Abstract
We assessed the immunovirological response to antiretroviral regimens containing maraviroc in HIV-infected viremic patients with viral tropism predicted by different assays. We selected antiretroviral treatment-experienced HIV-1-infected patients initiating regimens containing maraviroc after different phenotypic or genotypic viral tropism assays, with at least one HIV-1 RNA determination during follow-up. Survival analysis was employed to assess the virological response as time to HIV-1 RNA <50 copies/ml and immunological response as time to a CD4 cell count increase of ≥100/μl from baseline. Predictors of these outcomes were analyzed by multivariate Cox regression models. In 191 treatments with maraviroc, virological response was achieved in 65.4% and the response was modestly influenced by the baseline viral load and concomitant drug activity but not influenced by the type of tropism assay employed. Immunological response was achieved in 58.1%; independent predictors were baseline HIV-1 RNA (per log10 higher: HR 1.29, 95% CI 1.05–1.60) and concomitant therapy with enfuvirtide (HR 2.05, 0.96–4.39) but not tropism assay results. Of 17 patients with baseline R5-tropic virus and available tropism results while viremic during follow-up on maraviroc, seven (41%) showed a tropism switch to non-R5 virus. A significant proportion of experienced patients treated with regimens containing maraviroc achieved virological response. The tropism test type used was not associated with immunovirological response and concomitant treatment with enfuvirtide increased the chance of immunological response. More than half of virological failures with maraviroc were not accompanied by tropism switch.
Introduction
Six different classes of antiretroviral drugs have been developed that target HIV-1 replication at different stages. Among these, chemokine receptor 5 (CCR5) antagonists selectively inhibit the entry into host cells of CCR5-using (R5) HIV-1 strains by an allosteric mechanism after binding to the transmembrane CCR5 coreceptor cavity.1–6
Current European guidelines indicate a mandatory coreceptor tropism test in all cases in which a CCR5 antagonist is being considered as part of the subsequent regimen, such as virological failure, the need to change a successful regimen because of toxicity or inconvenience, and treatment of drug-naive patients in whom toxicity to common first-line treatments is expected.7
Several methodologies for determining HIV-1 coreceptor tropism are available, including genotypic and phenotypic approaches, but actually there is no diagnostic gold standard.7,8 Among the phenotypic tropism tests, the original Trofile assay (Monogram Biosciences, San Francisco, CA) allowed detection of CXCR4-using (X4) strains at a prevalence of ≥10% of the viral quasispecies. Since June 2008, a more sensitive assay version, the “enhanced sensitivity Trofile assay” (ESTA), has been introduced in clinical routine. This version increased the detection limit of minority X4 strains down to 0.3%.9–11 On the other hand, genotypic tropism testing is based on amplification and sequencing of the gp120 V3 loop region and its interpretation using several bioinformatic algorithms such as the position-specific scoring matrix (PSSM) and the geno2pheno[coreceptor] (G2P) system.12
Few observational studies have previously investigated the virological and immunological response to antiretroviral treatment (ART) regimens containing maraviroc in HIV-1-infected patients and their association with patient-related and virologic variables.13,14
Here we present the immunovirological outcome of patients undergoing maraviroc-based treatment in clinical practice where coreceptor tropism was determined by different assays. Correlates of immunological and virological responses were also analyzed and are presented here.
Materials and Methods
Study design and patients
We retrospectively examined HIV-1-infected patients initiating maraviroc-containing ART regimens between July 2005 and April 2011. These were all treatment-experienced patients, enrolled in the Antiretroviral Resistance Cohort Analysis (ARCA), a national observational cohort of HIV-1-infected patients followed by >100 clinical and laboratory units in Italy (www.hivarca.net). All patients were anonymous and were included in the ARCA database after signing an informed consent to provide their data for academic not-for-profit studies. The ARCA initiative is compliant with the Declaration of Helsinki and each participating center is subject to a local Ethics Committee that follows national (and where applicable European) regulations. Additional inclusion criteria for the study were availability of plasma HIV-1 RNA load within 120 days prior to maraviroc treatment initiation and of at least one HIV-1 RNA determined subsequent to maraviroc treatment initiation.
The following variables were retrieved for all enrolled patients from the information available in the ARCA database, using the date of maraviroc treatment initiation as the baseline time point: calendar year, age, gender, nation of birth, viral subtype, time since HIV diagnosis, baseline HIV-1 RNA, baseline CD4+ T cell count, nadir CD4+ T cell count, mode of HIV-1 transmission, time from the first HIV-1-positive antibody test to the first ART initiation, duration of prior antiretroviral exposure, number of previous antiretroviral drugs employed, and number of previous antiretroviral treatment lines employed.
Determination of viral tropism and calculation of the genotypic susceptibility to drugs accompanying maraviroc
All patients underwent testing for HIV-1 coreceptor tropism with the use of at least one of the following assays: a phenotypic assay, namely Trofile or ESTA, or a genotypic assay. Some patients underwent a combined viral tropism assessment using both a genotypic and a phenotypic assay. Genotypic analysis of the nucleotide sequence of env coding for the gp120 V3 region was performed in a single assay by population sequencing and results were interpreted using the clonal G2P prediction algorithm; the reported false positive rate (FPR) was used as quantitative output. The cut-off FPR for discriminating between R5 and X4 use was set at 10%, adopting a modification of the European tropism guidelines, following observations that this threshold would not significantly affect R5 detection accuracy even by single testing.12,15 X4 and dual/mixed (D/M) tropic viruses by Trofile or ESTA were cumulatively categorized as non-R5.
The baseline HIV-1 pol reverse transcriptase and protease genotype was processed by calculating the genotypic susceptibility score (GSS) using the latest available version from the Rega interpretation system (Rega 8.0.2) with respect to the antiretrovirals associated with maraviroc. We used the standard susceptible/intermediate/resistant categorization for all antiretrovirals used in combination with maraviroc, as by the output of Rega interpretation given by the HIVdb web-service (http://sierra2.stanford.edu/sierra/servlet/JSierra?action=hivalgs).
Individual drugs were assigned the following numerical susceptibility values, as suggested by the weighted Rega interpretation system: 0 for all drugs to which the virus was interpreted as resistant, 0.25 for intermediate resistant nevirapine and efavirenz, 0.5 for intermediate resistant nucleoside reverse transcriptase inhibitors (NRTIs), etravirine, and unboosted protease inhibitors (PIs), 0.75 for intermediate resistant boosted PIs, 1.0 for susceptible NRTIs, nonnucleoside reverse transcriptase inhibitors (NNRTIs), and unboosted PIs, and 1.5 for susceptible boosted PIs. Susceptibility to raltegravir and enfuvirtide was scored as 1 in case of first use of the drug or prior use in a virologically suppressive regimen and 0 in case of previous use but failure to suppress viral replication, conservatively assuming that the drugs had lost their antiviral activity. The arithmetic sum of the susceptibility scores of each drug associated with maraviroc was used to calculate the overall GSS of the cART regimen associated with maraviroc. Viral subtype was determined with automated BLAST analysis followed by manual phylogenetic analysis when the threshold of similarity to the best matching pure clade was below 95% or when the best matching clade was a circulating recombinant form (CRF). Unassigned subtypes were defined as undetermined.
Statistical analysis
Virological and immunological responses were assessed by survival analysis, as time to achieve an HIV-1 RNA of <50 copies/ml and time to achieve an increase of CD4+ ≥100 cells/μl from baseline. Factors analyzed by univariable and multivariable Cox regression models as possibly associated with virological and immunological response included baseline HIV-1 RNA (log10 transformed, as a continuous variable), the presence of non-R5 virus in at least one assay, type of tropism assay employed, nadir CD4+ T cell count, baseline CD4+ T cell count, GSS of concomitant regimens, and concomitant exposure to new antiretroviral drugs. Logistic regression was employed to analyze predictors of ever having a non-R5 tropism. Statistical analysis was performed by using SPSS version 18.0 (SPSS Inc., Chicago, IL).
Results
Baseline patients characteristics
A total of 191 ART regimens from 162 patients were eligible and analyzed. Table 1 summarizes patients' characteristics. One hundred and sixty-six patients (86.9%) carried viral subtype B, 16 (8.4%) non-B, and nine (4.7%) undetermined. The median (IQR) CD4 T cell count at baseline was 272/μl (130–442) and at nadir 78/μl (24–184) and the median HIV RNA at baseline was 4 log10 copies/ml (3.3–4.8).
Table 1.
Characteristics of the Study Population (n=191)
| Age (years)* |
46 (IQR 42–51) |
| Gender, n (%) |
Male 146 (76.4) |
| |
Female 45 (23.6) |
| Nationality, n (%) |
Italian 150 (78.5) |
| |
Non-Italian 4 (2.1) |
| |
Unknown 37 (19.4) |
| Risk group, n (%) |
Heterosexual 62 (32.5) |
| |
IDU 49 (25.7) |
| |
Homosexual/bisexual 41 (21.5) |
| |
Other/unknown 39 (20.3) |
| Viral subtype, n (%) |
B 166 (86.9) |
| |
Non-B 16 (8.4) |
| |
Undetermined 9 (4.7) |
| Time (years) since HIV diagnosis* |
16.4 (12.7–19.7) |
| Time (years) since starting cART* |
12.2 (7.6–15.2) |
| CD4 baseline cells count (cells/μl)* |
272 (130–442) |
| CD4 cells count nadir (cells/μl)* |
78 (24–184) |
| HIV RNA at baseline (log10 copies/ml)* |
4 (3.3–4.8) |
| Past antiretroviral drugs* |
11 (IQR 8–14) |
| Past treatment lines* |
10 (6–13) |
| GSS of the accompanying drugs, n (%) | |
| <1 |
29 (15.2) |
| 1–<2 |
69 (36.1) |
| ≥2 |
84 (44) |
| Unknown |
9 (4.7) |
| Median* | 1.75 (1–2.5) |
Values are expressed as n (%) except for *median (IQR).
IDU, intravenous drug use; cART, combined antiretroviral treatment; GSS, genotypic susceptibility score.
The cases with baseline HIV-1 RNA ≥100,000 copies/ml were 36/191 (18.9%). The viral tropism was predicted by Trofile in 23.6% (45 treatments), by ESTA in 48.7% (93 treatments), by genotyping in 13.1% (25 treatments), and by a combination of genotyping and phenotyping in 14.6% (28 treatments). The median number of previously employed antiretroviral drugs was 11 (IQR 8–14) and the median number of previous antiretroviral lines was 10 (IQR 6–13).
Type of antiretrovirals used concomitantly with maraviroc, their calculated activity, and results of premaraviroc viral tropism
The maraviroc-based ART regimens included NRTIs in 55.5% (106/191) of cases, NNRTIs in 26.2% (50/191, including etravirine in 44/191, 23%), PIs in 69.6% (133/191) (with ritonavir boosting in 121/191, 63.4%, including darunavir in 94/191, 49.2%), raltegravir in 59.2% (113/191), and enfuvirtide in 11% (21/191). The median (IQR) GSS of the accompanying regimen was 1.75 (1–2.5): 15.2% (29 patients) presented GSS <1, 36.1% (69) GSS 1 to <2, and 44% (84) GSS ≥2; GSS was unknown in 4.7%.9 The overall prevalences of R5, non-R5, and discordant strains were 92.2% (176/191), 4.7% (9/191), and 3.1% (6/191), respectively.
An R5 viral tropism was predicted in 176/191 (92.14%) of all cases analyzed; of these, 144/176 (81.8%) had baseline HIV-1 RNA <100,000 copies/ml and 32/176 (18.2%) had baseline HIV-1 RNA ≥100,000 copies/ml. In opposition, 144/155 (92.9%) with HIV-1 RNA <100,000 copies/ml and 32/36 (88.8%) with HIV-1 RNA ≥100,000 copies/ml were R5.
Overall, the R5 viral tropism was predicted for 45 of 47 isolates (95.7%) using Trofile, for 109 of 120 isolates (90.8%) using ESTA, and for 39 of 52 isolates (75%) using genotyping with G2P interpretation. Among the six discordant strains, five were classified as R5 tropic by ESTA and X4 tropic by G2P and one was classified as D/M tropic by ESTA and R5 by G2P.
The prevalence of non-R5 strains was 13.9% (11/79) among cases with baseline CD4+ cell count <50 cell/μl, 7.1% (2/28) in cases with CD4+ cell counts 50–100/μl, 4.8% (2/42) in cases with CD4+ cell count 101–200/μl, and 0% (0/42) among those with CD4+ cell counts >200/μl. The use of maraviroc in patients with non-R5 virus is partly justified by the deep salvage situation, whereby treating clinicians expected a residual antiviral activity and immunological activity of the drug, while lacking sufficiently active residual drug options. A lower nadir CD4+ cell count (p=0.01) and a lower baseline CD4+ cell count (p=0.016) were the only factors independently associated with non-R5 viral tropism by any assay.
Virological response and its predictors
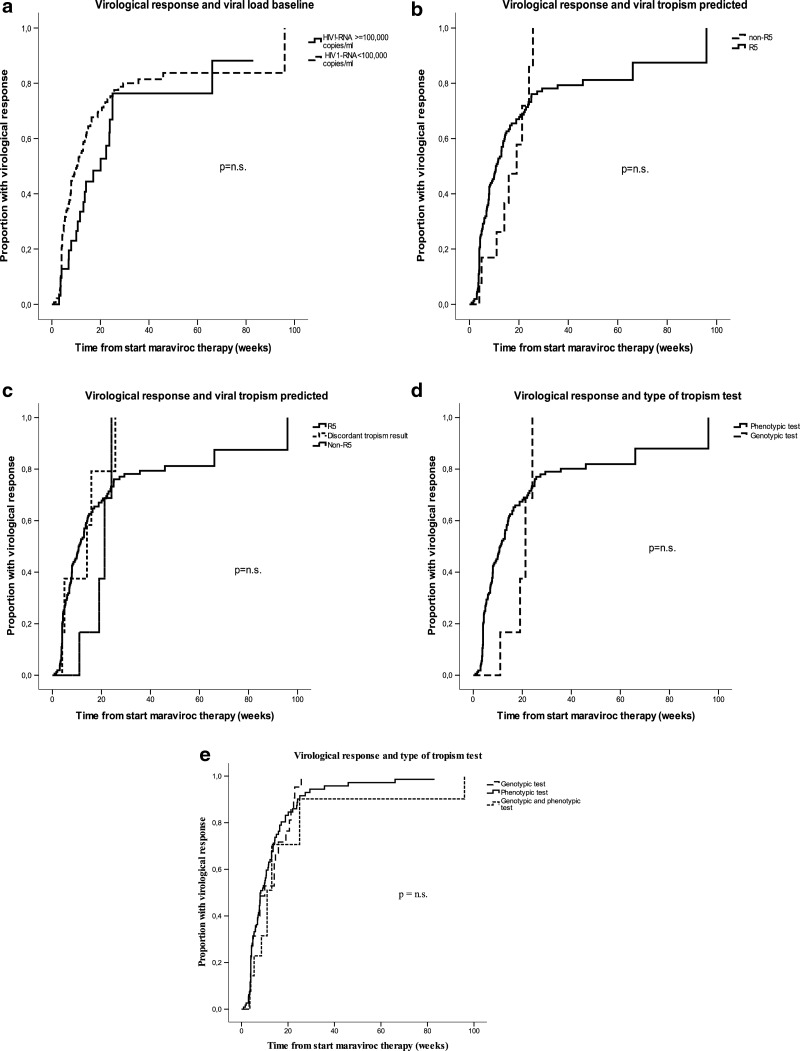
During a median follow-up of 8 weeks (IQR 4–19), virological success was achieved in 65.4% of cases (in 68.4% among those with baseline HIV-1 RNA <100,000 copies/ml and in 58.3% among those with baseline HIV-1 RNA ≥100,000 copies/ml). The estimated proportion achieving virological suppression at 24 weeks of treatment was 74% for patients with baseline HIV-1 RNA <100,000 copies/ml and 66% for patients with baseline HIV-1 RNA ≥100,000 copies/ml (see Fig. 1a). The estimated virological response was similar in patients whose virus was classified as R5 and in patients with non-R5 tropic strains by any assay (see Fig. 1b). Moreover, estimated proportions with 12-week response were higher, although not significantly, in patients with discordant tropism results using different assays as compared to patients with concordant non-R5 tropic strains (37% vs. 17%, see Fig. 1c). Interestingly, the estimated virological response was remarkably similar, regardless of whether the viral tropism was screened using a genotypic assay, a phenotypic assay, or both assays (see Fig. 1d and e and Table 2).
FIG. 1.
Kaplan–Meier plots showing the time to achieve a virological response (HIV-1 RNA <50 copies/ml) by (a) baseline viral load, (b,c) tropism tests results, and (d,e) type of tropism testing used to screen patients. p-values by the log-rank test are shown.
Table 2.
Univariable and Multivariable Cox Proportional Hazard Model Showing Relative Hazards for Virological Response, Fitted on the Whole Study Population (n=191)
|
Variable |
Univariable analysis |
Multivariable analysis |
||||
|---|---|---|---|---|---|---|
| RH | 95% CI | p | RH | 95% CI | p | |
| Baseline HIV-1 RNA (per log10 copies/ml higher) |
0.87 |
0.74–1.03 |
0.10 |
0.86 |
0.73–1.02 |
0.08 |
| Non-R5 tropism versus R5 |
1.16 |
0.59–2.30 |
0.65 |
n.e. |
n.e. |
n.e. |
| GSS of concomitant drugs (per unit increase) |
1.20 |
0.99–1.46 |
0.06 |
1.21 |
1.00–1.47 |
0.05 |
| Concomitant boosted PI use |
1.32 |
0.91–1.91 |
0.13 |
n.e. |
n.e. |
n.e. |
| Concomitant raltegravir use |
1.23 |
0.86–1.78 |
0.25 |
n.e. |
n.e. |
n.e. |
| Type of tropism assay | ||||||
| TROFILE (ref) |
1.00 |
|
0.60 |
n.e. |
n.e. |
n.e. |
| ESTA |
1.80 |
0.43–2.70 |
0.85 |
n.e. |
n.e. |
n.e. |
| Genotypic test |
0.55 |
0.20–1.51 |
0.25 |
n.e. |
n.e. |
n.e. |
| Genotypic and phenotypic test | 0.77 | 0.53–1.12 | 0.17 | n.e. | n.e. | n.e. |
RH, relative hazard; CI, confidence interval; ESTA, enhanced sensitivity trofile assay; GSS, genotypic susceptibility score; PI, protease inhibitor; n.e., not entered.
Multivariable analysis showed a borderline association of higher accompanying drugs GSS (HR per 1 higher 1.21; 95% CI 1.00–1.47) and lower baseline viral load (HR per 1 log copies/ml higher 0.86; 0.73–1.02) with virological response.
Immunological response and its predictors
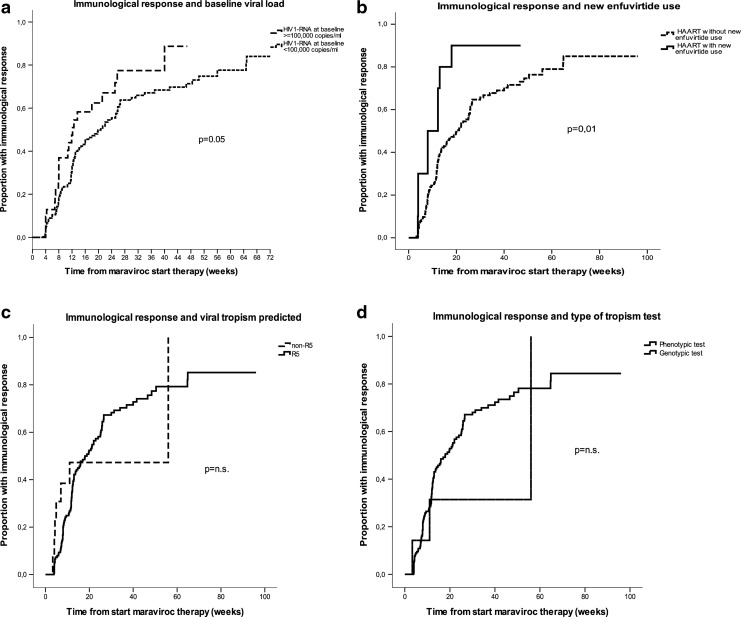
During a median follow-up of 12 weeks (IQR 7–25), immunological success was achieved in 58.1% of cases: 61.1% of those with baseline HIV-1 RNA ≥100,000 copies/ml and 57.4% of those with baseline HIV-1 RNA <100,000 copies/ml. After 24 weeks of treatment the estimated proportion of immunological responders was 56% among patients with baseline HIV-1 RNA <100,000 copies/ml and 67% among those with HIV-1 RNA ≥100,000 copies/ml (log-rank p=0.05, see Fig. 2a). We also noted a significantly shorter time to immunological response in patients first using enfuvirtide as concomitant treatment (Fig. 2b). The new treatment with enfuvirtide was used in 10/191 patients (5.2%): all these cases had R5 viral tropism. At week 24 the immunological response estimate was 90% among patients treated with enfuvirtide and 66% among those without enfuvirtide (log-rank p=0.01). Viral tropism assay types and results were not associated with time to immunological response (see Fig. 2c and d and Table 3). Multivariable Cox analysis revealed that higher baseline HIV-1 RNA (p=0.015 HR per log copies/ml higher 1.29, 95% CI 1.05–1.60) and concomitant therapy with enfuvirtide (p=0.06, HR 2.05, 0.96–4.39) were independently associated with shorter time to immunological response (see Table 3).
FIG. 2.
Kaplan–Meier plots showing the time to achieve an immunological response (CD4 cell count increase of >100 cells/μl from baseline) by (a) baseline viral load stratum, (b) enfuvirtide use, (c) tropism tests results, and (d) type of tropism testing used to screen patients. p-values by the log-rank test are shown.
Table 3.
Univariable and Multivariable Cox Proportional Hazard Model Showing Relative Hazards for Immunological Response, Fitted on the Whole Study Population (n=191)
|
Variable |
Univariable analysis |
Multivariable analysis |
||||
|---|---|---|---|---|---|---|
| RH | 95% CI | p | RH | 95% CI | p | |
| Baseline HIV-1 RNA (per log10 copies/ml higher) |
1.26 |
1.05–1.52 |
0.01 |
1.29 |
1.05–1.60 |
0.01 |
| Enfuvirtide first use |
2.30 |
1.16–4.58 |
0.01 |
2.05 |
0.96–4.39 |
0.06 |
| Baseline CD4 (per 100 cells/μl higher) |
1.05 |
0.97–1.14 |
0.19 |
n.e. |
n.e. |
n.e. |
| Non-R5 tropism vs. R5 |
1.08 |
0.50–2.33 |
0.77 |
n.e. |
n.e. |
n.e. |
| Type of tropism assay | ||||||
| Genotypic test (ref) |
1.00 |
|
0.55 |
n.e. |
n.e. |
n.e. |
| Phenotypic test |
1.21 |
0.44–3.32 |
0.70 |
n.e. |
n.e. |
n.e. |
| Genotypic and phenotypic test | 0.70 | 0.22–2.12 | 0.54 | n.e. | n.e. | n.e. |
RH, relative hazard; CI, confidence interval; n.e., not entered.
Evolution of viral tropism
After more than 1 month of maraviroc treatment at least one follow-up tropism test result was available for 20 treatments. These patients represent a subset of the patients with virological failure on maraviroc: no particular reason for genotyping this subset was found, except for the attitude of the site or clinician, since these patients were not different in terms of viral load, CD4, or GSS from the others who were failing (not shown).
The type of assays employed and the longitudinal tropism evolution results are summarized in Table 4. The baseline tropism was R5 in 17 (85%) cases, four of them by two different assays, and discordant in three cases (15%), all by different assays. The follow-up tropism was determined by a genotypic assay in 70% (14/20), by a phenotypic assay in 25% (5/20), and by a genotypic and phenotypic assay in 5% (1/20). The median time between the baseline and follow-up assays was 312 days (IQR 141.2–525.2) and the median level of viral load at the follow-up test was 3.8 log10 copies/ml (2.66–4.25). Changes in predicted coreceptor use were observed in 7 of 17 (41%) cases with R5 virus at baseline, which showed a tropism switch to non-R5 (see Table 4 for details).
Table 4.
Comparison of Viral Tropism Before and After Maraviroc Treatment (n=20)
| Baseline and follow-up assay used | Tropism evolution | Number of cases |
|---|---|---|
| ESTA→G2P |
R5→R5 |
4 |
| ESTA→ESTA |
R5→R5 |
1 |
| ESTA, G2P→G2P |
R5→R5 |
4 |
| G2P→ESTA |
R5→R5 |
1 |
| ESTA→G2P |
R5→non-R5 |
6 |
| ESTA→G2P, OTA |
R5→non-R5 |
1 |
| G2P, ESTA→ESTA | DISC→non-R5 | 3 |
ESTA, enhanced sensitivity Trofile assay; G2P, geno2pheno.
Discussion
In the present study we investigated the impact of the use of maraviroc-containing regimens in clinical practice in a population of HIV-1-infected patients with a long infection history and past use of several antiretroviral drug classes. Even if all but three patients included were screened for the presence of R5-tropic virus, non-R5 tropism was still found in a small proportion of the sample. This finding can be explained by the fact that in selected cases clinicians might have attempted to exploit a residual virological or immunological activity of maraviroc in the context of a salvage ART regimen. In line with other studies,3,16–21 the non-R5 viral tropism was correlated with a lower CD4 count at baseline and especially at nadir.
We found that a significant proportion of experienced patients treated with regimens containing maraviroc achieved a virological response. In agreement with previous observations,3 the virological response rate showed a tendency to be higher in the group with lower baseline viral load than in the group with higher baseline viral load,3 and in cases with a higher activity of the accompanying drugs, as estimated by the interpreted genotypic resistance assay results. Notably, the type of tropism assay used was not associated with virological response nor was it associated with immunological response. This confirms previous clinical trial-based observations suggesting that both genotypic and phenotypic R5 tropism are associated with virological response.22–25 In addition, in previous studies genotypic tropism tests were retrospectively validated, while here we show that selection of prospective maraviroc regimens using genotypic or phenotypic assays is associated with similar virological and immunological responses. In contrast with expectations, non-R5 tropism was not negatively associated with virological response. This might be due to the limited sample size of the category with non-R5 virus and might have been confounded by the relatively good activity of the accompanying drugs.
We found that more than half of experienced patients treated with regimens containing maraviroc achieved an immunological response. The immunological response rate was significantly higher in the group with higher baseline viral load. Interestingly, we observed a higher immunological response during concomitant treatment with enfuvirtide, both overall and when this drug was used for the first time (Table 3). To our knowledge, this is the first demonstration of an additive immunological effect of this drug in the context of maraviroc treatment. This observation is in line with an earlier in vitro study showing a positive correlation between low CCR5 density levels on CD4 T cells and increased sensitivity of R5 HIV-1 strains to enfuvirtide.22 Enfuvirtide and maraviroc have shown a potential for synergistic antiviral activity in vitro and in vivo. Nonetheless, we observed an immunological benefit but not a virological advantage when using this combination in vivo. In agreement with our finding, previous in vivo studies have demonstrated a significant immunological benefit with enfuvirtide, which was at least partly independent of its antiviral activity.26–29
Among patients with pure R5 strains before maraviroc therapy, half showed a tropism shift from R5 to non-R5 at failure. An expansion of preexisting X4 or dual-tropic (R5X4) variants during clinical use of small molecule CCR5 inhibitors has been described in clinical trials.30 This is in line with previous observations made using phenotypic assays and suggests that the potential use of the cheaper and more practical genotypic assays for interpreting the cause of virological failure with CCR5 antagonists and making informed decisions regarding their interruption deserves further evaluation. These results, however, must be interpreted with caution given the fact that tropism was determined only in part of the maraviroc failures, which cannot exclude selection bias, and that different assays for tropism determination were employed at baseline and during follow-up, although concordance between genotyping, and Trofile or ESTA is expected to be high (80% or higher).25,31
In conclusion, we observed significant virological and immunological responses with maraviroc-based regimens in antiretroviral-experienced patients. Any of the approved methods for determining R5 tropism is suitable for predicting virological and immunological responses in a real world setting. The potential for an additional immunological benefit of enfuvirtide in this context might be of clinical relevance and warrants a prospective evaluation.
Acknowledgments
Part of this study was supported by funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under the project “Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN)”—grant 223131 and by Programma Nazionale AIDS project number 40H94 to A.D.L. and 40H81 to M.Z.
Author Disclosure Statement
M.Z. has been a consultant to or has received research support or lecture fees from Abbott Pharmaceuticals, Abbott Molecular, Gilead Sciences, Janssen-Cilag, Merck Sharp and Dome, and ViiV Healthcare. A.D.L. has been a consultant to Abbott Virology, Gilead Sciences, Janssen-Cilag, and ViiV Healthcare and has received research support from ViiV Healthcare.
References
- 1.Clapham PR. and McKnight A: Cell surface receptors, virus entry and tropism of primate lentiviruses. J Gen Virol 2002;83:1809–1829 [DOI] [PubMed] [Google Scholar]
- 2.Moore JP, Kitchen SG, Pugach P, et al. : The CCR5 and CXCR4 co-receptors central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses 2004;20:111–126 [DOI] [PubMed] [Google Scholar]
- 3.Gulick RM, Lalezari J, Goodrich J, et al.: Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med 2008;359:1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Medicines Agency (EMA): www.ema.europa.eu/humandocs/…/celsentri.ht September18, 2007
- 5.US Food and Drug Administration (FDA): www.fda.gov/cder/foi/label/2007/022128lbl.pdf June8, 2007
- 6.AIDS info. Adult and Adolescent Guidelines: www.aidsinfo.nih.gov/Guidelines Accessed January14, 2012
- 7.Vandekerckhove , Verhofstede C, Demecheleer E, et al. : Comparison of phenotypic and genotypic tropism determination in triple-class-experienced HIV patients eligible for maraviroc treatment. J Antimicrob Chemother 2011;66:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chueca N, Garrido C, Alvarez M, et al. : Improvement in the determination of HIV-1 tropism using the V3 gene sequence and a combination of bioinformatic tools. J Med Virol 2009;81:763–767 [DOI] [PubMed] [Google Scholar]
- 9.Rose JD, Rhea AM, Weber J, et al. : Current tests to evaluate HIV-1 co-receptor tropism. Curr Opin HIV AIDS 2009;4(2):136–142 [DOI] [PubMed] [Google Scholar]
- 10.Reeves JD, Coakley E, Petropoulos CJ, et al. : An enhanced-sensitivity Trofile™ HIV co-receptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: A review of analytical and clinical studies. J Viral Entry 2009;3:94–102 [Google Scholar]
- 11.Trinh L, Han D, Huang W, et al. : Technical validation of an enhanced sensitivity Trofle HIV co-receptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Antivir Ther 2008;13(Suppl 3):A128 [Google Scholar]
- 12.Vandekerckhove LP, Wensing AM, et al. ; European Consensus Group on clinical management of tropism testing European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 2011;11(5):394–407 [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Mateos E, González-Serna A, Genebat M, et al. : Virological response after short-term CCR5 antagonist exposure in HIV-infected patients: Frequency of subjects with virological response and associated factors. Antimicrob Agents Chemother 2011;55(10):4664–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reuter S, Braken P, Jensen B, et al. : Maraviroc in treatment-experienced patients with HIV-1 infection experience from routine clinical practice. Eur J Med Res 2010;15(6):231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Luca A, Meini G, Rossetti B, et al. : HIV-1 co-receptor tropism evolution in naïve patients undergoing successful ART: Concordance of DNA vs RNA and triplicate vs singlicate sequencing. 10th European Meeting on HIV & Hepatitis Treatment Strategies & Antiviral Drug Resistance, Barcelona, Spain, March28–30, 2012. Abstract O17. [Google Scholar]
- 16.Pulido I, Machmach K, Romero-Sánchez MC, et al. : T-cell changes after a short-term exposure to maraviroc in HIV-infected patients are related to antiviral activity. J Infect 2012;64(4):417–423 [DOI] [PubMed] [Google Scholar]
- 17.Schuitemaker H, Koot M, Kootstra NA, et al. : Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol 1992;66:1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connor RI, Sheridan KE, Ceradini D, et al. : Change in co-receptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med 1997;185:621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenyo EM, Morfeldt-Manson L, Chiodi F, et al. : Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol 1988;62:4414–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koot M, Keet IP, Vos AH, et al. : Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4 cell depletion and progression to AIDS. Ann Intern Med 1993;118:681–688 [DOI] [PubMed] [Google Scholar]
- 21.Koot M, van Leeuwen R, de Goede REY, et al. : Conversion rate towards a syncytium-inducing (SI) phenotype during different stages of human immunodeficiency virus type 1 infection and prognostic value of SI phenotype for survival after AIDS diagnosis. J Infect Dis 1999;179:254–258 [DOI] [PubMed] [Google Scholar]
- 22.Richman DD. and Bozzette SA: The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis 1994;169:968–974 [DOI] [PubMed] [Google Scholar]
- 23.Michael NL, Chang G, Louie LG, et al. : The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med 1997;3:338–340 [DOI] [PubMed] [Google Scholar]
- 24.McGovern RA, Thielen A, Mo T, et al. : Population-based V3 genotypic tropism assay: A retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 2010;24(16):2517–2525 [DOI] [PubMed] [Google Scholar]
- 25.Swenson L, Dong W, Mo T, et al. : Use of cellular HIV DNA to predict virologic response to maraviroc: Performance of population-based and deep sequencing. Clin Infect Dis 201356:1569–1576 [DOI] [PubMed] [Google Scholar]
- 26.Heredia A, Gilliam B, De Vico A, et al. : CCR5 density levels on primary CD4 T cells impact the replication and enfuvirtide susceptibility of R5 HIV-1. AIDS 2007;21(10):1317–1322 [DOI] [PubMed] [Google Scholar]
- 27.Melby T, Despirito M, Demasi R, et al. : HIV-1 co-receptor use in triple-class treatment-experienced patients: Baseline prevalence, correlates, and relationship to enfuvirtide response. J Infect Dis 2006;194(2):238–246 [DOI] [PubMed] [Google Scholar]
- 28.Svicher V, Aquaro S, D'Arrigo R, et al. : Specific enfuvirtide-associated mutational pathways in HIV-1 Gp41 are significantly correlated with an increase in CD4(+) cell count, despite virological failure. J Infect Dis 2008;197(10):1408–1418 [DOI] [PubMed] [Google Scholar]
- 29.Melby TE, Despirito M, Demasi RA, et al. : Association between specific enfuvirtide resistance mutations and CD4 cell response during enfuvirtide-based therapy. AIDS 2007;21(18):2537–2539 [DOI] [PubMed] [Google Scholar]
- 30.Westby M, et al. : Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol 2006;80:4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prosperi MCF, Bracciale L, Fabbiani M, et al.: Comparative determination of HIV-1 co-receptor tropism by Enhanced Sensitivity Trofile, gp120 V3-loop RNA and DNA genotyping. Retrovirology 2010;7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]