Abstract
The notoriously low fidelity of HIV-1 replication is largely responsible for the virus's rapid mutation rate, facilitating escape from immune or drug control. The error-prone activity of the viral reverse transcriptase (RT) is predicted to be the most influential mechanism for generating mutations. The low fidelity of RT has been successfully exploited by nucleoside and nucleotide analogue reverse transcriptase inhibitors (NRTIs) that halt viral replication upon incorporation. Consequently, drug-resistant strains have arisen in which the viral RT has an increased fidelity of replication, thus reducing analogue incorporation. Higher fidelity, however, impacts on viral fitness. The appearance of compensatory mutations in combination with higher fidelity NRTI resistance mutations and the subsequent reversion of NRTI-resistant mutations upon cessation of antiretroviral treatment lend support to the notion that higher fidelity exacts a fitness cost. Potential mechanisms for reduced viral fitness are a smaller pool of mutant strains available to respond to immune or drug pressure, slower rates of replication, and a limitation to the dNTP tropism of the virus. Unraveling the relationship between replication fidelity and fitness should lead to a greater understanding of the evolution and control of HIV.
Introduction
RNA viruses commonly exist as quasispecies, harboring enormous genetic diversity, primarily as a result of low replication fidelity. This diversity allows them to adapt to differing environments and to pressure from immune responses, antiviral drugs, and vaccines.1 Low replication fidelity is important for the survival of many RNA viruses. A poliovirus mutant with increased fidelity of replication was unable to adapt to adverse growth conditions2 and a mutant arbovirus with decreased genetic diversity was also attenuated.3 Herein, we discuss the fitness costs that arise from increased replication fidelity of HIV and the possible mechanisms underpinning these costs.
HIV-1 has a remarkably low fidelity of replication, resulting in rapid mutation and, consequently, the ability to rapidly escape control by the immune system, antiretroviral drugs, and vaccines.4 The sequences of HIV-1 genomes vary greatly, both between infected individuals and within an infected patient.5,6 The low fidelity of HIV replication is a result of the error-prone nature of the reverse transcriptase (RT), as well as numerous other potential sources of variation discussed below. The HIV RT lacks the proofreading ability of cellular polymerases and, despite sharing the structural elements of high-fidelity polymerases,7 it has a fidelity that is considerably lower than cellular RNA polymerases and also lower than other retroviral RTs.8,9 HIV RT's relatively high affinity for dNTPs is likely to underpin its error-prone polymerization.10
The low fidelity of HIV RT can be exploited with nucleoside and nucleotide reverse transcriptase inhibitors (referred to here collectively as NRTIs), which are analogues of natural nucleosides and nucleotides. NRTIs are less effective against host DNA and RNA polymerases, which have higher fidelity. Resistance to NRTIs is a significant challenge to the effective treatment of HIV, and many different NRTI-resistant strains of HIV-1 have been characterized.11 It is not surprising that among them are RTs that have a higher fidelity of replication, incorporating less of the NRTI than of natural nucleosides. Higher fidelity, however, comes at a cost to the virus, which is the main subject of this review.
Sources of Genetic Variation in HIV
The error-prone activity of RT is the most pertinent source of sequence variation to this review; however, there are a number of other potential sources of HIV-1 mutations. During reverse transcription, recombination occurs when RT transfers between the two RNA templates present in each virion, which leads to insertions or deletions at the point of transfer as well as recombinant viruses.12 Another source of error occurs after reverse transcription, when the viral genome is replicated by cellular RNA polymerases that make errors, albeit at a much lower rate than RT.8 Members of the APOBEC3 family of cellular proteins, particularly APOBEC3G, can also make mutations in the HIV-1 genome. Furthermore, the very large population of HIV-1 in an infected individual (estimated at 10.3×109 HIV virions/day) is expected to exacerbate these effects.13
The APOBEC3 family of cellular proteins inhibits retroviral pathogenesis by hypermutating the ssDNA copy or by blocking reverse transcription. APOBEC3G is the family member that most potently inhibited HIV-1 replication, at least under certain conditions.14 This cellular cytidine deaminase is incorporated into HIV virions where it ultimately leads to G-to-A mutations in the daughter genomic copies of the virus. In the absence of vif, multiple G-to-A mutations of HIV-1 cripple the virus.14 Vif, however, reduces the activity of APOBEC3G by promoting its ubiquitinization and degradation. The extent to which APOBEC3G contributes to genetic variation in HIV during the course of an infection is currently controversial, with some studies indicating that it contributes to variation by a sublethal level of mutagenesis,15 whereas other data are consistent with an “All or Nothing” phenomenon.16
Previously, the process of reverse transcription has been predicted to be the most error-prone step in the HIV replication cycle;17 however, these studies occurred prior to the characterization of APOBEC3G. This review focuses on the effects of higher fidelity RT mutants on viral fitness, but we note that the activity of APOBEC3G will likely have important consequences for viral fitness that should be better understood in the near future.
Reverse Transcription of HIV
The RT enzymes of retroviruses are unique among polymerases in that they use either an RNA or DNA template to make a DNA copy, culminating in a double-stranded DNA copy of the RNA genome of the virus. The RT of HIV performs two enzymatic activities: polymerization of DNA from template and degradation of the RNA template, performed by its RNase H domain. Unlike many eukaryote cellular polymerases, HIV RT contains no intrinsic proofreading capability.
The process of HIV reverse transcription, summarized here, has been the subject of recent, comprehensive reviews.4 HIV RT uses the genomic RNA (plus strand) and a cellular tRNA primer to synthesize the first strand of DNA. An RNA/DNA duplex is thus created, which is a substrate for the RNase H domain of RT. Once the first strand of DNA is synthesized, almost all the genomic RNA will be degraded, with the exception of two purine-rich sequences (polypurine tracts) that are resistant to RNase H cleavage. These short segments of RNA serve as primers for the synthesis of the second DNA strand and are eventually replaced with a DNA copy, finally resulting in the production of a double-stranded DNA copy of the virus.
The RT enzyme of HIV is a heterodimer of two subunits made from the same gene. The larger p66 subunit contains the two catalytic domains (polymerase and RNase H) and the smaller p51 subunit is believed to play a structural role.18 HIV-1 RT crystal structures have been useful in determining which regions of the RT directly influence polymerization and where incoming dNTPs are added to the newly synthesized DNA strand.18 The polymerase domain is often compared to a hand, consisting of “fingers,” “palm,” and “thumb”-like structures (Fig. 1). The end of the primer is positioned near the active site (the palm) where three negatively charged residues (D110, D185, and D186) interact with Mg2+ ions associated with the incoming dNTP. Specific amino acid residues within the palm, fingers, and thumb regions are involved in processes such as template binding, primer binding, and dNTP interactions. Pertinent to this review are the residues K65, L74, V148, and Q151, involved in dNTP interactions, M184, which directly interacts with the primer, and E89, involved in template interactions (Fig. 1).18 Specific mutations of these residues have been associated with a higher fidelity of replication (Table 1).
FIG. 1.
Detail of the structure of wild-type HIV-1 reverse transcriptase cross-linked to dsDNA and AZTppppA66 generated from the NCBI Structure database (MMDB ID:85000) using Cn3D. The main diagram shows the polymerase active site in detail, with the full structure in the insert. The amino acid residues described in Table 1 are in yellow. The green line depicts the position of the primer and the orange line the template. The upper left side pink domain depicts the “fingers,” the lower central pink domain is the “palm,” containing the active site, and the gray domain is labeled the “thumb.” Color images available online at www.liebertpub.com/aid
Table 1.
HIV-1 Reverse Transcriptase Mutants with Higher Fidelity
| Mutation in RT | NRTI Resistance | Assay and fidelity measure | Fidelity increase compared to WT |
|---|---|---|---|
| K65R |
AZT, 3TC, ddl, ddC, TDF11 |
Reporter gene assay: Misincorporation, lacZα reporter |
8-fold22 |
| |
|
Cell-based assay: Misincorporation, GFP reporter, measured by flow cytometry |
1.51-fold35 |
| |
|
Kinetics assay: Misinsertion of all incorrect nucleotides opposite A on a DNA template |
2.5- to 3.6-fold51 |
| |
|
Kinetics assay: Extension of mismatched G-T and A-G on a DNA template |
0.6- to 3-fold51 |
| M184V |
3TC, ddl, ddC11 |
Reporter gene assay: Misincorporation, lacZα reporter |
1.5- to 2.5-fold69 |
| |
|
Cell-based assay: Misincorporation, GFP reporter, measured by flow cytometry |
1.33-fold35 |
| |
|
Kinetics assay: Misinsertion of all incorrect nucleotides opposite C on a DNA template |
3.2-fold (average)70 |
| |
|
Kinetics assay: Misinsertion of T opposite G on a DNA template |
6.2-fold56 |
| M184I |
3TC, ddI11 |
Reporter gene assay: Misincorporation, lacZα reporter |
4-fold69 |
| |
|
Cell-based assay: Misincorporation, GFP reporter, measured by flow cytometry |
1.45-fold35 |
| |
|
Kinetics assay: Misinsertion of G opposite T on a DNA template |
8-fold56 |
| V148I |
dTTP analogues71 |
Reporter gene assay: Misincorporation, lacZα reporter |
8.7-fold45 |
| |
|
Cell-based assay: Misincorporation with virus particles, lacZα reporter |
5.7-fold45 |
| |
|
Cell-based assay: Misincorporation, GFP reporter, measured by flow cytometry |
1.96-fold35 |
| |
|
Kinetics assay: Extension of mismatched G-T on a DNA template |
24.4-fold45 |
| E89G |
ddG, AZT, 3TC11,72 |
Reporter gene assay: Misincorporation, lacZα reporter |
1.4- to 2-fold73 |
| |
|
Kinetics assay: Misinsertion of each possible incorrect nucleotide on a DNA template |
2- to 18.6-fold74 |
| L74V |
ddI, ddC11 |
Reporter gene assay: Misincorporation, lacZα reporter |
3.5- to 4.8-fold22,73 |
| |
|
Cell-based assay: Misincorporation, GFP reporter, measured by flow cytometry |
1.08-fold35 |
| Q151Na |
Multidrug resistant52 |
Reporter gene assay: Misincorporation, lacZα reporter |
13-fold45,46 |
| |
|
Cell-based assay: Misincorporation with virus particles, lacZα reporter |
3.8-fold45 |
| |
|
Kinetics assay: Extension of mismatched G-T on a DNA template |
15.9-fold45 |
| Kinetics assay: Misinsertion of all incorrect nucleotides opposite C and A on an RNA template | 8- to 26-fold52 |
Q151 is an important example of a higher fidelity RT mutant, but uncommon as an NRTI-resistant mutant.
AZT, 3′-azido-3′-deoxythymidine; 3TC, lamivudine (2′,3′-dideoxy-3′-thiacytidine); ddI, didanosine (2′,3′-dideoxyinosine); ddC, zalcitabine (2′,3′-dideoxycytidine); TDF, tenofovir, ddGTP: 2′,3′-dideoxyguanosine; WT, wild type; RT, reverse transcriptase.
Higher Fidelity as a Mechanism of NRTI Resistance
The low fidelity of HIV RT is exploited during antiretroviral therapy by the use of NRTIs. These drugs mimic natural nucleosides, but usually lack the 5′-OH group11 that is required for addition of the next nucleotide. While these drugs are quite effective in combination therapy, drug resistance can emerge during treatment if taken intermittently or as mono- or dual-therapy.19 Drug resistance is mediated by two general mechanisms: the removal of NRTIs from the newly synthesized DNA strand (pyrophosphorolysis) and an increase in the selectivity for correct dNTPs, which in some cases significantly change replication fidelity.20,21
NRTI resistance, mediated by increasing selectivity for dNTPs, can occur through increased rigidity in the RT's active site that decreases its affinity for analogues.22 This leads to increased selectivity for correct dNTPs, or a general reduction in dNTP affinity,10,22 resulting in a higher fidelity of replication. Many higher fidelity NRTI-resistant mutants have been characterized and their mechanisms of resistance explored (Table 1). The best characterized of these mutants is K65R, which commonly arises during treatment with zalcitabine (ddC), didanosine (ddI), lamivudine (3TC), or tenofovir (TDF)11 and has an estimated 8-fold increase in fidelity compared to the same HIV strain with no mutation at K65, in reporter gene assays.23 Table 1 also includes the Q151N mutant of RT that rarely arises in vivo, but is a well-characterized example of a mutant with higher fidelity. In terms of molecular biology, the consequences of increased fidelity in vitro include (1) a reduction in the amount of NRTI incorporated during polymerization, (2) a reduction in incorrect natural nucleotide incorporation, and generally (3) a decrease in the efficiency of reverse transcription. Selectivity increases fidelity, but the predicted cost of fidelity is viral fitness.
It is interesting to note that the process of pyrophosphorolysis, which removes incorporated NRTIs, may also have an impact on replication fidelity. During in vitro assays, pyrophosphorolysis was able to significantly influence the fidelity and selectivity of HIV-1 RT.24 As a consequence of pyrophosphorolysis, lesions are produced that need to be repaired or bypassed. Although not the main focus of this review, it should be noted that the repair and bypass processes at these lesions play a significant role in determining the ability of HIV RT to misincorporate nucleotides.24
Analyzing Fidelity Using in Vitro Assays
The overall fidelity of an enzyme involves the interaction of different factors that are defined in Table 2. The most important of these are misinsertion (insertion of an incorrect nucleotide or nucleotide analogue) and mismatched extension (polymerization past a misinsertion). A number of in vitro assays can provide specific information about these processes.
Table 2.
Definitions of key terms
| Term | Definition |
|---|---|
| Misinsertion |
The addition of the incorrect nucleotide during polymerization |
| Mismatched extension |
The polymerization past an incorrect nucleotide |
| Misincorporation |
Addition of the incorrect nucleotide and polymerization past it without repair |
|
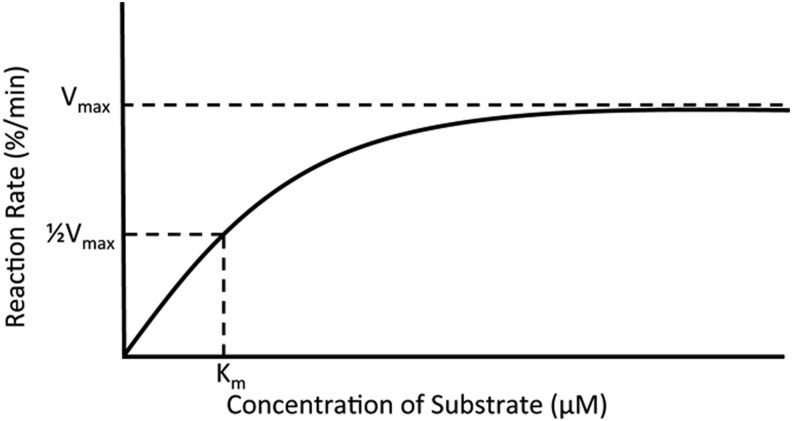
Vmax |
The maximum reaction rate of an enzyme in %/min (see Fig. 2) |
|
Km |
Substrate concentration required for ½ Vmax (see Fig. 2) |
| ƒins |
Efficiency of misinsertion; higher ƒins indicates a lower fidelity24 |
| Processivity |
The length of time the enzyme remains associated with the template/primer72 |
| Fitness |
Capacity of the virus to replicate (produce infectious progeny)1; this may be in the context of a given environment, relative to other strains of the virus |
| Fitness cost | The reduction in the replicative capacity of a virus associated with a particular mutation, relative to unmutated virus |
There are three broad categories of in vitro assay that have been used to quantitatively assess RT fidelity. First, there are assays that determine catalytic constants (i.e., Vmax, Km) and the efficiency of insertion or extension at a given nucleotide, referred to here as kinetic assays. Second, there are assays that use reporter gene constructs to assess the frequency of mutation, which we refer to as reporter gene assays. Third, there are “cell-based” assays that transfect cells with reporter genes and virus to measure the frequency of mutation. It should also be noted that qualitative biochemical assays have also been used, where the ability of RT to polymerize in the absence of a specific dNTPs is measured by gel electrophoresis. These assays do not produce quantitative values and have mostly been superseded by the ones discussed below.
Kinetics assays are cell free and combine a template, primer, and purified RT, to which dNTPs are added, allowing polymerization. Two kinetic parameters are determined from these reactions: Vmax, in %/min, which is defined as the maximum reaction rate of the enzyme, and Km, in μM, defined as the substrate concentration at which 1/2Vmax occurs (Fig. 2). These constants can be used to calculate ƒins, the efficiency of misinsertion (also referred to as frequency of misinsertion or fidelity of misinsertion), at a given nucleotide by the following equation:
 |
FIG. 2.
Hypothetical saturation curve for an enzyme reaction that gives the relationship between the catalytic constants Vmax and Km.
In this equation, the frequency of misinsertion is calculated as the ratio of the efficiency of insertion of the wrong nucleotide (W) compared to the right nucleotide (R).25 The efficiency of misinsertion is used as a measure of fidelity, where a high ƒins indicates a low fidelity. Insertion efficiency beyond a mismatched template/primer can be quantified using similar methods.26 The efficiency of misinsertion and efficiency of extension beyond a misinsertion contribute to fidelity. However, fidelity is multifactorial and cannot be calculated by these two factors alone.
Reporter gene assays measure misincorporation during polymerization of an entire gene. The most common reporter gene assay is the M13-based forward mutation lacZα assay. The RT of interest is combined with bacteriophage DNA that has a section of single-stranded DNA over the length of the reporter gene (lacZα).27 The enzyme polymerizes over the gap and the resulting products are transfected into bacteria. Error-free synthesis is observed by the dark blue color of these colonies. Light blue or colorless colonies are analyzed by sequencing their lacZα gene to detect mutations.28 However, only mutations that result in the loss of LacZα protein function are reported, since errors that do not interfere with the function of lacZα are not selected for.
Cell-based assays employ reporter gene constructs in a way similar to the cell-free reporter gene assays described above. In these assays, cell lines are transfected with the reporter gene construct, such as lacZα, as part of the vector virus sequence.29 Cells are then analyzed for phenotype changes by observation of color change (as above), or by flow cytometry in the case of fluorescent reporters. Once again, these assays detect only changes that affect report gene function. However, they have the advantage of including viral and host cell factors that interact with the RT complex, affecting its function.30
While these assays provide important quantitative data on the fidelity of specific mutant RTs, there are limitations to these techniques. All of these assays employ templates that are not reflective of the HIV genome. Kinetic and cell-free reporter gene assays use purified RT where viral and host cell factors that are part of the in vivo RT complex are absent.30 This is perhaps reflected by studies showing that the purified RT has a higher mutation rate than whole virus in vivo.31
The range of methods and reagents (primers, templates, reporter genes) in these assays varies widely, which may explain the variation in fidelity data for a given mutant, as described in Table 1. Despite this variation, these assays have an encouraging consensus; experiments using each assay suggested that the RT mutants described in the table have a higher fidelity than “wild-type” (unmutated) RT.
Lower Viral Fitness: The High Cost of Fidelity
The capacity of a virus to produce infectious progeny (replicate), in comparison to another strain of the same virus, is termed the “fitness” of the virus. Where a mutation reduces the relative replication capacity of a virus this is referred to as a “fitness cost.” Fitness costs can be measured in a number of increasingly sophisticated ways. On a simple level, replication kinetics of HIV variants in cell lines can be compared, although this is relatively insensitive to subtle fitness costs. A competition assay, where two viruses are seeded into tissue culture for a period of time and the “winning” viral strain is detected at the end of the culture, is a fairly sensitive assay, particularly if performed in primary blood CD4 T cells rather than cell lines.32 An effective but expensive method to assess fitness is to observe the reversion of particular mutations in vivo in macaque infection experiments with SIV or chimeric SHIV viruses.33 These experiments more accurately reflect the total fitness impact of any given mutation. One final in vivo method that is useful for testing the fitness cost of drug-resistant mutants involves monitoring the decay of drug-resistant virus in patients who have stopped treatment with a specific antiretroviral.34,35 The main advantage of these studies is that they translated readily to clinical situations because they involved humans. However, only drug-resistant mutants can be tested in this fashion, whereas compensatory mutations also need to be taken into account.
There is evidence that altering the fidelity of the RT, by either increasing or decreasing nucleotide selectivity, is detrimental to viral fitness. It has become increasingly clear that increases in fidelity always come with a fitness cost, as higher fidelity mutants replicated more slowly in cell lines.35,36 In vivo studies in both humans and SIV-infected macaques showed that the M184V/I,34,37 K65R,35 and E89G38 mutants were less fit than “wild-type” viruses and often reverted to wild-type in the absence of drug treatment. The in vivo fitness costs of the M184I and M184V are particularly interesting, as the M184I mutation appeared early but transiently during 3TC treatment and was replaced by the M184V mutant.37 The M184I mutant had a higher fidelity than M184V (Table 1) and these data indicate that it had a greater fitness cost compared to the lower fidelity M184V. A comprehensive comparison of the fitness cost associated with changes to HIV fidelity found a significant positive correlation between fidelity and fitness cost for high-fidelity HIV mutants, and interestingly, showed that low fidelity was also associated with lower viral fitness.36
Compensatory mutations are additional changes to the viral genome that are predicted to offset the fitness costs of an initial mutation. They often arise in the presence of selection pressures such as immune responses38 or drug treatment39 and are associated with the presence of known drug escape mutants. Compensatory mutations do not confer resistance themselves, but they can restore partial replicative fitness.40 Examples of this are the A62V and S68G mutations that increase the fitness of the K65R mutant of RT.40 Many higher fidelity mutants are associated with compensatory mutations,39,41 supporting the prediction that increased fidelity comes at a cost.
It is likely that the level of HIV-1 replication fidelity that achieves optimal fitness is actually a balance, whereby too high fidelity as well as too low fidelity decreases fitness. It is predicted that mutation rates of RNA viruses such as HIV sit near a critical “error threshold” considered to be near a theoretical “extinction threshold.”42,43 The effect of lowering fidelity further is predicted to make the genome unstable, producing too many nonviable HIV genomes. Certainly there is evidence that too many mutations can decrease the fitness of a quasispecies, as demonstrated in studies of the aforementioned APOBEC3G. In the absence of Vif, HIV could not suppress the activity of APOBEC3G and underwent hypermutation, crippling the virus.44 Dapp et al.45 demonstrated a negative correlation between mutation frequency and viral fitness for low-fidelity mutant HIV strain and that even relatively small increases in mutation rates decreased fitness. Further evidence comes from the Q151N mutation, which has a very high fidelity compared to other RT mutants (Table 1).46 RT carrying Q151N has 13-fold lower incorporation of incorrect endogenous nucleotides47 during lacZα reporter gene assays. This is likely due to its 15.9-fold decreased ability to extend past a mismatch in kinetics assays.46 Q151N was created in a mutagenesis study, based on the Q151M NRTI-resistant mutation that has a small increase in fidelity. The Q151N version is also drug resistant, but rarely arises in patients, perhaps indicating that too high fidelity is untenable.
Potential Mechanisms Underpinning the Fitness Costs of Higher Fidelity
Studies of the fitness of higher fidelity mutants, together with the assays describing changes to RT fidelity outlined above, have provided insights into the mechanisms behind the fitness costs of higher fidelity. We discuss three potential mechanisms for the decreased fitness of higher fidelity HIV-1: (1) a smaller pool of mutations available to facilitate escape from immune or drug control, (2) a reduced processivity and/or rate of replication, and (3) a lowered affinity for dNTPs that may limit the tropism of the virus.
A reduced pool of mutations available to facilitate escape from immune responses and from drug treatments is the logical outcome of an increase in fidelity of replication, although a demonstration that this occurs in vivo is currently lacking. A large pool of mutations would certainly be an advantage in the presence of drugs or a healthy immune system, and it makes vaccination against HIV very difficult. Many studies that tracked the escape of HIV during infection from immune pressures such as neutralizing antibodies, CTL responses, and antibody-dependent cellular cytotoxicity showed that specific mutations are commonly associated with escape from these immune pressures.48–50 Similarly, a plethora of drug-resistant mutations consistently arises during treatment.11 It is logical to predict that a virus that produces a smaller pool of mutations due to higher fidelity will have a reduced capacity to overcome these pressures.
A high-fidelity mutant may not have reduced fitness in hosts with depleted immune systems. Supporting this point is the discovery of a naturally occurring higher fidelity RT mutant, V148I, which is in a conserved region of the simian immunodeficiency virus.51 This V148I mutation arose in the absence of drug treatment during the late stage of infection where immune pressure was diminished due to CD4 T cell depletion and clonal exhaustion. It is possible that under these conditions, a higher fidelity of replication may reduce the number of nonviable mutant viruses produced.
A second potential mechanism underpinning the reduced fitness of higher fidelity RT mutants is a reduction in the rate of replication and/or processivity of the RT. An RT mutant that is more selective for correct dNTPs is likely to polymerize DNA more slowly. Numerous studies that determined a lower Vmax or Km (Table 2) for many higher fidelity RTs supported this prediction.52,53 Furthermore, many higher fidelity mutant RTs have been shown to have a lower processivity,47,54,55 which is defined as the length of time the enzyme remains associated with the nucleic acids. Processivity, however, is not a direct measure of the rate of replication, and strand transfer can occur when the RT dissociates, continuing polymerization.12 Our understanding of the relationship between fidelity and processivity is evolving. Although there is evidence for a negative relationship between processivity and fidelity of M184 mutants,56,57 similar studies with RTs harboring L74V and E89G found no such relationship.58 Furthermore, emerging evidence suggests that the apparent processivity defects of K65R, M184, and Q151N may be due to the altered dNTP affinity of these mutants.55,59,60
Finally, a reduction in dNTP affinity of some higher fidelity mutants is proposed to limit the tropism of HIV, thereby reducing the fitness of HIV in vivo. The increased selectivity of high fidelity mutants is often modulated by active site changes that lead to a decrease in dNTP affinity.22,61 The consequences of this have been analyzed in the context of both polymerization kinetics and virus tropism. Cell-based assays have shown that the Q151N and M184V/I mutants had severely reduced growth in primary cells with low dNTP concentrations.56,60 The K65R mutant also had a lower affinity for dNTPs, but its ability to infect cells with lower dNTP concentrations remains poorly characterized. Since HIV-1 can target cells such as macrophages that contain low dNTP levels, a lower dNTP affinity is predicted to limit viral tropism in vivo, thereby reducing the fitness of the virus. In this manner, the lower dNTP affinity associated with higher fidelity may have a specific selective disadvantage, in addition to the general impact on the rate of replication.
The Next Generation of Fidelity Analysis: High-Throughput Sequencing
While assays that measure the biochemical properties of RT and the cell-based LacZα assays quantifying mutation rates have provided important mechanistic information, a measure that analyzes total misincorporation during HIV replication in vivo would greatly contribute to our understanding of the relationship between fidelity and fitness. Prior to the emergence of high-throughput “Next-Generation Sequencing” technologies, the direct detection of viral genome mutations involved cloning and sequencing individual genomes, a process that is time consuming, expensive and yields limited data, in that only a very small proportion of genotypes present in the plasma of a patient, for example, would be analyzed. High-throughput sequencing now provides the means to take a “snapshot” of the entire viral population at a given time.62,63 Sequencing platforms such as Illumina, SOLiD, and Ion Torrent generate millions of sequencing reads per run, providing the depth necessary to theoretically analyze all of the HIV genomes present in a patient sample.64 These techniques are currently revolutionizing the study of HIV sequence diversity, with applications ranging from drug resistance monitoring to exploring the total antibody response,64,65 but they have yet to be applied to questions of viral fidelity in published studies.
There are technical challenges that need to be overcome to accurately measure the replication fidelity of different HIV strains in vivo. If higher fidelity mutants are less fit, then they will likely produce fewer viral particles than wild-type virus. Consequently, they may produce fewer mutations as a result of decreased replication rather than an increase in fidelity. Similarly, fitness costs increase the likelihood of reversion, especially in vivo, and a mixed population of WT and RT fidelity mutants would complicate fidelity measurements. Nevertheless, high-throughput sequencing has enormous potential and promises a much greater exploration of the genomic population of higher fidelity mutants and is the key to increasing our understanding of the cost of fidelity.
High-Fidelity Viruses as Live-Attenuated Vaccines
Live attenuated vaccines have proven effective in the prevention of many viral diseases such as smallpox, polio, and measles.42 Early vaccine strains were derived empirically through passaging in cell lines; however, rational attenuation of viruses is now a common approach. RNA viruses that harbor extensive genetic diversity are particularly challenging candidates as they are more likely to escape the attenuation through mutation, and low-frequency mutants may escape vaccine-elicited immune responses. Attenuation of RNA viruses by increasing their replication fidelity is one possible strategy for designing live-attenuated viruses.42 One example is a poliovirus variant with increased replication fidelity, which as a live-attenuated vaccine successfully protected mice from lethal doses of poliovirus.66 An interesting alternative to high-fidelity live-attenuated vaccine design is a severe acute respiratory syndrome (SARS)-coronavirus vaccine candidate that has a reduced fidelity of replication, sufficient to attenuate the virus while affording protection in a mouse model.67
It is possible that a mutant HIV strain with increased replication fidelity could make an effective live-attenuated vaccine. However, use of a live-attenuated HIV vaccine in humans seems unlikely to become a reality due to safety and efficacy concerns. Regardless, studies of live-attenuated SIV vaccines in macaques have provided useful information on the types of immune responses necessary to prevent SIV infection,68,69 making an important contribution to the understanding of effective vaccine-induced immunity to HIV.
Conclusions
A higher fidelity of HIV-1 replication is predicted to come at a cost to the fitness of the virus. Overall, the level of HIV RT fidelity is likely to be a balance in which too high fidelity leads to a reduction in viral fitness but too low fidelity is also detrimental to the propagation of the virus. Manipulation of viral fidelity is therefore a possible route for controlling the virus. Understanding the relationship between fidelity and viral fitness will lead to a better appreciation of HIV evolution and the potential for controlling the virus. This will certainly be enhanced by the application of high-throughput sequencing to study the fidelity of replication in vivo.
Acknowledgments
We thank Dr. Con Sonza, Dr. Matthew Parsons, and Ms. Caroline Fernandez for advice on the manuscript. This work was supported by NHMRC fellowships (1013221 to W.R.W. and 1041832 to S.J.K.), the NHMRC program grant 510448, and an Early Career Researcher grant from the University of Melbourne.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Lauring AS, Frydman J, and Andino R: The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 2013;11(5):327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, and Andino R: Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006;439(7074):344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffey LL, Beeharry Y, Borderia AV, Blanc H, and Vignuzzi M: Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci USA 2011;108(38):16038–16043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu WS. and Hughes SH: HIV-1 reverse transcription. Cold Spring Harb Perspect Med 2012;2(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin L, Liu L, Sun YJ, et al.: High-resolution deep sequencing reveals biodiversity, population structure, and persistence of HIV-1 quasispecies within host ecosystems. Retrovirology 2012;9:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibo D. and Birch C: Increasing diversity of human immunodeficiency virus type 1 subtypes circulating in Australia. AIDS Res Hum Retroviruses 2012;28(6):578–583 [DOI] [PubMed] [Google Scholar]
- 7.Scott JG, Tae Woo K, Michael AP, Eric TK, and Vinayaka RP: Site-directed mutagenesis in the fingers subdomain of HIV-1 reverse transcriptase reveals a specific role for the ß3-ß4 hairpin loop in dNTP selection. J Mol Biol 2007;365:38–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neil PK, Sun G, Yu H, Ron Y, Dougherty JP, and Preston BD: Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J Biol Chem 2002;277(41):38053–38061 [DOI] [PubMed] [Google Scholar]
- 9.Redmond PS, Miles PD, and Johnson M: The origin of genetic diversity in HIV-1. Virus Res 2012;169(2):415–429 [DOI] [PubMed] [Google Scholar]
- 10.Weiss KK, Bambara RA, and Kim B: Mechanistic role of residue Gln151 in error prone DNA synthesis by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) Pre-steady state kinetic study of the Q151N HIV-1 RT mutant with increased fidelity. J Biol Chem 2002;277(25):22662–22669 [DOI] [PubMed] [Google Scholar]
- 11.Menendez-Arias L: Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res 2008;134(1–2):124–146 [DOI] [PubMed] [Google Scholar]
- 12.Delviks-Frankenberry K, Galli A, Nikolaitchik O, Mens H, Pathak VK, and Hu WS: Mechanisms and factors that influence high frequency retroviral recombination. Viruses 2011;3(9):1650–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perelson AS, Neumann AU, Markowitz M, Leonard JM, and Ho DD: HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science 1996;271(5255):1582–1586 [DOI] [PubMed] [Google Scholar]
- 14.Goila-Gaur R. and Strebel K: HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 2008;5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadler HA, Stenglein MD, Harris RS, and Mansky LM: APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol 2010;84(14):7396–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armitage AE, Deforche K, Chang C-H, Wee E, et al.:. APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete “all or nothing” phenomenon. PLoS Genet 2012;8(3)1553–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menendez-Arias L: Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog Nucleic Acid Res Mol Biol 2002;71:91–147 [DOI] [PubMed] [Google Scholar]
- 18.Le Grice SF: Human immunodeficiency virus reverse transcriptase: 25 years of research, drug discovery, and promise. J Biol Chem 2012;287(49):40850–40857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bershteyn A. and Eckhoff PA: A model of HIV drug resistance driven by heterogeneities in host immunity and adherence patterns. BMC Syst Biol 2013;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isel C, Ehresmann C, Walter P, Ehresmann B, and Marquet R: The emergence of different resistance mechanisms toward nucleoside inhibitors is explained by the properties of the wild type HIV-1 reverse transcriptase. J Biol Chem 2001;276(52):48725–48732 [DOI] [PubMed] [Google Scholar]
- 21.Arts EJ. and Hazuda DJ: HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 2012;2(4):a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris D, Kaushik N, Pandey PK, Yadav PN, and Pandey VN: Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J Biol Chem 1998;273(50):33624–33634 [DOI] [PubMed] [Google Scholar]
- 23.Shah FS, Curr KA, Hamburgh ME, et al.: Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem 2000;275(35):27037–27044 [DOI] [PubMed] [Google Scholar]
- 24.Sharma B, Crespan E, Villani G, and Maga G: The balance between the rates of incorporation and pyrophosphorolytic removal influences the HIV-1 reverse transcriptase bypass of an abasic site with deoxy-, dideoxy-, and ribonucleotides. Proteins 2008;71(2):715–727 [DOI] [PubMed] [Google Scholar]
- 25.Echols H. and Goodman MF: Fidelity mechanisms in DNA replication. Annu Rev Biochem 1991;60:477–511 [DOI] [PubMed] [Google Scholar]
- 26.Goodman MF, Creighton S, Bloom LB, and Petruska J: Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol 1993;28(2):83–126 [DOI] [PubMed] [Google Scholar]
- 27.Loeb LA. and Kunkel TA: Fidelity of DNA synthesis. Annu Rev Biochem 1982;51:429–457 [DOI] [PubMed] [Google Scholar]
- 28.Bebenek K. and Kunkel TA: Analyzing fidelity of DNA polymerases. Methods Enzymol 1995;262:217–232 [DOI] [PubMed] [Google Scholar]
- 29.Mansky LM, Le Rouzic E, Benichou S, and Gajary LC: Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J Virol 2003;77(3):2071–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warrilow D, Tachedjian G, and Harrich D: Maturation of the HIV reverse transcription complex: Putting the jigsaw together. Rev Med Virol 2009;19(6):324–337 [DOI] [PubMed] [Google Scholar]
- 31.Mansky LM. and Temin HM: Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol 1995;69(8):5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marozsan AJ, Moore DM, Lobritz MA, et al.: Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. Virology 2005;79(11):7121–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh L, Batten CJ, Petravic J, Davenport MP, and Kent SJ: In vivo fitness costs of different Gag CD8 T-cell escape mutant simian-human immunodeficiency viruses for macaques. Virology 2007;81(10):5418–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paredes R, Sagar M, Marconi VC, et al.: In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J Virol 2009;83(4):2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber J, Chakraborty B, Weberova J, Miller MD, and Quinones-Mateu ME: Diminished replicative fitness of primary human immunodeficiency virus type 1 isolates harboring the K65R mutation. J Clin Microbiol 2005;43(3):1395–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dapp MJ, Heineman RH, and Mansky LM: Interrelationship between HIV-1 fitness and mutation rate. J Mol Biol 2012;425(1):41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuurman R, Nijhuis M, van Leeuwen R, et al.: Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J Infect Dis 1995;171(6):1411–1419 [DOI] [PubMed] [Google Scholar]
- 38.Newstein MC. and Desrosiers RC: Effects of reverse-transcriptase mutations M184V and E89G on simian immunodeficiency virus in rhesus monkeys. J Infect Dis 2001;184(10):1262–1267 [DOI] [PubMed] [Google Scholar]
- 39.Trevino A, de Mendoza C, Caballero E, et al.: Drug resistance mutations in patients infected with HIV-2 living in Spain. J Antimicrob Chemother 2011;66(7):1484–1488 [DOI] [PubMed] [Google Scholar]
- 40.Svarovskaia ES, Feng JY, Margot NA, et al.: The A62V and S68G mutations in HIV-1 reverse transcriptase partially restore the replication defect associated with the K65R mutation. J Acquir Immune Defic Syndr 2008;48(4):428–436 [DOI] [PubMed] [Google Scholar]
- 41.Cong ME, Heneine W, and Garcia-Lerma JG: The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J Virol 2007;81(6):3037–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauring AS, Jones JO, and Andino R: Rationalizing the development of live attenuated virus vaccines. Nat Biotechnol 2010;28(6):573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dapp MJ, Patterson SE, and Mansky LM: Back to the future: Revisiting HIV-1 lethal mutagenesis. Trends Microbiol 2013;21(2):56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lecossier D, Bouchonnet F, Clavel F, and Hance AJ: Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 2003;300(5622):1112. [DOI] [PubMed] [Google Scholar]
- 45.Dapp MJ, Heineman RH, and Mansky LM: Interrelationship between HIV-1 fitness and mutation rate. J Mol Biol 2013;425(1):41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss KK, Chen R, Skasko M, Reynolds HM, et al.: A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry 2004;43(15):4490–4500 [DOI] [PubMed] [Google Scholar]
- 47.Weiss KK, Isaacs SJ, Tran NH, Adman ET, and Kim B: Molecular architecture of the mutagenic active site of human immunodeficiency virus type 1 reverse transcriptase: Roles of the beta 8-alpha E loop in fidelity, processivity, and substrate interactions. Biochemistry 2000;39(35):10684–10694 [DOI] [PubMed] [Google Scholar]
- 48.Richman DD, Wrin T, Little SJ, and Petropoulos CJ: Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA 2003;100(7):4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung AW, Isitman G, Navis M, et al.: Immune escape from HIV-specific antibody-dependent cellular cytotoxicity (ADCC) pressure. Proc Natl Acad Sci USA 2011;108(18):7505–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goulder PJ. and Watkins DI: HIV and SIV CTL escape: Implications for vaccine design. Nat Rev Immunol 2004;4(8):630–640 [DOI] [PubMed] [Google Scholar]
- 51.Diamond TL, Souroullas G, Weiss KK, et al.: Mechanistic understanding of an altered fidelity simian immunodeficiency virus reverse transcriptase mutation, V148I, identified in a pig-tailed macaque. J Biol Chem 2003;278(32):29913–29924 [DOI] [PubMed] [Google Scholar]
- 52.Garforth SJ, Domaoal RA, Lwatula C, et al.: K65R and K65A substitutions in HIV-1 reverse transcriptase enhance polymerase fidelity by decreasing both dNTP misinsertion and mispaired primer extension efficiencies. J Mol Biol 2010;401(1):33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaushik N, Talele TT, Pandey PK, Harris D, Yadav PNS, and Pandey VN: Role of glutamine 151 of human immunodeficiency virus type-1 reverse transcriptase in substrate selection as assessed by site-directed mutagenesis. Biochemistry 2000;39(11):2912–2920 [DOI] [PubMed] [Google Scholar]
- 54.Sharma PL, Nettles JH, Feldman A, Rapp K, and Schinazi RF: Comparative analysis of in vitro processivity of HIV-1 reverse transcriptases containing mutations 65R, 74V, 184V and 65R+74V. Antiviral Res 2009;83(3):317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H-T, Martinez-Cajas JL, Ntemgwa ML, et al.: Effects of the K65R and K65R/M184V reverse transcriptase mutations in subtype C HIV on enzyme function and drug resistance. Retrovirology 2009;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Back NKT, Nijhuis M, Keulen W, et al.: Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J 1996;15(15):4040–4049 [PMC free article] [PubMed] [Google Scholar]
- 57.Oude Essink BB, Back NK, and Berkhout B: Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res 1997;25(16):3212–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Avidan O. and Hizi A: The processivity of DNA synthesis exhibited by drug-resistant variants of human immunodeficiency virus type-1 reverse transcriptase. Nucleic Acids Res 1998;26(7):1713–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao L, Hanson MN, Balakrishnan M, et al.: Apparent defects in processive DNA synthesis, strand transfer, and primer elongation of Met-184 mutants of HIV-1 reverse transcriptase derive solely from a dNTP utilization defect. J Biol Chem 2008;283(14):9196–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amie SM, Noble E, and Kim B: Intracellular nucleotide levels and the control of retroviral infections. Virology 2013;436(2):247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silverman AP, Garforth SJ, Prasad VR, and Kool ET: Probing the active site steric flexibility of HIV-1 reverse transcriptase: Different constraints for DNA- versus RNA-templated synthesis. Biochemistry 2008;47(16):4800–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gall A, Ferns B, Morris C, et al.: Universal amplification, next-generation sequencing and assembly of HIV-1 genomes. J Clin Microbiol 2012; 50(12):3838–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beerenwinkel N, Gunthard HF, Roth V, and Metzner KJ: Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front Microbiol 2012;3:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gall A, Ferns B, Morris C, et al.: Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J Clin Microbiol 2012;50(12):3838–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu J, Ofek G, Yang Y, et al.: Mining the antibodyome for HIV-1-neutralizing antibodies with next-generation sequencing and phylogenetic pairing of heavy/light chains. Proc Natl Acad Sci USA 2013;110(16):6470–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vignuzzi M, Wendt E, and Andino R: Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med 2008;14(2):154–161 [DOI] [PubMed] [Google Scholar]
- 67.Graham RL, Becker MM, Eckerle LD, Bolles M, Denison MR, and Baric RS: A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med 2012;18(12):1820–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alpert MD, Harvey JD, Lauer WA, et al.: ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS Pathog 2012;8(8):e1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manrique J, Piatak M, Lauer W, et al.: Influence of mismatch of Env sequences on vaccine protection by live attenuated simian immunodeficiency virus. J Virol 2013;87(13):7246–7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rezende LF, Drosopoulos WC, and Prasad VR: The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Res 1998;26(12):3066–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wainberg MA, Drosopoulos WC, Salomon H, et al.: Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 1996;271(5253):1282–1285 [DOI] [PubMed] [Google Scholar]
- 72.Jamburuthugoda VK, Guo D, Wedekind JE, and Kim B: Kinetic evidence for interaction of human immunodeficiency virus type 1 reverse transcriptase with the 3’-OH of the incoming dTTP substrate. Biochemistry 2005;44(31):10635–10643 [DOI] [PubMed] [Google Scholar]
- 73.Quan Y, Inouye P, Liang C, Rong L, Gotte M, and Wainberg MA: Dominance of the E89G substitution in HIV-1 reverse transcriptase in regard to increased polymerase processivity and patterns of pausing. J Biol Chem 1998;273(34):21918–21925 [DOI] [PubMed] [Google Scholar]
- 74.Jonckheere H, De Clercq E, and Anne J: Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur J Biochem 2000;267(9):2658–2665 [DOI] [PubMed] [Google Scholar]
- 75.Drosopoulos WC. and Prasad VR: Increased polymerase fidelity of E89G, a nucleoside analog-resistant variant of human immunodeficiency virus type 1 reverse transcriptase. J Virol 1996;70(7):4834–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]