.9 |
| Missing |
13 |
1.8 |
17 |
1.4 |
|
Debulking status | ||||
| Optimal | 470 | 65.0 | 689 | 58.2 |
| Suboptimal | 179 | 24.8 | 348 | 29.4 |
| Missing |
74 |
10.2 |
146 |
12.3 |
|
Data source | ||||
| GSE14764 (Affy U133A)a | 68 | 5.7 | ||
| GSE18520 (Affy U133 Plus 2.0)b | 53 | 4.5 | ||
| GSE26712 (Affy U133A)c | 185 | 15.6 | ||
| GSE9891 (Affy U133 Plus 2.0)d | 213 | 29.5 | 216 | 18.3 |
| PMID17290060 (Affy U133A)e | 116 | 9.8 | ||
| TCGA (Affy HT U133A)f |
510 |
70.5 |
545 |
46.1 |
|
Events | ||||
| Alive NED | 232 | 32.1 | 299 | 25.3 |
| Alive with progression | 138 | 19.1 | 190 | 16.1 |
| Alive with unknown progression status | 63 | 5.3 | ||
| Deaths | 353 | 48.8 | 631 | 53.3 |
Table 4. Relationship between the expression of candidate genes and progression-free survival or overall survival in women with invasive serous ovarian cancer and public Affymetrix gene expression data.
|
Progression-free survival (N=723) |
Overall survival (N=1183) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Unadjusted modelling |
Adjusted modellingb |
Unadjusted modelling |
Adjusted modellingb |
||||||||
| Gene (Affymetrix probe set identifier) | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P |
|
Continuous variablesa | ||||||||||||
| MMP1 (204475_at) | 1.001 | 0.944–1.062 | 0.964 | 0.995 | 0.938–1.056 | 0.875 | 0.998 | 0.950–1.048 | 0.923 | 0.997 | 0.948–1.047 | 0.893 |
| MMP10 (205828_at) | 1.151 | 0.945–1.402 | 0.163 | 1.185 | 0.964–1.458 | 0.108 | 1.066 | 0.881–1.291 | 0.509 | 1.080 | 0.889–1.312 | 0.438 |
| CTH (206085_s_at and 217127_at) | 0.893 | 0.714–1.116 | 0.320 | 0.898 | 0.717–1.124 | 0.347 | 0.860 | 0.709–1.043 | 0.126 | 0.871 | 0.718–1.057 | 0.162 |
|
COL11A1 (204320_at and 37892_at) |
1.078 |
1.035–1.124 |
0.0003 |
1.053 |
1.010–1.097 |
0.015 |
1.070 |
1.035–1.106 |
0.00007 |
1.059 |
1.024–1.094 |
0.001 |
|
Categorised COL11A1c | ||||||||||||
| Low | 1.000 | 1.000 | 1.000 | 1.000 | ||||||||
| High | 1.534 | 1.261–1.866 | 0.000018 | 1.237 | 1.058–1.448 | 0.008 | 1.287 | 1.100–1.505 | 0.002 | 1.373 | 1.126–1.674 | 0.002 |
Abbreviations: CI=confidence interval; HR, hazard ratio; MMP=matrix-degrading metalloproteinases.
HR and 95% CI estimated for each unit increase in expression of each gene.
Adjusted for tumour stage categorised as early vs advanced.
Categorised at the median as low (⩽6.8722) or high (>6.8722).
Significant P-values <0.05 were bolded.
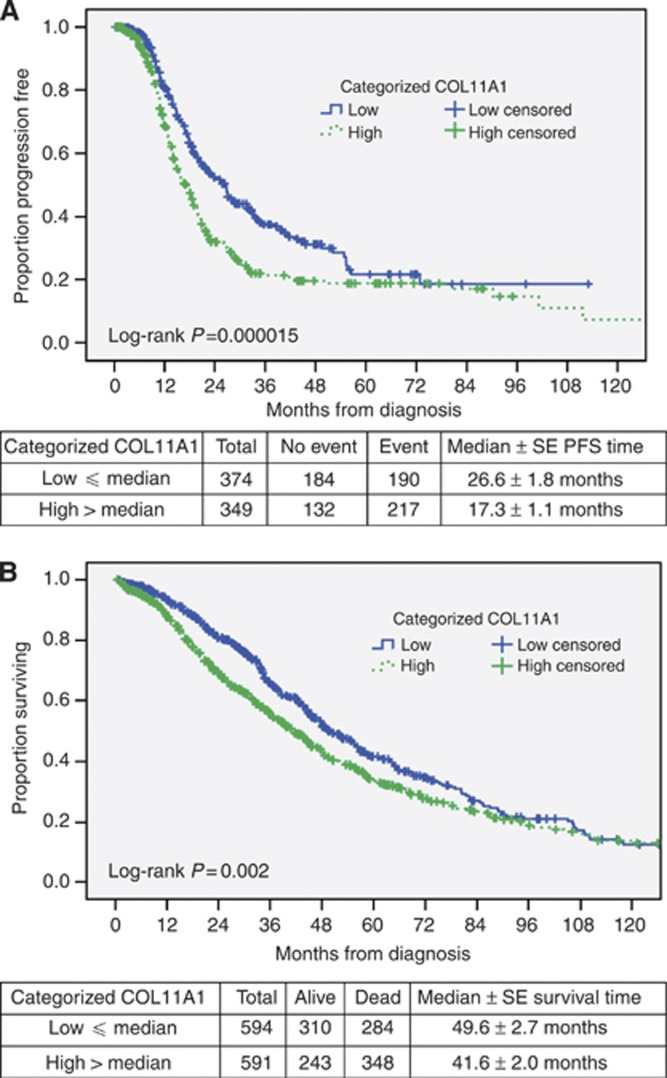
Analyses were then performed with expression levels categorised at the median. Women with primary tumors with high (>6.8722) relative to low (⩽6.8722) transcript expression values of COL11A1 had worse PFS (Figure 5A, log-rank P=1.5 × 10−5) and OS (Figure 5B, log-rank P=0.002). High COL11A1 expression was also associated with an increased risk of disease progression (unadjusted HR=1.534, 95% CI=1.261–1.866, P=2 × 10−5) and death (unadjusted HR=1.287, 95% CI=1.1–1.505, P=0.002). The association between categorised expression of COL11A1 and PFS or OS remained statistically significant even after adjusting for tumour stage (Table 5, adjusted HR=1.237, 95% CI=1.058–1.448, P=0.008 or adjusted HR=1.373, 95% CI=1.126–1.674, P=0.002, respectively). Women with high COL11A1 expression were also shown to have an increased chance of having advanced-stage disease (odds ratio=3.978, 95% CI=2.261–6.998, P=1.7 × 10−6).
Figure 5.
Kaplan–Meier plots for PFS (A) and OS (B) for women with low (⩽ median) or high (>median) tumour expression of COL11A1. There were 723 women with 138 events (recurrences) included in the PFS analysis and 1183 women included in the OS analysis with 631 deaths.
Table 5. Relationship between the categorised COL11A1 expression and tumour stagea,b.
| |
Categorised
COL11A1 |
|
|---|---|---|
| Tumour stage | Low⩽6.8722 | High>6.8722 |
| Early stage (I+II) |
59 (78.7%) |
16 (21.3%) |
| Advanced stage (III+IV) | 533 (48.1%) | 575 (51.9%) |
Fisher's exact test P=2.67 × 10−7.
Mantel–Haenszel common odds ratio=3.978, 95% confidence interval=2.261–6.998, P=1.7 × 10−6.
Discussion
Development of drug resistance represents a major obstacle in treating ovarian cancer patients and ultimately results in disease progression and death. This obstacle underscores the clinical need for biomarkers to identify and monitor patients that develop drug resistance to more effectively triage patients to therapies that reverse resistance and/or have distinct mechanisms of action. We have conducted a LC-MS/MS analysis of secreted proteins harvested from human EOC cell lines, including A2780, A2780-CP20, CaOV3, OVCAR3 and OV90, that identified 16 significantly differentially abundant proteins from platinum-sensitive vs platinum-resistant cells. Thirteen proteins were elevated and three proteins were reduced in platinum-resistant compared with platinum-sensitive EOC cells. Among these, MMP1, MMP10, CTH and COL11A1 were observed to be elevated in the secretome from platinum-resistant compared with platinum-sensitive EOC cells, and verified by immunoblot analysis.
Matrix-degrading metalloproteinases have major roles in cancer biology including proteolysis of ECM-releasing growth factors and generation of angiogenesis inhibitors, as well as processing and activation of signalling molecules to control tumour cell growth, survival, invasion and metastasis (Bauvois, 2012). Urinary MMPs have been shown to predict ovarian cancer in patients with normal CA-125 levels (Coticchia et al, 2011). In addition, several MMP transcript expression levels have been indicated to be associated to platinum-resistant head and neck cancer cell lines (Ansell et al, 2009). Matrix-degrading metalloproteinase-1 is known to activate protease-activated receptor-1, a G-protein-coupled receptor that promotes angiogenesis and metastasis in peritoneal mouse models of ovarian cancer (Agarwal et al, 2008). Salivary MMP1 levels have been shown to be elevated in oral squamous cell carcinomas (Stott-Miller et al, 2011). Matrix-degrading metalloproteinase-10 is involved in tumour initiation, cancer cell migration, cancer stem cell maintenance and expansion (Regala et al, 2011; Justilien et al, 2012). Matrix-degrading metalloproteinase-10 gene expression has been shown to be highly expressed in platinum-resistant (A2780-CP70 and A2780-CP200) when compared with platinum-sensitive cells (A2780) (Solar and Sytkowski, 2011). CTH, an enzyme that catalyses the last step in the trans-sulfuration pathway from methionine to cysteine, is regulated by the PI3K/Akt pathway (Yin et al, 2012). The trans-sulfuration pathway links methionine metabolism to the production of redox-modulating proteins and has been suggested to have a role in tumour development by altered redox equilibrium (Rosado et al, 2007). A patent has been filed by Abnova and the Japan National Cancer Center on CTH as an in vitro biomarker distinguishing ovarian cancer patients with poor prognosis (Tesshi Yamada and Wilber, 2012). Further, CTH has been suggested to be a provocative drug target where preliminary data demonstrates increased sensitisation of CTH-overexpressing cells to platinum when CTH is inhibited (Tesshi Yamada and Wilber, 2012). Elevated levels of gene and protein expression of CTH have also been reported in a multi-drug-resistant oesophageal squamous carcinoma cell line (EC109/CDDP) when compared with its parent line EC109 (Wen et al, 2010). COL11A1 overexpression has been shown to be associated with multiple advanced-stage cancers including ovarian carcinoma (Kim et al, 2010). COL11A1, along with other ECM proteins, could contribute to ECM remoulding and tumorigenesis in ovarian cancer promoting tumour cell survival and migration. Together, these observations suggest that this subset of 16 proteins, including MMP1, MMP10, CTH, and COL11A1, may hold potential as circulating biomarkers of platinum resistance, and warrant further verification in clinically relevant serum samples from recurrent/resistant and non-recurrent EOC patients.
Finally, high COL11A1 was associated with advanced stage, worse PFS and worse OS in women with invasive serous ovarian cancer as assessed from publicly available transcript expression data with clinical outcomes. The secretome from human ovarian cancer cell lines represents a rich source of candidate secreted or shed proteins with translational potential and clinical relevance as candidate circulating biomarkers. Well annotated tumour tissues, sera and/or plasma from EOC patients are being assembled to further interrogate the clinical relevance of protein levels of COL11A1 and the other candidate biomarkers from our in vitro secretome analysis as markers of cisplatin resistance and poor outcome. The ultimate goal will be to develop a panel of circulating biomarkers that can be deployed to identify/monitor platinum resistance, and triage-resistant patients to new standards of care that abate resistance and enhance outcome.
Acknowledgments
This study was funded by award W81XWH-11-2-0131 from the United States Army Medical Research and Materiel Command.
Disclaimer
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army, Department of the Air Force or the Department of Defense.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Agarwal A, Covic L, Sevigny LM, Kaneider NC, Lazarides K, Azabdaftari G, Sharifi S, Kuliopulos A. Targeting a metalloprotease-PAR1 signaling system with cell-penetrating pepducins inhibits angiogenesis, ascites, and progression of ovarian cancer. Mol Cancer Ther. 2008;7:2746–2757. doi: 10.1158/1535-7163.MCT-08-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- Ansell A, Jerhammar F, Ceder R, Grafstrom R, Grenman R, Roberg K. Matrix metalloproteinase-7 and -13 expression associate to cisplatin resistance in head and neck cancer cell lines. Oral Oncol. 2009;45:866–871. doi: 10.1016/j.oraloncology.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Bauvois B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochim Biophys Acta. 2012;1825:29–36. doi: 10.1016/j.bbcan.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC, Boyd J, Birrer MJ. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis JE, Moreira JM, Cabezon T, Gromov P, Friis E, Rank F, Gromova I. Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients: toward dissecting the molecular circuitry of epithelial-adipocyte stromal cell interactions. Mol Cell Proteomics. 2005;4:492–522. doi: 10.1074/mcp.M500030-MCP200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Yin Y, Gao G, Li S, Jiang Y, Gu X, LUO J. SPD--a web-based secreted protein database. Nucleic Acids Res. 2005;33:D169–D173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coticchia CM, Curatolo AS, Zurakowski D, Yang J, Daniels KE, Matulonis UA, Moses MA. Urinary MMP-2 and MMP-9 predict the presence of ovarian cancer in women with normal CA125 levels. Gynecol Oncol. 2011;123:295–300. doi: 10.1016/j.ygyno.2011.07.034. [DOI] [PubMed] [Google Scholar]
- Denkert C, Budczies J, Darb-Esfahani S, Gyorffy B, Sehouli J, Konsgen D, Zeillinger R, Weichert W, Noske A, Buckendahl AC, Muller BM, Dietel M, Lage H. A prognostic gene expression index in ovarian cancer - validation across different independent data sets. J Pathol. 2009;218:273–280. doi: 10.1002/path.2547. [DOI] [PubMed] [Google Scholar]
- Dressman HK, Berchuck A, Chan G, Zhai J, Bild A, Sayer R, Cragun J, Clarke J, Whitaker RS, Li L, Gray J, Marks J, Ginsburg GS, Potti A, West M, Nevins JR, Lancaster JM. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J Clin Oncol. 2007;25:517–525. doi: 10.1200/JCO.2006.06.3743. [DOI] [PubMed] [Google Scholar]
- Faca VM, Ventura AP, Fitzgibbon MP, Pereira-Faca SR, Pitteri SJ, Green AE, Ireton RC, Zhang Q, Wang H, O'briant KC, Drescher CW, Schummer M, Mcintosh MW, Knudsen BS, Hanash SM. Proteomic analysis of ovarian cancer cells reveals dynamic processes of protein secretion and shedding of extra-cellular domains. PLoS One. 2008;3:e2425. doi: 10.1371/journal.pone.0002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, Watts JD, Lin B, Zhang H, Moritz RL, Aebersold R. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10:M110 006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- Ganzfried BF, Riester M, Haibe-Kains B, Risch T, Tyekucheva S, Jazic I, Wang XV, Ahmadifar M, Birrer MJ, Parmigiani G, Huttenhower C, Waldron L. CuratedOvarianData: clinically annotated data for the ovarian cancer transcriptome. Database. 2013;2013:bat013. doi: 10.1093/database/bat013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gortzak-Uzan L, Ignatchenko A, Evangelou AI, Agochiya M, Brown KA, Onge ST, Kireeva P, Schmitt-Ulms I, Brown G, Murphy TJ, Rosen J, Shaw B, Jurisica P, Kislinger, T. I. A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J Proteome Res. 2008;7:339–351. doi: 10.1021/pr0703223. [DOI] [PubMed] [Google Scholar]
- Gunawardana CG, Kuk C, Smith CR, Batruch I, Soosaipillai A, Diamandis EP. Comprehensive analysis of conditioned media from ovarian cancer cell lines identifies novel candidate markers of epithelial ovarian cancer. J Proteome Res. 2009;8:4705–4713. doi: 10.1021/pr900411g. [DOI] [PubMed] [Google Scholar]
- Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- Heintz AP, Odicino F, Maisonneuve P, Quinn MA, Benedet JL, Creasman WT, Ngan HY, Pecorelli S, Beller U. Carcinoma of the ovary. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;95 (Suppl 1:S161–S192. doi: 10.1016/S0020-7292(06)60033-7. [DOI] [PubMed] [Google Scholar]
- Hoskins ER, Hood BL, Sun M, Krivak TC, Edwards RP, Conrads TP. Proteomic analysis of ovarian cancer proximal fluids: validation of elevated peroxiredoxin 1 in patient peripheral circulation. PLoS One. 2011;6:e25056. doi: 10.1371/journal.pone.0025056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien V, Regala RP, Tseng IC, Walsh MP, Batra J, Radisky ES, Murray NR, Fields AP. Matrix metalloproteinase-10 is required for lung cancer stem cell maintenance, tumor initiation and metastatic potential. PLoS One. 2012;7:e35040. doi: 10.1371/journal.pone.0035040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuk C, Kulasingam V, Gunawardana CG, Smith CR, Batruch I, Diamandis EP. Mining the ovarian cancer ascites proteome for potential ovarian cancer biomarkers. Mol Cell Proteomics. 2009;8:661–669. doi: 10.1074/mcp.M800313-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Guo S, LI Q, Yang L, Xia Z, Zhang L, Huang Z, Zhang N. Phosphoglycerate dehydrogenase induces glioma cells proliferation and invasion by stabilizing forkhead box M1. J Neurooncol. 2013;111:245–255. doi: 10.1007/s11060-012-1018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marengo E, ROBOTTI E, Bobba M, Gosetti F. The principle of exhaustiveness vs the principle of parsimony: a new approach for the identification of biomarkers from proteomic spot volume datasets based on principal component analysis. Anal Bioanal Chem. 2010;397:25–41. doi: 10.1007/s00216-009-3390-8. [DOI] [PubMed] [Google Scholar]
- Mcdonald JH. Handbook of Biological Statistics. Sparky House Publishing: Baltimore, MD, USA; 2009. pp. 160–164. [Google Scholar]
- Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, Ozbun L, Samimi G, Brady J, Randonovich M, Pise-Masison CA, Barrett JC, Wong WH, Welch WR, Berkowitz RS, Birrer MJ. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi-Takada E, Nakazawa K, Wakabayashi M, Mori A, Ichimura S, Yasugi T, Ushijima T. Silencing of the UCHL1 gene in human colorectal and ovarian cancers. Int J Cancer. 2006;119:1338–1344. doi: 10.1002/ijc.22025. [DOI] [PubMed] [Google Scholar]
- Regala RP, Justilien V, Walsh MP, Weems C, Khoor A, Murray NR, Fields AP. Matrix metalloproteinase-10 promotes Kras-mediated bronchio-alveolar stem cell expansion and lung cancer formation. PLoS One. 2011;6:e26439. doi: 10.1371/journal.pone.0026439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado JO, Salvador M, Bonatto D. Importance of the trans-sulfuration pathway in cancer prevention and promotion. Mol Cell Biochem. 2007;301:1–12. doi: 10.1007/s11010-006-9389-y. [DOI] [PubMed] [Google Scholar]
- Sedlaczek P, Frydecka I, Gabrys M, VAN Dalen A, Einarsson R, Harlozinska A. Comparative analysis of CA125, tissue polypeptide specific antigen, and soluble interleukin-2 receptor alpha levels in sera, cyst, and ascitic fluids from patients with ovarian carcinoma. Cancer. 2002;95:1886–1893. doi: 10.1002/cncr.10917. [DOI] [PubMed] [Google Scholar]
- Seliger B, Handke D, Schabel E, Bukur J, Lichtenfels R, Dammann R. Epigenetic control of the ubiquitin carboxyl terminal hydrolase 1 in renal cell carcinoma. J Transl Med. 2009;7:90. doi: 10.1186/1479-5876-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Solar P, Sytkowski AJ. Differentially expressed genes associated with cisplatin resistance in human ovarian adenocarcinoma cell line A2780. Cancer Lett. 2011;309:11–18. doi: 10.1016/j.canlet.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Sole S, Petegnief V, Gorina R, Chamorro A, Planas AM. Activation of matrix metalloproteinase-3 and agrin cleavage in cerebral ischemia/reperfusion. J Neuropathol Exp Neurol. 2004;63:338–349. doi: 10.1093/jnen/63.4.338. [DOI] [PubMed] [Google Scholar]
- Stott-Miller M, Houck JR, Lohavanichbutr P, Mendez E, Upton MP, Futran ND, Schwartz SM, Chen C. Tumor and salivary matrix metalloproteinase levels are strong diagnostic markers of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2011;20:2628–2636. doi: 10.1158/1055-9965.EPI-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng PN, Hood BL, Sun M, Dhir R, Conrads TP. Differential proteomic analysis of renal cell carcinoma tissue interstitial fluid. J Proteome Res. 2011;10:1333–1342. doi: 10.1021/pr101074p. [DOI] [PubMed] [Google Scholar]
- Teng PN, Rungruang BJ, Hood BL, Sun M, Flint MS, Bateman NW, Dhir R, Bhargava R, Richard SD, Edwards RP, Conrads TP. Assessment of buffer systems for harvesting proteins from tissue interstitial fluid for proteomic analysis. J Proteome Res. 2010;9:4161–4169. doi: 10.1021/pr100382v. [DOI] [PubMed] [Google Scholar]
- Tesshi Yamada KH, Wilber Huang.2012Use of overexpression of cystathionine gamma lysase as a prognostic, diagnostic and therapeutic target for cancer. Japan, Taiwan patent application 20120058204.
- The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew YE, Haviv I, Gertig D, Defazio A, Bowtell DD. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- Wen J, Zheng B, Hu Y, Zhang X, Yang H, LI Y, Zhang CY, Luo KJ, Zang X, Li YF, Guan XY, Fu JH. Comparative proteomic analysis of the esophageal squamous carcinoma cell line EC109 and its multi-drug resistant subline EC109/CDDP. Int J Oncol. 2010;36:265–274. [PubMed] [Google Scholar]
- Xiang T, Li L, Yin X, Yuan C, Tan C, SU X, Xiong L, Putti TC, Oberst M, Kelly K, Ren G, Tao Q. The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PLoS One. 2012;7:e29783. doi: 10.1371/journal.pone.0029783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Zhao C, Li Z, Mei C, Yao W, Liu Y, Li N, Qi J, Wang L, Shi Y, Qiu S, Fan J, Zha X. Sp1 is involved in regulation of cystathionine gamma-lyase gene expression and biological function by PI3K/Akt pathway in human hepatocellular carcinoma cell lines. Cell Signal. 2012;24:1229–1240. doi: 10.1016/j.cellsig.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Yu J, Tao Q, Cheung KF, Jin H, Poon FF, Wang X, Li H, Cheng YY, Rocken C, Ebert MP, Chan AT, Sung JJ. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48:508–518. doi: 10.1002/hep.22343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.