Abstract
Using the feline immunodeficiency virus (FIV) model for AIDS lentivirus infection, we previously demonstrated that Treg cells from FIV-infected cats up-regulate membrane-associated tumor growth factor beta (mTGF-ß) during the course of infection and that activated T lymphocytes up-regulate TGF-ß receptor II (TGF-ßRII) during the course of infection. Furthermore, we have demonstrated that autologous coculture of Tregs with Th cells from FIV-infected cats leads to suppression of interleukin (IL)-2 production and loss of proliferation in a TGF-ß-dependent fashion. Nuclear factor of activated T cells (NFAT) 2 has been identified as integral to effector Th cell maturation and function by promoting IL-2 transcription. Therefore, we questioned whether NFAT2 expression might be altered by TGF-β signaling. Feline NFAT2 exon sequences were identified based upon sequence homology to human and murine NFAT2. Following stimulation, IL-2 and NFAT2 mRNA levels were similarly increased in both FIV− and FIV+ cats. Activated CD4+CD25− cells from both FIV− and FIV+ cats cocultured with autologous CD4+CD25+ cells or treated with TGF-β demonstrated decreased IL-2 production; however, NFAT2 mRNA levels were unaffected. Although NFAT2 mRNA levels were unaffected, chromatin immunoprecipitation (ChIP) for NFAT2 indicated decreased NFAT2 binding at the IL-2 promoter in suppressed Th cells. These data suggest that TGF-β-mediated Treg cell suppression of IL-2 transcription is modulated through alterations in NFAT2 binding to the IL-2 promoter.
Introduction
Feline immunodeficiency virus (FIV) causes acquired immunodeficiency syndrome (AIDS) in its natural host, the domestic cat. We previously demonstrated that FIV infection phenotypically and functionally activates immunosuppressive CD4+CD25+ T regulatory (Treg) cells during both the acute and chronic stage of infection.1,2 Activated CD4+CD25+ Treg cells progressively up-regulate membrane-bound tumor growth factor beta (mTGF-β) during the course of infection and mediate suppressor function by engagement of TGF-β receptor II (TGF-βRII) on the surface of activated CD4+ and CD8+ target cells.3,4 While there is comparatively little information regarding the intracellular events that occur in lymphocyte targets following interaction with activated Treg cells, we have sought to understand in more detail the molecular events occurring in activated CD4+ and CD8+ effector cells following interaction with lentivirus-activated Treg cells.
The nuclear factor of activated T cells (NFAT) is one of the primary families of transcription factors known to modulate cytokine gene expression in lymphocytes.5 In naive T cells, NFAT1 and NFAT2 proteins are the primary regulators of CD4+ Th helper (Th) cell activation and effector function.6,7 During T cell activation, preexisting NFAT1 activates NFAT2 expression inducing a self-sustaining positive autoregulatory loop to maintain interleukin (IL)-2 production.6,8–11
During FIV infection, Th cells display an activated phenotype, yet have compromised effector function. We have found that activated Th cells receive both stimulatory and inhibitory signals, leading to a complex integration of intracellular signaling events.2,12 We therefore asked which Th effector cell transcription factors might be affected by the convergence of T cell activation signals and inhibitory TGF-β signals. NFAT2 has been identified as integral to effector Th cell maturation and function following Th activation.6,13 In contrast, inhibitory TGF-β signaling in effector cell targets leads to Smad phosphorylation and inhibition of IL-2 production through Foxp3-dependent and -independent pathways.14–16 Evidence suggests that the balance between activation signaling through NFAT2 and suppressive signaling through the TGF-ß/SMAD pathway may be integral to the modulation of Th IL-2 transcription under these circumstances.16,17 The object of this study was to identify feline NFAT2 in CD4+ lymphocytes and determine what role it plays in TGF-β-dependent suppression of effector cell targets. Here we report a decrease in IL-2 mRNA and protein, but no change in NFAT2 mRNA, in activated Th cells from FIV+ cats or FIV− cats cocultured with autologous Treg cells. Although the overall amount of NFAT mRNA was not altered, our results suggest that TGF-β-mediated suppression reduces NFAT2 binding to the IL-2 promoter in Th effector cells.
Materials and Methods
Cats
Specific pathogen-free (SPF) cats were obtained from Liberty Research, Inc. (Waverly, NY) and housed in the Laboratory Animal Resource Facility at the College of Veterinary Medicine, North Carolina State University. FIV-infected cats were housed separately from FIV-negative control cats. Protocols were approved by the North Carolina State University Institutional Animal Care and Use Committee.
FIV infection
The NCSU1 isolate of FIV was originally obtained from a naturally infected cat and has been described in detail elsewhere.18,19 Virus was grown as a single tissue culture passage in FCD4E cells (an IL-2-dependent feline CD4+ cell line) as described previously.19 Cats were infected with 1×105 TCID50 of cell-free virus and controls were sham inoculated with an equal volume of cell-free CD4E tissue culture medium. At the time of this study, FIV+ cats had been infected for at least 1 year.
Lymph node biopsies and purification of lymphocyte subsets
Lymph nodes were removed following euthanasia or surgical excision as described previously.1 Lymph node cells were processed into single cell suspensions and stained with anti-CD4-biotin (and secondary PerCP streptavidin), anti-CD8-PE, and anti-CD25-FITC and sorted into CD4+CD25− and CD4+CD25+ populations using a MoFlo high-speed cell sorter (Beckman Coulter, Inc., Brea, CA). Sorted populations were greater than 95% pure. For suppression assays, CD4+CD25− target cells were reconstituted with a CD4+ and CD8+ lymphocyte-depleted population prior to coculture with CD4+CD25+ lymphocytes.
Cell culture and activation
T cells were cultured in CTL media [RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 0.01 M HEPES, 0.075% NaBicarb, 100 units/ml Pen/100 (g/ml Strep, 5×10–5 M 2-mercaptoethanol (2-ME)]. To activate lymphocytes, cells were cultured at 1×106/ml and treated with ionomycin (I, 1.5 μM) alone, phorbol myristate acetate (PMA) (20 ng/ml) plus ionomycin (PMA/I, 1.5 μM), concanavalin A (Con A) (5 μg/ml), or Con A (5 μg/ml) plus ionomycin (1.5 μM) for the indicated length of time. Nontreated cells were used as controls.
NFAT2 cDNA PCR
Feline NFAT2 exon fragments were found in GenBank. Total mRNA was extracted from 106 FACS-purified lymphocytes using the RNeasy plus Mini Kit (Qiagen) and eluted in a final volume of 50 μl RNase-free water per reaction. Of the RNA 20 μl was reverse transcribed into cDNA using random primers and reverse transcribed according to the manufacturer's instructions (Promega). Reactions were incubated at 25°C for 10 min, 42°C for 15 min, 95°C for 5 min, 5°C for 5 min, and 42°C for 60 min before holding at 5°C. Polymerase chain reaction (PCR) primers were designed to amplify regions with exon junctions and are listed in Table 1.
Table 1.
Primers used in Polymerase Chain Reaction for Identification of Feline Nuclear Factor of Activated T Cells 2 Exon Conjunctions
| Forward | Reverse | |
|---|---|---|
| ex3-ex4 |
5′-catccggaagggcgccttctgc-3′ |
5′-gagggcacctgccagtccagg-3′ |
| ex4-ex5 |
5′-ctacgagacggaggggagccg-3′ |
5′-cacggtcttgcccgtgatccg-3′ |
| ex5-ex6 |
5′-ccgtgtccaccaccagtcacgag-3′ |
5′-cctttgcgcagctcgatgtccg-3′ |
| ex6-ex7 |
5′-ccctgcaagtggcctcgaacc-3′ |
5′-gaccatcttcttcccgccgagg-3′ |
| ex7-ex9 | 5′-tcctgtccggccataactttctgc-3′ | 5′-ggcggtatctcgaccaccaacg-3′ |
Reverse transcription and real-time PCR
Relative quantification of IL-2 and NFAT2 mRNA was achieved by reverse transcription and real-time PCR. A Quantitect SYBR Green PCR kit (Qiagen) was used for quantification of IL-2 and NFAT2 mRNA levels. Reactions were run in triplicate in 96-well plates. All reactions were carried out using identical cycling conditions as follows: denatured at 95°C for 20 s, annealed at 58°C for 20 s, and elongated at 72°C for 30 s with 40 cycles. The GAPDH mRNA level was used as the control. For Figs. 2 and 3, data were calculated using the delta delta Ct method. Specific primers for feline IL-2, NFAT2, and GAPDH amplification are shown in Table 2.
FIG. 2.
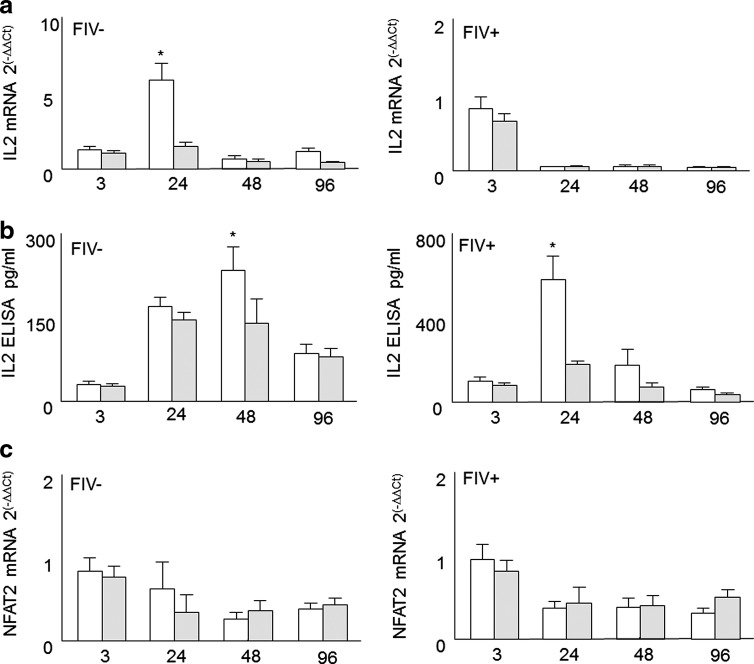
Evaluation of interleukin (IL)-2 and NFAT2 levels in T helper cells cocultured with Treg cells from feline immunodeficiency virus (FIV)− and FIV+ cats. CD4+CD25− and CD4+CD25+ T cells were purified as described in Materials and Methods. CD4+CD25− cells were stimulated with Con A (5 μg/ml) for 1 h and autologous CD4+CD25− cells (control, white bars) or CD4+CD25+ cells (suppressor cells, gray bars) were added to the culture. Relative IL-2 and NFAT2 mRNA expression levels were assessed using real time RT-PCR and culture supernatant was collected for IL-2 ELISA. (a) CD4+CD25− IL-2 mRNA was reduced in FIV− cats (left plate) after a 24 h coculture with autologous CD4+CD25+ cells. (b) CD4+CD25− IL-2 production was reduced in FIV− (left plate) and FIV+ (right plate) cats after a 48 h coculture with autologous CD4+CD25+ cells. (c) CD4+CD25− NFAT2 mRNA expression levels appear unaltered by autologous CD4+CD25+ coculture. For IL-2 and NFAT2 mRNA, expression of the housekeeping gene GAPDH was used as the control, and results were calculated as 2–ΔΔCt. Error bars indicate the mean+SEM for each time point. Data are representative of four independent experiments; asterisks=p<0.05.
FIG. 3.
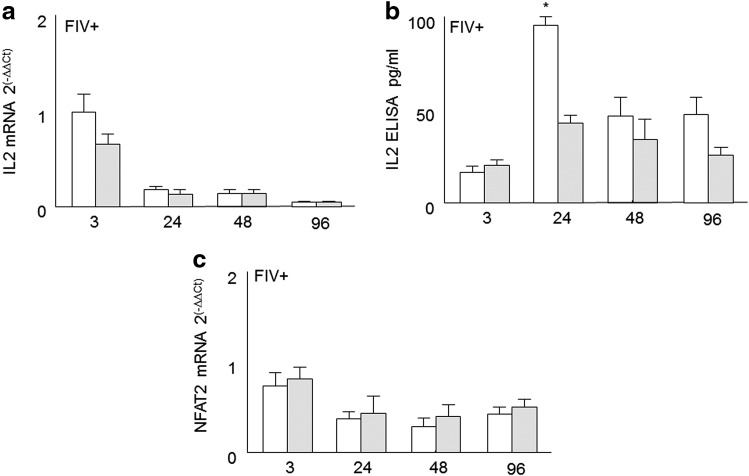
Evaluation of IL-2 and NFAT2 levels in T helper cells treated with tumor growth factor (TGF)-β. CD4+CD25− T cells from FIV+ cats were purified as described in Materials and Methods. CD4+CD25− cells were stimulated with Con A (5 μg/ml) for 1 h (white bars) and then soluble TGF-β (10 ng/ml, gray bars) was added to the culture. Relative IL-2 and NFAT2 mRNA expression levels were assessed using real time RT-PCR and culture supernatant was collected for IL-2 ELISA. (a, b) IL-2 mRNA and ELISA kinetics for TGF-β-suppressed CD4+CD25− T cells are similar to that of the CD4+CD25− cells cocultured with CD4+CD25+ cells in Fig. 2. (c) Also similar to Fig. 2, CD4+CD25− NFAT2 mRNA expression levels appear unaltered by TGF-β treatment. For IL-2 and NFAT2 mRNA, expression of the housekeeping gene GAPDH was used as the control, and results were calculated as 2–ΔΔCt. Bars represent the mean+SEM for six measurements at each time point; asterisks=p<0.05.
Table 2.
Primers Used in Real-Time Polymerase Chain Reaction for Quantitation of Feline Interleukin-2 and Nuclear Factor of Activated T Cells 2 mRNA Levels
| Gene | Forward | Reverse |
|---|---|---|
|
IL-2 |
5′-acagtgcacctgcttcaagctct-3′ |
5′-cctggagagtttggggttctcagg-3′ |
|
NFAT2 |
5′-acccctaccagtgggccagg-3′ |
5′-gaggcttgggctgcacctcg-3′ |
| GAPDH | 5′-ggagaaggctggggctcac-3′ | 5′-ggtgcaggaggcattgctga-3′ |
Purification of peripheral blood mononuclear cells
Approximately 28 ml of blood was collected via jugular venipuncture into vacutainer tubes containing EDTA anticoagulant. Blood was spun at 1,500 rpm for 10 min and plasma was removed. Peripheral blood mononuclear cells (PBMCs) were separated using Histopaque-1077 (Sigma-Aldrich) following the manufacturer's instructions.
CD4+CD25+ Treg cell suppression assay
Cats were euthanized and peripheral lymph nodes were collected and processed into a single cell suspension. Lymphocytes were stained with anti-CD4-biotin-streptavidin PerCP, anti-CD25-FITC, and anti-CD8-PE. CD4+CD25− and CD4+CD25+ T cell populations and CD4−CD8− cells were purified by high-speed sorting. CD4+CD25− cells (2×106) (target cells) were reconstituted with CD4−CD8− cells at a 1:1 ratio and treated with Con A (5 μg/ml) for 1 h. Cells were pelleted at 1,500 rpm for 10 min and then resuspended in 2 ml CTL media. Approximately 2×106 CD4+CD25+ cells (suppressor cells) were stained with DiD Vybrant stain (Molecular Probes) and added to the culture. A control culture consisting of CD4+CD25− cells (target cells) and CD4−CD8−-depleted cells was cocultured with DiD Vybrant-stained CD4+CD25− cells. Cells were cultured for various times (3 h, 1 day, 2 days, and 4 days) and DiD+ cells were removed by high-speed cell sorting. Then 106 DiD− cells were collected and RNA was extracted and subjected to real-time PCR for measurement of IL-2 and NFAT2 mRNA levels. Culture supernatant was collected for quantification of IL-2 protein by enzyme-linked immunosorbent assay (ELISA).
TGF-β suppression assay
Cats were euthanized and peripheral lymph nodes were collected and processed into a single cell suspension. Lymphocytes were stained with anti-CD4-biotin-streptavidin PerCP, anti-CD25-FITC, and anti-CD8-PE. CD4+CD25− and CD4+CD25+ T cell populations and CD4−CD8− cells were purified by high-speed sorting. CD4+CD25− cells (1×106) were cultured in 1 ml CTL media with Con A (5 μg/ml) for 1 h followed by the addition of recombinant human TGF-β (10 ng/ml). The control consisted of Con A-stimulated CD4+CD25− cells cultured in the absence of TGF-β. Cells were cultured for various times (3 h, 6 h, 1 day, 2 days, 3 days, and 4 days) and then 106 cells from each group were harvested for RNA extraction and real-time PCR. Culture supernatant was collected for quantification of IL-2 protein by ELISA. In some experiments, cells were collected after 3 h of culture and fixed for a chromatin immunoprecipitation (ChIP) assay.
IL-2 ELISA
Culture supernatants from the Treg cell suppression assays and TGF-β suppression assays were analyzed for IL-2 production using the DuoSet ELISA Development kit (R&D systems) according to the manufacturer's instructions. Following development, the optical density of each well was determined using a microplate reader set to 450 nm using KC junior software (BioTek). Each sample was analyzed in triplicate and all values for IL-2 were determined using a linear regression based upon optical density of wells with known concentrations of IL-2 standard.
Chromatin immunoprecipitation assay
For each cat, approximately 2×107 CD4+CD25− Th cells were collected by high-speed cell sorting. Lymphocytes were stained with anti-CD4-PE and anti-CD25-FITC. Cells were untreated, treated with Con A (5 μg/ml) for 1 h, or treated with Con A (5 μg/ml) for 1 h and then TGF-β1 (10 ng/ml) for another hour. To maintain intracellular calcium and promote NFAT2 nuclear translocation, ionomycin (1.5 μM) was then added to both Con A-stimulated groups and cells were cultured for 3 more h, harvested, and fixed by the addition of 37% formaldehyde (1% final concentration) and then incubated on a shaker for 10 min at room temperature. Glycine (2.5 M) was added to the cells at a 1% final concentration and the cells were shaken for an additional 5 min at room temperature. Cells were centrifuged at 1,100 rpm for 10 min at 4°C, washed once with 10 ml phosphate-buffered saline (PBS), and repelleted. Then 5 μl of 100 mM PMSF and 5 μl of 100×PIC were added and the pellet was stored at −80°C before use.
Frozen cell pellets from two cats were pooled, resuspended in 1 ml ice-cold lysis buffer (100 μl 1 M Tris, pH 7.5; 30 μl 1 M MgCl2; 20 μl 5 M NaCl; 500 μl 10% NP40; 9.35 ml H2O) plus 5 μl each of 100 mM PMSF and 100×PIC, and incubated on ice for 30 min. The cells were transferred to 1.7-ml microfuge tubes and the nuclei were centrifuged for 10 min at 5,000 rpm at 4°C. The supernatant was removed and the nuclei were resuspended to a final concentration of 0.5 mg/ml in MNase buffer (500 μl 1 M Tris, pH 7.5; 40 μl 1 M MgCl2; 10 μl 1 M NaCl; 3.33 ml 1 M sucrose; 50 μl 1 M Na-butyrate; 6.1 ml H2O) plus 2.5 μl each of 100 mM PMSF and 100×PIC, and incubated for 5 min at 37°C. Nuclei were digested by adding 10 U/ml MNase and incubating for 7 min at 37°C. Then 20 μl/ml 0.5 M EDTA was added to stop digestion and the nuclei were put on ice for 10 min then pelleted at 4°C for 10 min at maximum speed. The supernatant was aliquoted and stored at −80°C before use.
Five microliters each of protein A and protein G magnetic beads (Invitrogen) were used for each immunoprecipitation. Beads were washed twice in 100 μl 1×RIPA/ssDNA (1 ml 10×RIPA; 1 ml 5 mg/ml salmon sperm DNA; 8 ml PCR grade water), resuspended in 50 μl 1×RIPA/ssDNA plus 0.5 μl each of 100 mM PMSF and 100×PIC, and incubated with separate antibodies for 1 h at 4°C. Either 10 μg antihuman NFAT2 (sc-13033X, Santa Cruz) or 2 μg of control rabbit IgG was used. Antibody-bead conjugates were washed three times in 100 μl 1×RIPA/ssDNA, then resuspended in 80 μl 1×RIPA/ssDNA plus 1 μl each of 100 mM PMSF and 100×PIC and aliquoted to different tubes. To each tube 20 μl chromatin was added and incubated with constant agitation for 2 h at 4°C. Conjugates were washed four times in 180 μl 1×RIPA, twice in 180 μl TE (pH 8.0), and resuspended in 100 μl fresh elution buffer (100 mM NaHCO3/1% SDS), followed by 15 min of light vortexing. Tubes were placed on a magnet to retain the antibody-bead complexes and medium containing the immunoprecipitate was transferred to fresh tubes to which 2 μl 5 M NaCl was added. For reversal of cross-linking, tubes were incubated at 95°C for 15 min. Then 2 μl of 1 mg/ml proteinase K was added and the tubes were incubated at 37°C for 1 h. DNA was purified using buffer PN (Qiagen) and buffer PE (Qiagen), and was washed in 60 μl of water. Isolated DNA samples were then subjected to real-time PCR using primers for the detection of NFAT binding to the feline IL-2 promoter (Table 3).
Table 3.
Primers Used in Chromatin Immunoprecipitation Assay for Detection of Feline Nuclear Factor of Activated T Cells 2 Binding to the Feline Interleukin-2 Promoter
| Forward | Reverse | |
|---|---|---|
| Set 1 |
5′-catccattcagtcagtttatggggg-3′ |
5′-ctggaaaaatcttatgggggtgtc-3′ |
| Set 2 |
5′-aactgccacctaagtgtgggc-3′ |
5′-tacctgtgtggcaaaaagcattacc-3′ |
| Set 3 | 5′-agccacttttgtatccccaccccc-3′ | 5′-agcccacacttaggtggcagttg-3′ |
Statistical analysis
Data were analyzed using a Mann–Whitney–Wilcoxon analysis for nonparametric data and p values of less than 0.05 were considered significant.
Results
Identification of feline NFAT2
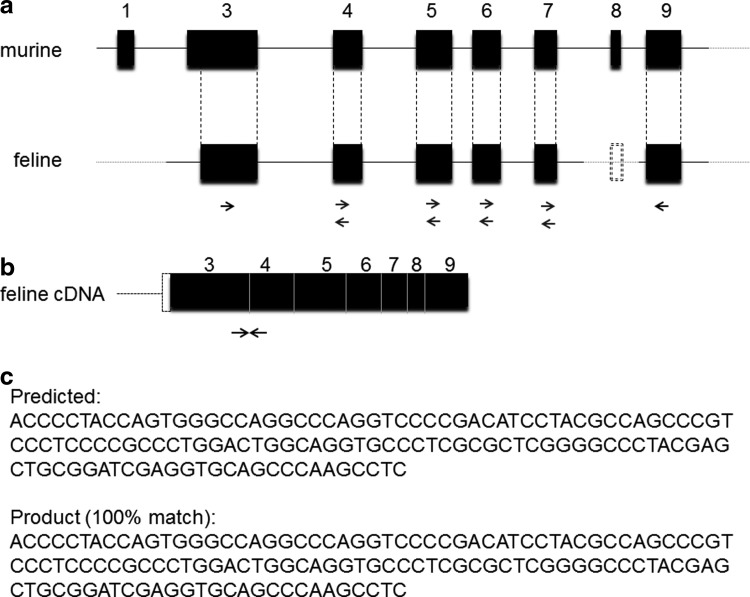
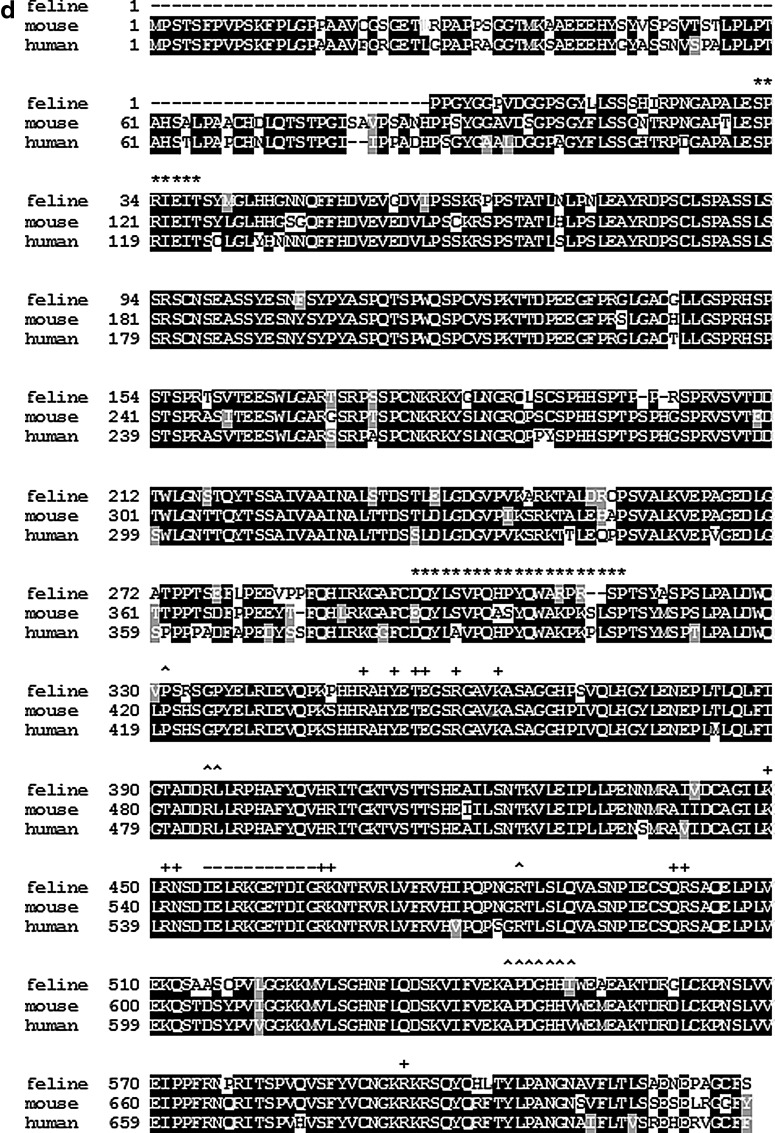
As NFAT2 is a major transcriptional activator of the IL-2 promoter, and because Treg cells suppress IL-2 transcription in CD4+ Th target cells, it is possible that Treg cell-mediated suppression of IL-2 transcription in Th target cells is due to the inhibition of NFAT2 expression. To address this question, it was necessary to first identify NFAT2 in the cat. The NFAT2 gene in humans and mice has at least 11 exons and the inducible NFAT2 isoform contains 8 exons.5 Based upon the published nucleotide sequences of human and mouse NFAT2 (NCBI Reference Sequence: NM_172390.1 and NM_001164109.1), feline exon fragments that share high sequence homology to the human and mouse genes were found. As illustrated in Fig. 1a, exon conjunctions were then identified by PCR using primers (Table 1) that span the different exons. The nucleotide sequence data were then derived from overlapping 5′-3′ and 3′-5′ nucleotide fragments. After identifying the exon conjunctions shown in Fig. 1a, reverse transcription PCR was used to amplify a partial feline NFAT2 cDNA sequence (primer sequence, Table 2). The schematic structure of the identified partial feline NFAT2 cDNA is shown in Fig. 1b. As shown in Fig. 1c, the real-time PCR product sequence displays 100% sequence homology with the predicted feline cDNA sequence. This primer set was then used for detecting NFAT2 by real time RT-PCR in the following experiments. To further confirm that the gene we identified is the NFAT2 homolog, protein alignment was performed for the predicted feline sequence, the mouse, and human sequences. Although the 1–85 amino acid sequence at the N-terminal of the target protein is missing, alignment shows that this feline protein shares a high degree of homology with human and mouse NFAT2. Isoforms of NFAT proteins are variable at N- and C-termini, yet contain a highly conserved core region, which contains calcineurin docking sites and Fos/Jun binding sites (Fig. 1d).20 These functionally critical motifs in feline NFAT2 are identical or highly conserved when compared to human and mouse NFAT2, suggesting that the protein encoded by the gene we identified in cats is feline NFAT2.
FIG. 1.
Identification of feline nuclear factor of activated T cells 2 (NFAT2) gene sequences. (a) The inducible NFAT2/A isoform contains eight exons. Based on the published cDNA sequence of human and mouse NFAT2/A, homologous feline NFAT2 exons (partial ex3, ex4, ex5, ex6, ex7, and ex9) were found in GenBank. The exon junctions and ex8 were identified using PCR primers designed to span adjacent exons. Primers are shown below each exon (black arrows). Primer sequences are listed in Table 1. (b) Schematic representation of the feline NFAT2 region amplified by real time PCR. The primer sequences listed in Table 2 are indicated by black arrows. (c) The real time PCR product sequence exhibits 100% homology with the predicted sequence. This primer set was then used for all subsequent assessments of NFAT2 by real time RT-PCR. (d) Protein alignment of predicted partial feline NFAT2 with human and mouse NFAT2. Identical regions are shaded black and similar regions are shaded gray. Above the protein sequences, the two calcineurin-binding regions are denoted by an asterisk (*), residues that participate in DNA contact are denoted by a plus (+), and residues that interact with Fos and Jun are denoted by (^) and (−), respectively.
IL-2 mRNA and cytokine production are decreased but NFAT2 mRNA expression is unchanged in Th cells from both FIV− and FIV+ cats following autologous Treg coculture
It is well established that coculture of mitogen-stimulated Th targets with autologous Treg cells from FIV-infected cats results in decreased IL-2 mRNA and protein production.1,2 Based upon these findings, we hypothesized that coculture of stimulated Th targets with autologous Treg cells would also lead to a reduction in NFAT2 mRNA. Th/Treg coculture experiments were performed to analyze IL-2 mRNA production, IL-2 protein secretion, and NFAT2 mRNA production in parallel. As shown in Fig. 2a, IL-2 mRNA was decreased in FIV− cats after 24 h of Treg coculture (left, p<0.05). In FIV+ cats, IL-2 mRNA was somewhat decreased after 3 h of Treg coculture but did not reach significance. As shown in Fig. 2b, activated Th cells from FIV+ cats have the capacity to rapidly produce IL-2, but this ability is dramatically inhibited in vitro after 24 h of Treg coculture. Most importantly and contrary to our hypothesis, Fig. 2c demonstrates that there was no consistent difference between control cultures and Th/Treg cocultures in NFAT2 mRNA expression at any of the time points tested.
IL-2 mRNA and protein production are decreased but NFAT2 mRNA expression is unchanged in Th cells following TGF-β treatment
Several studies, including those in this laboratory, have demonstrated that Treg suppression of T helper cells is mediated by mTGF-β on the Treg cell and TGF-β receptor (TGF-βRII) on the target cell.3,4,15,21,22 Following Con A stimulation, TGF-βRII is unregulated on CD4+CD25− Th cells.3 To address the role of TGF-β/TGF-βRII signaling, and to mimic TGF-ß-mediated Treg suppression, an in vitro suppression assay of Con A-activated CD4+CD25− T helper cells was performed using recombinant human TGF-β (10 ng/ml). IL-2 and NFAT2 mRNA were quantified at the time points indicated following TGF-β treatment. As shown in Fig. 3a and b, treatment with TGF-β resulted in decreased IL-2 mRNA and protein secretion. As shown in Fig. 3c, the increase in NFAT2 mRNA expression was similar in the Con A/TGF-β1-treated T helper cells compared to the T helper cells treated with Con A only. More importantly, TGF-β treatment resulted in similar kinetics demonstrated in Treg coculture experiments and is consistent with previous evidence that Treg suppression of T helper cells is mediated by mTGF-β. However, just as with the Th/Treg cocultures, TGF-β1 treatment did not decrease NFAT2 mRNA expression.
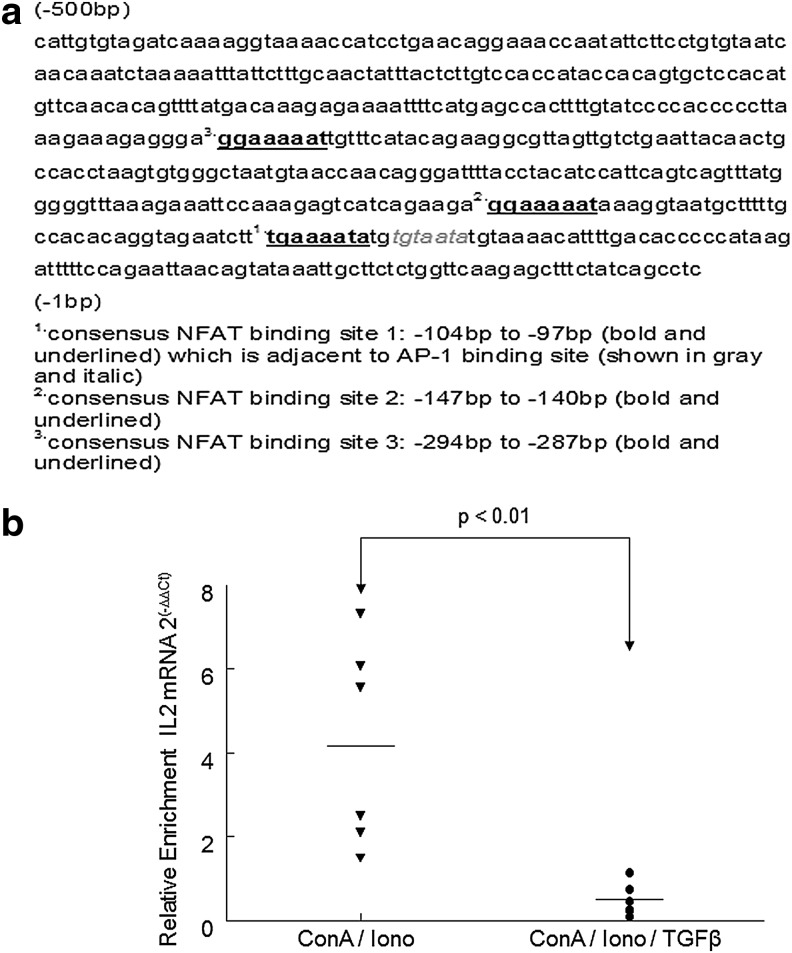
NFAT2 binding to the IL-2 promoter in activated Th cells is reduced following TGF-β treatment
NFAT2 mRNA expression was unaltered by either Treg coculture or treatment with TGF-β. This suggests that other factors, including inhibition of nuclear translocation and alteration of NFAT2 binding at the IL-2 promoter, might modulate NFAT2 activity. Chen et al.16 reported that TCR-stimulated NFAT2 nuclear translocation is reduced following TGF-β treatment; however, they also demonstrated that translocation was reestablished by maintaining intracellular Ca2+ concentrations through concurrent treatment with PMA/I. Therefore, our investigations focused on how NFAT2 binding to the IL-2 promoter might be altered by TGF-β signaling. As shown in Fig. 4a, we identified potential NFAT binding sites within the feline IL-2 promoter based upon human and murine sequence analysis. ChIP was then utilized to assess NFAT2 binding at the IL-2 promoter. Prior to ChIP, the cells were first treated with Con A to promote up-regulation of TGF-βRII and then with ionomycin to maintain intracellular calcium and promote NFAT2 nuclear translocation. A TGF-β-mediated suppressor assay was then performed as described in Fig. 3. Control Th cells and TGF-β-treated Th cells were fixed, chromatin was harvested, and NFAT2 binding to the IL-2 promoter was analyzed by ChIP. As shown in Fig. 4b, binding of NFAT2 to the IL-2 promoter in non-TGF-β-treated (stimulated) cells is greater than 2-fold that of TGF-β-treated (suppressed) cells.
FIG. 4.
The level of NFAT2 binding to the IL-2 promoter is higher in activated T helper cells than in suppressed T helper cells. (a) There are three predicted NFAT binding sites between −500 bp and −1 bp 5′ upstream of the feline IL-2 promoter. The predicted NFAT binding sites are underlined and shown in bold. The first NFAT binding is adjacent to the AP-1 binding site, which is denoted by gray and italic. (b) CD4+CD25− T helper cells were purified as described in Materials and Methods. Cells were either untreated (normalized value=1, not shown), treated with Con A (5 μg/ml) for 1 h (black triangles), or treated with Con A (5 μg/ml) for 1 h and then TGF-β1 (10 ng/ml) for another hour (black circles). To maintain intracellular calcium and promote NFAT2 nuclear translocation, ionomycin (I, 1.5 μM) was then added to both Con A-stimulated groups and cells were cultured for 3 more hours, harvested, and fixed. In CD4+CD25− lymphocytes, NFAT2 binding to the IL-2 promoter was reduced to basal levels following suppression with TGF-β. For ChIP analysis NFAT2 immunoprecipitation was performed followed by IL-2 PCR as described in Materials and Methods. Dots indicate independent experiments (n=6) from FIV+ cats and the horizontal line represents the mean for relative IL-2 enrichment.
Discussion
We have previously reported that FIV infection phenotypically and functionally activates CD4+CD25+ Treg cells and that these cells mediate suppressor function against CD4+ Th cells and CD8+ CTLs through a TGF-β-dependent pathway.1,3,4 Following coculture with lentivirus-activated Treg cells, CD4+ and CD8+ lymphocyte targets exhibit reduced proliferative capacity, late G1 cell cycle arrest, and decreased production of IL-2 and interferon (IFN)-γ, a profile consistent with anergy.2,4,17 Using the FIV model for AIDS lentivirus infection, our current investigations are directed toward developing an understanding of the transcription events that lead to Treg cell-induced lymphocyte anergy and the failure to differentiate into effector/memory populations following interaction with activated Treg cells. Recently, we have focused on the modulation of NFAT2, which is integral to Th development and differentiation.
Although the induction of Foxp3 in Th target cells through the TGF-β signaling pathway is implicated as a negative regulator of NFAT2 expression, TGF-β-dependent, Foxp3-independent pathways have been identified as well.23,24 There are multiple pathways leading to Foxp3-independent modulation of NFAT2 at the IL-2 promoter, including alterations of iCa2+, inhibition of nuclear translocation, decreased binding at the promoter site, and decreased transcription of NFAT2.16,25 Chen et al.16 demonstrated that increased iCa2+ following TCR stimulation promotes NFAT2 nuclear translocation and that TGF-β-mediated inhibition of Itk (a Tec kinase) activity reduces iCa2+, resulting in decreased NFAT2 nuclear translocation. However, Chen et al.16 also reported that if PMA/I were used to maintain intracellular Ca2+, TGF-β-mediated inhibition of NFAT2 nuclear translocation was reversed. Based upon these studies and our own preliminary findings, we hypothesized that Treg-mediated TGF-β signaling in target cells might reduce NFAT2 mRNA expression and thus IL-2 mRNA and protein production. We first had to identify the feline NFAT2 gene and Fig. 1 demonstrates that the feline NFAT2 exons share a high degree of homology with the human and mouse. As shown in Fig. 2, although NFAT2 mRNA, IL-2 mRNA, and IL-2 protein were increased following Th stimulation, Treg cells inhibited IL-2 but did not alter NFAT2 mRNA expression. Similarly, as shown in Fig. 3, soluble TGF-β failed to reduce NFAT2 mRNA expression.
To further address the potential role of TGF-β suppression of NFAT2 function we explored the possibility of altered NFAT2 binding to the IL-2 promoter. Stimulated Th cells were subjected to a soluble TGF-β suppression assay and analyzed by ChiP. As expected, Fig. 4 demonstrates that there was increased NFAT binding to the IL-2 promoter following Th stimulation. More importantly, Fig. 4b shows that NFAT2 binding to the IL-2 promoter was decreased by TGF-β treatment. As reduced NFAT2 binding following TGF-β suppression may have been due to competition from other repressive transcription factors or by epigenetic changes at the IL-2 promoter, we are currently investigating competitive binding by other transcription factors and are assessing histone acetylation and DNA methylation status at the IL-2 promoter.
Acknowledgments
This work was supported in part by funding from the following sources: 1K08AI074445-01A1 (J. Fogle) and R01 AI080288 (M. Tompkins). The authors thank Ms. Janet Dow and Ms. Deb Anderson for their excellent technical assistance and Dr. Jeff Yoder and Dr. Mike Sikes for their help with primer design and ChIP assays.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Mexas AM, Fogle JE, Tompkins WA, et al. : CD4+CD25+ regulatory T cells are infected and activated during acute FIV infection. Vet Immunol Immunopathol 2008;126:263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vahlenkamp T, Tompkins M, and Tompkins W: Feline immunodeficiency virus (FIV) infection phenotypically and functionally activates immunosuppressive CD4+CD25+ T regulatory (Treg) cells. J Immunol 2004;172:4752–4761 [DOI] [PubMed] [Google Scholar]
- 3.Petty CS, Tompkins MB, and Tompkins WA: Transforming growth factor-beta/transforming growth factor-betaRII signaling may regulate CD4+CD25+ T-regulatory cell homeostasis and suppressor function in feline AIDS lentivirus infection. J Acquir Immune Defic Syndr 2008;47:148–160 [DOI] [PubMed] [Google Scholar]
- 4.Fogle JE, Mexas AM, Tompkins WA, et al. : CD4(+)CD25(+) T regulatory cells inhibit CD8(+) IFN-gamma production during acute and chronic FIV infection utilizing a membrane TGF-beta-dependent mechanism. AIDS Res Hum Retroviruses 2010;26:201–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macian F: NFAT proteins: Key regulators of T-cell development and function. Nat Rev Immunol 2005;5:472–484 [DOI] [PubMed] [Google Scholar]
- 6.Chuvpilo S, Jankevics E, Tyrsin D, et al. : Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity 2002;16:881–895 [DOI] [PubMed] [Google Scholar]
- 7.Serfling E, Chuvpilo S, Liu J, et al. : NFATc1 autoregulation: A crucial step for cell-fate determination. Trends Immunol 2006;27:461–469 [DOI] [PubMed] [Google Scholar]
- 8.Chuvpilo S, Avots A, Berberich-Siebelt F, et al. : Multiple NF-ATc isoforms with individual transcriptional properties are synthesized in T lymphocytes. J Immunol 1999;162:7294–7301 [PubMed] [Google Scholar]
- 9.Zhou B, Cron RQ, Wu B, et al. : Regulation of the murine Nfatc1 gene by NFATc2. J Biol Chem 2002;277:10704–10711 [DOI] [PubMed] [Google Scholar]
- 10.Hogan PG, Chen L, Nardone J, et al.: Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 2003;17:2205–2232 [DOI] [PubMed] [Google Scholar]
- 11.Serfling E, Klein-Hessling S, Palmetshofer A, et al. : NFAT transcription factors in control of peripheral T cell tolerance. Eur J Immunol 2006;36:2837–2843 [DOI] [PubMed] [Google Scholar]
- 12.Vahlenkamp T, Bull M, Dow J, et al. : B7+CTLA4+ T cells engage in T-T cell interactions that mediate apoptosis: A model for feline immunodeficiency-induced T cell depletion. Vet Immunol Immunopathol 2004;98:203–214 [DOI] [PubMed] [Google Scholar]
- 13.Koltsova EK, Ciofani M, Benezra R, et al.: Early growth response 1 and NF-ATc1 act in concert to promote thymocyte development beyond the beta-selection checkpoint. J Immunol 2007;179:4694–4703 [DOI] [PubMed] [Google Scholar]
- 14.Lutz M. and Knaus P: Integration of the TGF-beta pathway into the cellular signalling network. Cell Signal 2002;14:977–988 [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K, Kitani A, Fuss I, et al. : TGF-beta 1 plays an important role in the mechanism of CD4+CD25+ regulatory T cell activity in both humans and mice. J Immunol 2004;172:834–842 [DOI] [PubMed] [Google Scholar]
- 16.Chen CH, Seguin-Devaux C, Burke NA, et al. : Transforming growth factor beta blocks Tec kinase phosphorylation, Ca2+ influx, and NFATc translocation causing inhibition of T cell differentiation. J Exp Med 2003;197:1689–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogle JE, Tompkins WA, and Tompkins MB: CD4+CD25+ T regulatory cells from FIV+ cats induce a unique anergic profile in CD8+ lymphocyte targets. Retrovirology 2010;7:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tompkins MB, Nelson PD, English RV, et al. : Early events in the immunopathogenesis of feline retrovirus infections. J Am Vet Med Assoc 1991;199:1311–1315 [PubMed] [Google Scholar]
- 19.Davidson MG, Rottman J, English RV, et al. : Feline immunodeficiency virus predisposes cats to acute generalized toxoplasmosis. Am J Pathol 1993;143:1486–1497 [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, Takeuchi A, and Sharma S: Characterization of a new isoform of the NFAT (nuclear factor of activated T cells) gene family member NFATc. J Biol Chem 1996;271:20914–20921 [DOI] [PubMed] [Google Scholar]
- 21.Chen ML, Pittet MJ, Gorelik L, et al. : Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-beta signals in vivo. Proc Natl Acad Sci USA 2005;102:419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Annunziato F, Cosmi L, Liotta F, et al. : Phenotype, localization, and mechanism of suppression of CD4(+)CD25(+) human thymocytes. J Exp Med 2002;196:379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettelli E, Dastrange M, and Oukka M: Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci USA 2005;102:5138–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaeth M, Schliesser U, Muller G, et al. : Dependence on nuclear factor of activated T-cells (NFAT) levels discriminates conventional T cells from Foxp3+ regulatory T cells. Proc Natl Acad Sci USA 2012;109:16258–16263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torgerson TR, Genin A, Chen C, et al.: FOXP3 inhibits activation-induced NFAT2 expression in T cells thereby limiting effector cytokine expression. J Immunol 2009;183:907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]