Abstract
Mammalian adult hematopoietic stem cells (HSCs) reside in the hypoxic bone marrow microenvironment and display a distinct metabolic phenotype compared with their progenitors. It has been proposed that HSCs generate energy mainly through anaerobic glycolysis in a pyruvate dehydrogenase kinase (Pdk)–dependent manner. Cited2 is an essential regulator for HSC quiescence, apoptosis, and function. Herein, we show that conditional deletion of Cited2 in murine HSCs results in elevated levels of reactive oxygen species, decreased cellular glutathione content, increased mitochondrial activity, and decreased glycolysis. At the molecular level, Cited2 deficiency significantly reduced the expression of genes involved in metabolism, such as Pdk2, Pdk4, and lactate dehydrogenases B and D (LDHB and LDHD). Cited2-deficient HSCs also exhibited increased Akt signaling, concomitant with elevated mTORC1 activity and phosphorylation of FoxOs. Further, inhibition of PI3/Akt, but not mTORC1, partially rescued the repression of Pdk4 caused by deletion of Cited2. Altogether, our results suggest that Cited2 is required for the maintenance of adult HSC glycolytic metabolism likely through regulating Pdk2, Pdk4, LDHB, LDHD, and Akt activity.
Introduction
Mammalian adult hematopoietic stem cells (HSCs) primarily reside in the hypoxic bone marrow microenvironment, and preferentially utilize anaerobic glycolysis to obtain energy and maintain quiescence, self-renewal, and function [1–3]. Compared with more differentiated hematopoietic progenitor cells, HSCs contain fewer mitochondria and display lower mitochondrial membrane potential (ΔΨm) [3–6]. Mitochondria are the major source of reactive oxygen species (ROS), which are detrimental to HSCs when their levels are too high [7], and experimental data have shown that HSCs have much lower levels of ROS than differentiated progenitor cells [4,8]. While the mechanisms that control HSC quiescence and self-renewal have been extensively investigated, the underlying regulation of glycolytic metabolism in these cells is just beginning to be understood.
Studies in the mouse have revealed several factors implicated in the metabolic regulation of HSCs. Hypoxia of bone marrow microenvironment stabilizes the hypoxia inducible factor HIF-1α, which regulates the quiescence and function of HSCs in a dose-dependent manner and maintains HSC metabolism through Cripto/GRP78 signaling [9,10]. In addition, various studies showed that homeobox protein Meis1 is required for optimal transcriptional activation of HIF-1 [3,11,12]. Recently, Takubo et al. revealed that activation of pyruvate dehydrogenase kinase (Pdk) by HIF-1 inhibits pyruvate dehydrogenase (PDH) activity, reduces supply of acetyl-CoA to mitochondria, and thus results in the maintenance of glycolytic flow in HSCs [13]. Further, glycolytic metabolism governed by Pdk acts as a cell cycle checkpoint that modulates HSC quiescence and function.
Multiple lines of evidence also suggest critical roles of Akt, mTOR, and FoxOs signaling pathways in HSC homeostasis by tightly regulating ROS levels [14–24]. mTOR is negatively regulated by the adenosine monophosphate-activated protein kinase, which can be activated by the tumor suppressor Lkb1. Loss of Lkb1 results in more HSC cycling and rapid HSC depletion, unexpectedly, not through mTOR but possibly by peroxisome proliferator activated receptor gamma coactivator (PGC)-1α [25–27]. In addition, promyelocytic leukemia (PML)–peroxisome proliferator-activated receptor (PPAR)-δ pathway for fatty acid oxidation is reported to control the asymmetric division of HSCs [28]. Deletion of PPAR-δ or PML as well as inhibition of fatty acid oxidation by etomoxir, a pharmacological inhibitor of mitochondrial β-oxidation of long-chain fatty acids, results in the symmetric commitment of HSC daughter cells, whereas PPAR-δ activation by GW-501516 increases asymmetric cell division. Further, PTPMT1, a phosphatase and tensin homolog-like mitochondrial phosphatase, is required for HSC differentiation and mitochondrial metabolism, and phosphatidylinositol phosphate substrates of PTPMT1 directly enhance fatty-acid-induced activation of mitochondrial uncoupling protein 2 [29].
Cited2 is a cytokine-inducible gene, which plays various roles during mouse development [30–37]. In particular, Cited2 is essential for hematopoiesis in fetal liver and adult bone marrow [38–40]. Loss of Cited2 in differentiating mouse embryonic stem cells leads to disturbance in differentiation toward various cell types, including hematopoietic cells [41]. Fetal liver cells from Cited2-deleted murine embryos give rise to reduced number of colonies in the colony-forming unit assay and display markedly compromised reconstitution capacity in transplantation studies [38]. In adult mouse bone marrow, Cited2 is highly expressed in long-term (LT-) HSCs but much less abundant in short-term (ST-) HSCs and multipotent progenitors [39,42]. Further, conditional deletion of Cited2 in adult mouse results in loss of quiescence, increased apoptosis, and impaired HSC function. Cited2 is dispensable for the maintenance of committed myeloid and lymphoid cells, suggesting a crucial role of Cited2 primarily in HSCs [39]. Mechanistically, deletion of Cited2 significantly elevates the activity of p53 and decreases the expression of HSC-quiescence-related genes, such as p57 and Hes1 mediated by enhanced activity of HIF-1 [39,40]. Altogether, these independent studies suggest a crucial role of Cited2 in hematopoiesis and the maintenance of HSC function. However, the role of Cited2 in the metabolic regulation of HSCs is unclear. In the present study, by using a conditional knockout (KO) strategy, we showed that loss of Cited2 in adult HSCs results in attenuated glycolytic metabolism and elevated mitochondrial activity, thus suggesting a critical role of Cited2 in HSC metabolic regulation.
Materials and Methods
Mice
As described in our previous work [40], the Cited2fl/+ mouse line, the HIF-1αfl/+ mouse line, and the Mx1-Cre transgenic mouse line (The Jackson Laboratory) were maintained on the C57/BL6 background and bred to generate Cited2fl/fl (referred to as wild-type, WT), Cited2fl/fl;Mx1-Cre (referred to as KO), and Cited2fl/fl:HIF-1αfl/fl;Mx1-Cre (referred to as double KO) mice. Genotyping of mice was performed by PCR using genomic DNA isolated from mouse tails with the following primers: Cited2fl/fl (forward) 5′-GTC TCA GCG TCT GCT CGT TT-3′, Cited2fl/fl (reverse) 5′-CTG CTG CTG CTG GTG ATG AT-3′; HIF-1αfl/fl (forward) 5′-ATA TGC TCT TAT GAA GGG GCC TAT GGA GGC-3′, HIF-1αfl/fl (reverse) 5′-GAT CTT TCC GAG GAC CTG GAT TCA ATT CCC-3′; and Cre (forward) 5′-GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG-3′, Cre (reverse) 5′-GAG TGA ACG AAC CTG GTC GAA ATC AGT GCG-3′. Mice at the age of 5–6 weeks were treated every other day with five doses of 16 mg/kg body weight polyinosinic-polycytidylic acid (Sigma-Aldrich) via intraperitoneal injection and analysis was performed 2–3 weeks after the last injection. All animal studies were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University (CWRU).
Antibodies
Rat monoclonal antibodies (mAbs) against Ter119, B220, CD3, Gr-1, Mac-1, c-Kit, Sca-1, Flt3, and CD34 were purchased from BD Biosciences, or eBiosciences. Cocktail of biotin-conjugated mAbs against lineage markers was purchased from Miltenyi Biotec. Rabbit polyclonal antibody against phospho-PDH-E1α (Ser293) was purchased from EMD Millipore. Phycoerythrin-conjugated rabbit mAbs against phospho-S6 Ribosomal protein (Ser235/236) and phospho-Akt (Thr308), rabbit polyclonal antibodies against phospho-FoxO1 (Ser256) and phospho-FoxO3a (Ser253), and rabbit IgG isotype control were purchased from Cell Signaling Technology.
Flow cytometry
Flow cytometry analysis was performed on an LSR II (BD Biosciences), and cells were sorted using FACSAria (BD Biosciences). In some cases of HSC sorting, magnetic lineage depletion (autoMACS separator; Miltenyi Biotec) was used, followed by additional fluorescent surface marker staining.
For intracellular staining and flow cytometry analysis, cells were fixed and permeabilized using nuclear factor fixation and permeabilization buffer set purchased from BioLegend, Inc., stained with respective antibodies according to the manufacturer's instructions, and then analyzed on LSR II flow cytometer (BD Biosciences).
Analysis of ROS, mitochondrial mass, and membrane potential
For measurement of intracellular ROS levels, cells were incubated at 37°C in the dark for 20 min with 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA; Invitrogen), and then analyzed by flow cytometry. DCFDA rapidly diffuses through the cell membrane as a colorless probe. Upon cleavage of the acetate groups by intracellular esterases and oxidation, the nonfluorescent DCFDA is converted to the highly fluorescent 2′,7′-dichlorofluorescein.
For measurement of mitochondrial mass, cells were incubated at 37°C in the dark for 15 min with 50 nM MitoTracker Green (Invitrogen), and then analyzed with flow cytometry. MitoTracker Green is a nonproton gradient sensitive dye that binds to the mitochondrial lipid membrane independent of mitochondrial membrane potential [43].
For measurement of mitochondrial membrane potential, cells were incubated at 37°C in the dark for 20 min with 100 nM tetramethylrhodamine methyl ester (TMRM; ImmunoChemistry Technologies) and then analyzed by flow cytometry.
Determination of cellular glutathione
Monochloromobimane is nonfluorescent until conjugated, more selectively reacts with glutathione (GSH), and has been extensively used for the measurement of GSH by flow cytometry [44,45]. Bone marrow cells were harvested and incubated in Iscove's modified Dulbecco's media complemented with 15% fetal bovine serum (FBS; Hyclone) and 50 μM monochlorobimane (Sigma-Aldrich) at room temperature in the dark for 30 min. Then, bone marrow cells were washed once with phosphate-buffered saline solution supplemented with 2% FBS, stained with various phenotypic surface markers, and analyzed by flow cytometry.
Determination of mitochondrial DNA copy number
Total DNA (mitochondrial and genomic DNA) was extracted from cells using DNeasy Blood & Tissue Kit (Qiagen). The copy number of mitochondrial DNA (estimated by the ND1 gene) was measured by quantitative real-time PCR and normalized to genomic DNA (estimated by the 18S rRNA gene).
Measurement of glucose uptake and intracellular ATP content
For flow cytometric analysis of glucose uptake, bone marrow cells were preincubated for 30 min at 37°C in glucose-free Dulbecco's modified Eagle's medium (DMEM) with 50 μM 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG; Invitrogen) and stained with surface markers. Intracellular adenosine triphosphate (ATP) was measured using the Luciferase ATP Determination Kit (Sigma-Aldrich) following the manufacturer's instructions.
Determination of lactate production
Lactate production was measured by gas chromatography–mass spectrometry using a modification of the previously described methods [46]. Sorted cells (5×104) were cultured in 5% CO2 at 37°C for 12 h in 80 μL of glucose-free DMEM supplemented with 10 mM D-[13C6] glucose (Cambridge Isotope Labs, Inc.) and 6% dialyzed FBS (Hyclone). Then, the samples were analyzed for glucose-derived lactate amount on an Agilent 5973 mass spectrometer, linked to a Model 6890 gas chromatograph at the Mouse Metabolic Phenotyping Center of CWRU. Concentration of lactate was calculated using a standard curve of lactate relative to internal standard d6-BHB. The final results are presented as the 13C-lactate abundance per 1×104 cells. General chemicals were purchased from Sigma-Aldrich.
Determination of mitochondrial
Mitochondrial nicotinamide adenine dinucleotide+hydrogen (NADH) was determined by the method described previously [3]. In brief, fresh bone marrow cells were stained directly with surface markers to define LT- and ST-HSCs, and then analyzed on LSR II flow cytometer (BD Biosciences) to set the baseline NADH fluorescence gate using the UV440/40 nm channel. Next, cells were treated with 2 μM antimycin A (AMA; Sigma-Aldrich) at room temperature for 5 min and analyzed for NADH fluorescence.
Electron microscopy
Sorted cells were pelleted and fixed with 2.5% glutaraldehyde, and then embedded as previously described [47]. Standard transmission electron microscopy ultrastructural analysis was performed at Electron Microscopy Facility of CWRU.
Gene expression analysis
RNA was isolated from sorted cells using the PicoPure RNA Isolation Kit (Applied Biosystems, Invitrogen) and cDNA was prepared with the GoScript Reverse Transcription System (Promega) according to the manufacturer's instructions. Real-time quantitative PCR was performed using FastStart SYBR Green Master (Roche) and CFX Connect™ Real-Time PCR Detection System (Bio-Rad). Data were normalized by the abundance of an internal control, 18S RNA. Sequences of primers used are available upon request.
Statistics
The significance of differences was determined by two-tailed Student's t-test. P<0.05 was considered statistically significant (ns: not significant; *P<0.05; **P<0.01).
Results
Loss of Cited2 alters HSC mitochondrial activity and metabolism
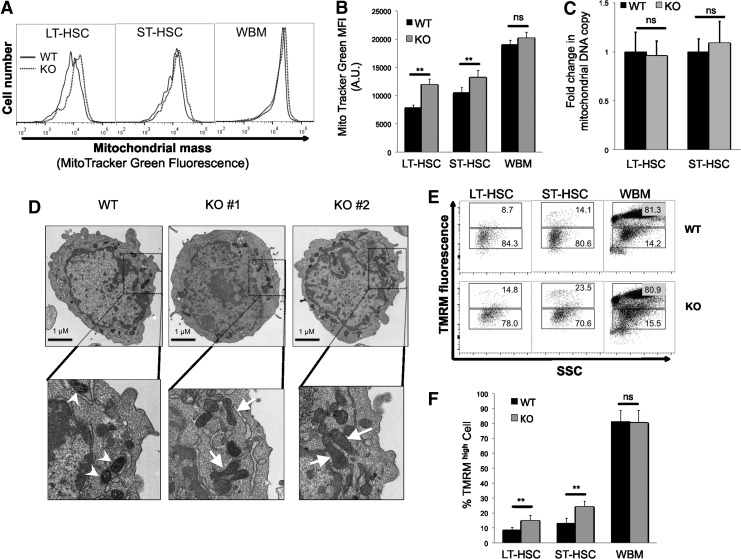
We have shown previously that Cited2 is essential for the regulation of HSC apoptosis, quiescence, and function in adult mice [40]. To determine its role in the maintenance of HSC metabolism, we examined metabolism-related parameters in LT-HSCs (Flt3− CD34− Lineage− Sca-1+ c-kit+; Flt3−CD34− LSK) and ST-HSCs (Flt3−CD34+ LSK) of Cited2 KO mice. First, mitochondrial mass was determined by staining cells with MitoTracker Green that localizes to mitochondria regardless of mitochondrial membrane potential and has been used extensively [3,16]. As shown in Fig. 1A and B, deletion of Cited2 resulted in a significant increase in mitochondrial mass in both LT- and ST-HSCs but not in the whole bone marrow (WBM) cells. We further examined whether deletion of Cited2 affects mitochondrial copy numbers in HSCs by real-time PCR using primers for a mitochondrial gene and a genomic sequence. No significant difference was observed in mitochondrial DNA copy number between Cited2 KO and WT HSCs (Fig. 1C).
FIG. 1.
Cited2 knockout (KO) hematopoietic stem cells (HSCs) display altered mitochondrial activity. (A) Representative plot of MitoTracker Green staining on long-term (LT-) HSCs, short-term (ST-) HSCs, and whole bone marrow (WBM) analyzed by flow cytometry. (B) Quantitative analysis of MitoTracker Green median fluorescence intensity (MFI) (mean±SD, n=5). AU, arbitrary unit. (C) Quantitative analysis of mitochondrial DNA copy number relative to genomic DNA (mean±SD, n=4). (D) Electron microscopy images of HSCs. One wild-type (WT) and two KO representative HSCs are shown. Note that the mitochondria indicated by white arrowheads in the WT HSCs were small and dark, while a number of mitochondria in the KO HSCs indicated by white arrows were markedly elongated and brighter. Bottom panels are enlarged images of the square areas from upper panels. Scale bars=1 μm. (E) Representative plot of tetramethylrhodamine methyl ester (TMRM) staining on LT-HSCs, ST-HSCs, and WBM analyzed by flow cytometry. SSC, side scatter. (F) Quantitative analysis of cells that exhibit high levels of TMRM stain in LT-HSCs, ST-HSCs, and WBM (mean±SD, n=5). Asterisks are explained in Materials and Methods Statistics section.
To reveal the morphological changes of mitochondria in Cited2 KO HSCs, we sorted fresh Flt3−LSK cells (containing LT- and ST-HSCs) and performed electron microscopy ultrastructural analysis. Consistent with the observations by Warr et al. [48], mitochondria in WT HSCs were mostly small, round or oval, and dark (Fig. 1D). However, Cited2 KO HSCs displayed markedly elongated and brighter mitochondria, similar to that in older WT HSCs (20–24 months of age) observed previously [48]. The differences in mitochondrial size and structure may partly explain the increased mitochondrial mass despite similar mitochondrial DNA copy number in Cited2 KO versus WT HSCs.
HSCs have been shown to exhibit low mitochondrial potential and low rates of mitochondrial respiration. Staining with fluorescent dye TMRM showed that, in contrast to WBM cells, more than 80% of WT HSCs displayed low mitochondrial membrane potential (Fig. 1E, F), which is in agreement with previous findings [3]. However, Cited2 KO HSCs displayed a significant population of cells with heightened mitochondrial potential compared with the WT control.
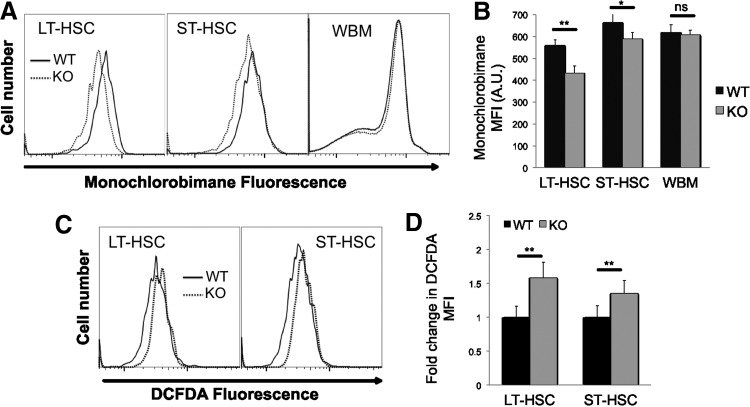
GSH is a tripeptide, which has multiple biological roles, including protection against ROS. We found that cellular GSH content was significantly decreased in Cited2 KO LT- and ST-HSCs, compared with WT controls (Fig. 2A, B). In the meantime, ROS levels in Cited2 KO HSCs were significantly higher than in WT controls (Fig. 2C, D). Together, these results suggest that depletion of Cited2 in HSCs increases mitochondrial activity and reduces GSH content, leading to the elevated level of ROS.
FIG. 2.
Cited2 KO HSCs display decreased glutathione content and elevated cellular levels of reactive oxygen species. (A) Representative plot of monochlorobimane staining on LT-HSCs, ST-HSCs, and WBM. (B) Quantitative analysis of monochlorobimane MFI (mean±SD, n=3). (C) Representative plot of dichlorodihydrofluorescein diacetate (DCFDA) staining on LT-HSCs and ST-HSCs. (D) Quantitative analysis of fold changes in DCFDA staining (mean±SD, n=6). The fold changes were calculated based upon DCFDA MFI of KO relative to that of WT HSCs. Asterisks are explained in Materials and Methods Statistics section.
Loss of Cited2 attenuates HSC glycolytic metabolism
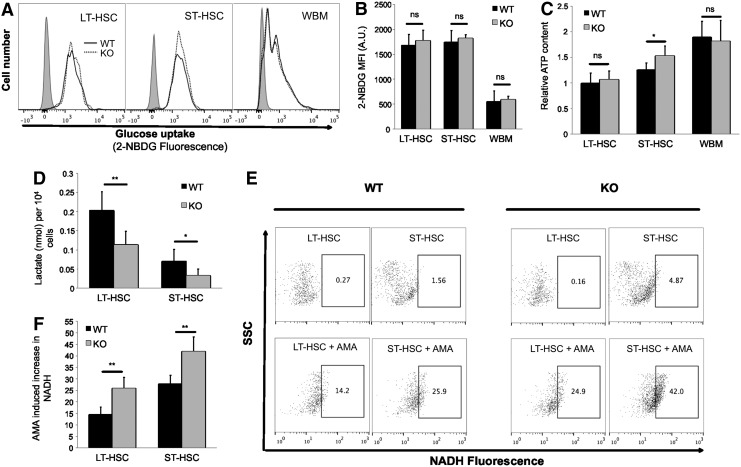
To further understand the metabolic changes in Cited2 KO HSCs, we measured glucose uptake using fluorescent indicator 2-NBDG. In contrast to the increased mitochondrial activity, glucose uptake was unchanged in Cited2-deleted HSCs (Fig. 3A, B), which is similar to HSCs with HIF-1α deficiency [13]. In addition, the ATP content in Cited2 KO LT-HSCs was maintained at the normal level; however, Cited2 KO ST-HSCs displayed moderately increased ATP levels (Fig. 3C).
FIG. 3.
Loss of Cited2 decreases glycolysis in HSCs. (A) Representative plot of 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) fluorescence on LT-HSCs, ST-HSCs, and WBM. Gray-filled histogram indicates control without 2-NBDG. (B) Quantitative analysis of 2-NBDG MFI on cells (mean±SD, n=3). (C) Relative intracellular adenosine-triphosphate (ATP) contents in various cell populations. The ATP level in WT LT-HSCs was set as one, and the ATP levels in other cell populations were presented as the relative values to that of WT LT-HSCs (mean±SD, n=6). (D) Glucose-derived 13C-lactate production per 1×104 cells (mean±SD, n=4). (E) Representative plot of cellular NADH: flow cytometry profiles of cells before (upper panel) and after (lower panel) treatment with antimycin A (AMA). (F) Percentage of cells that display increased nicotinamide adenine dinucleotide+hydrogen (NADH) fluorescence in response to AMA treatment (mean±SD, n=6). Asterisks are explained in Materials and Methods Statistics section.
Previous work showed that HSCs prefer the utilization of glycolysis than oxidative phosphorylation (OXPHOS) to generate energy [3,11,13]. Therefore, we continued to examine the glycolytic flux as determined by glucose-derived 13C-lactate production. As shown in Fig. 3D, the rate of 13C-lactate production was significantly lower in both LT- and ST-HSCs that lack Cited2 than in WT controls. To further confirm this finding, we treated WT and Cited2 KO HSCs with 2 μM AMA, a specific inhibitor of mitochondrial electron transport chain, for 5 min as described elsewhere [3]. The principle of this assay is that inhibition of the complex III of mitochondria electron transport chain by AMA results in NADH accumulation only in cells that rely on mitochondrial respiration for NADH production. We found that Cited2 KO HSCs displayed increased NADH after AMA treatment, compared with the WT control, indicating that mitochondrial respiration was increased in Cited2 KO HSCs (Fig. 3E, F). Taken together, these results indicate that loss of Cited2 attenuates HSCs' glycolytic metabolism while simultaneously enhancing their overall OXPHOS.
Loss of Cited2 affects the expression of metabolism-related genes
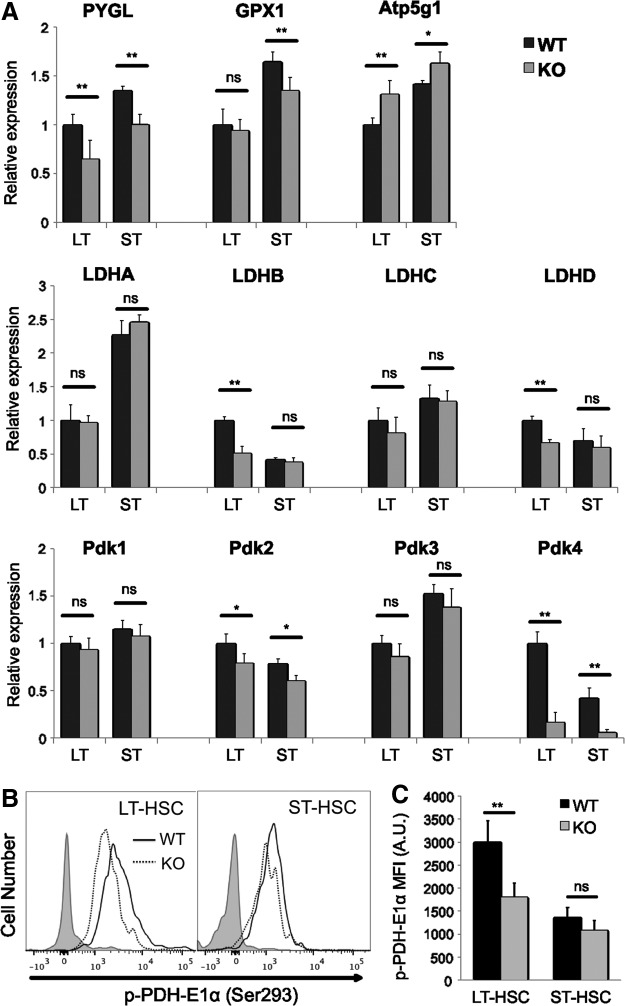
To further understand the metabolic dysregulation of HSCs that lack Cited2, we performed quantitative real-time PCR to determine the expression of genes involved in glucose metabolism (Fig. 4A). Lactate dehydrogenase (LDH) converts pyruvate to lactate and is highly expressed in HSCs [13]. We found that the expression of both LDHB and LDHD was significantly reduced in Cited2 KO LT-HSCs but not in ST-HSCs. The expression of the other two LDH family members, LDHA and LDHC, was not affected. Pdk is a kinase that phosphorylates and thereby inactivates PDH, an enzyme that converts pyruvate into acetyl-CoA. In LT-HSCs, Pdk2 and Pdk4 are highly expressed and act as major regulators of glycolysis. Glycolytic metabolism governed by Pdk acts as a cell cycle checkpoint that modulates HSC function [13]. PDH-E1α, the E1α subunit of PDH complex, contains the E1 active site and plays a key role in the function of the PDH complex. Meanwhile, previous studies showed that PDH-E1α is more phosphorylated in LT-HSCs than in their differentiated progeny [13]. Notably, we found that loss of Cited2 significantly reduced the expression of Pdk2 (∼20% decrease) and Pdk4 (∼85% decrease) in both LT- and ST-HSCs. Accordingly, the phosphorylation of PDH-E1α was significantly decreased in Cited2 KO LT-HSCs (Fig. 4B, C).
FIG. 4.
Loss of Cited2 affects expression profile of genes involved in metabolism. (A) Gene expression analysis of HSCs by real-time quantitative PCR. The results are presented as expression levels relative to that of WT LT-HSCs (mean±SD, n=6). LT: LT-HSCs; ST: ST-HSCs. (B) Representative plot of p-PDH-E1α (Ser293) staining with flow cytometry analysis. (C) Quantitative analysis of p-PDH-E1α (Ser293) MFI on cells. The gray-filled histograms represent the isotype IgG control. Asterisks are explained in Materials and Methods Statistics section.
The expression of PYGL (phosphorylase, glycogen, liver), a regulator of glycolysis in epiblast stem cells [49], was significantly decreased in Cited2 KO HSCs (Fig. 4A). In addition, GPX1 (glutathione peroxidase 1), which encodes an antioxidant enzyme, was dowregulated in Cited2 KO ST-HSCs (Fig. 4A). In agreement with the increased mitochondrial activity, the expression of Atp5g1 (ATP synthase lipid-binding protein, mitochondrial) was significantly higher in Cited2 KO HSCs (Fig. 4A).
Loss of Cited2 affects Akt/mTOR/FoxOs activities
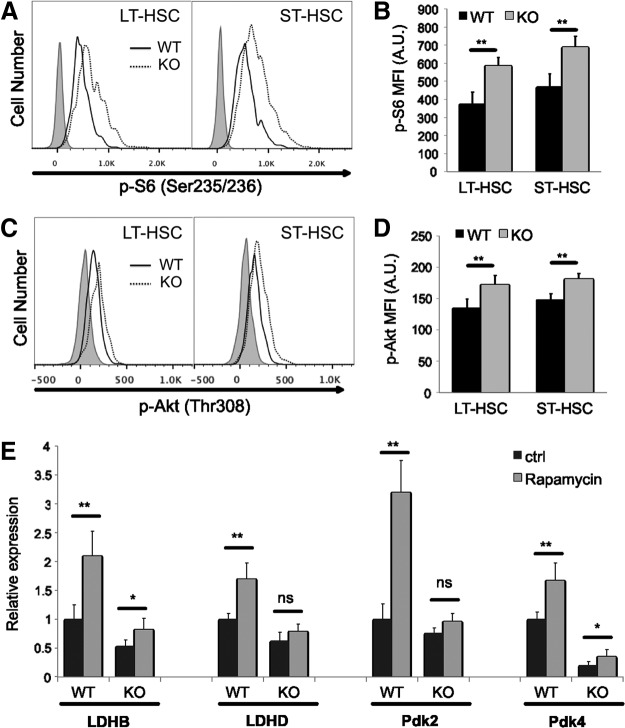
mTOR signaling pathway is a well-studied nutrient/energy/redox sensor controlling protein synthesis and mitochondrial function. To assess the mTORC1 activity in Cited2 KO HSCs, we examined phosphorylation of the S6 ribosomal protein, an indicator of the mTORC1 activity [50]. As shown in Fig. 5A and B, S6 phosphorylation was significantly higher in Cited2 KO HSCs than WT controls. In addition, phosphorylation of Akt, an upstream positive regulator of mTORC1, was significantly increased as well (Fig. 5C, D). We then tested whether the elevated mTORC1 signaling contributes to the dysregulation of metabolism-related genes in Cited2 KO HSCs. Fresh bone marrow cells from WT and KO mice were cultured under hypoxia (2% O2) with or without 20 nM rapamycin (Sigma-Aldrich) for 6 h. LT-HSCs were then sorted for gene expression analysis. As shown in Fig. 5E, inhibition of the mTORC1 activity by rapamycin increased the expression of LDHB, LDHD, Pdk2, and Pdk4 in WT, but to a lesser extent in Cited2 KO LT-HSCs, suggesting that elevated mTORC1 activity may not be the major contributor to the downregulation of glycolysis-related genes in Cited2 KO LT-HSCs.
FIG. 5.
Cited2 KO HSCs display elevated Akt-mTOR signaling activity. (A) Representative plot of p-S6 (Ser235/236) staining. (B) Quantitative analysis of p-S6 MFI on cells. (C) Representative plot of p-Akt (Thr308) staining. (D) Quantitative analysis of p-Akt MFI on cells. (E) Relative expression of genes determined by real-time quantitative PCR. Fresh bone marrow cells were cultured at 37°C under hypoxia (2% O2) for 6 h in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 50 ng/mL recombinant murine stem cell factor (rmSCF), 20 ng/mL recombinant murine interleukin-3 (rmIL-3), and 50 ng/mL recombinant human interleukin-6 (rhIL-6; R&D Systems), with or without 20 nM rapamycin. LT-HSCs were then sorted for RNA extraction and gene expression analysis. The results are shown as mean±SD (n=4). Asterisks are explained in Materials and Methods Statistics section.
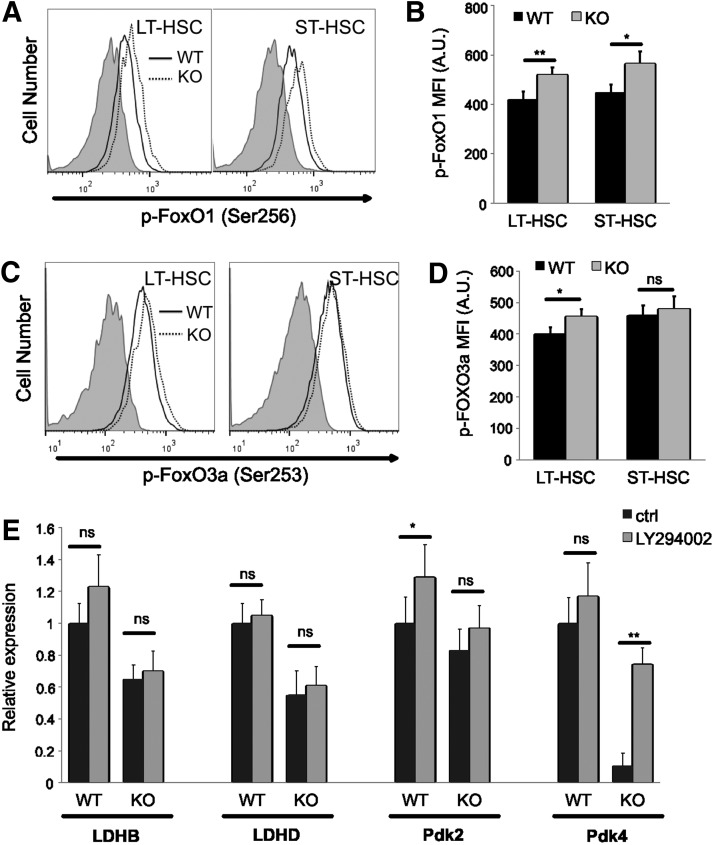
FoxO transcription factors are critical for the maintenance of HSCs [23,24]. Phosphorylation of FoxOs by Akt causes FoxOs' nuclear export, thus lowering their transcriptional activity. Pdk4 is a known target of FoxO1 and FoxO3a [51–55]. To understand the alterations of FoxOs in Cited2 KO HSCs, the levels of gene expression as well as phosphorylation of FoxO1 and FoxO3a were examined by real-time PCR and intracellular staining with flow cytometry, respectively. Loss of Cited2 did not significantly affect the mRNA levels of FoxO1 and FoxO3a in HSCs (data not shown). However, concomitant with the increased phosphorylation of Akt and S6 (Fig. 5A–D), Cited2 KO HSCs exhibited markedly increased phosphorylation of FoxO1 and moderately increased phosphorylation of FoxO3a (Fig. 6A–D). Further, treatment of HSCs with PI3K/Akt inhibitor LY294002 partially rescued the expression of Pdk4 although did not significantly affect the expression of LDHB, LDHD, and Pdk2 (Fig. 6E). Together, these findings suggest that the downregulation of Pdk4 in Cited2 KO HSCs is likely mediated by the inactivation of FoxOs caused by the elevated Akt activity.
FIG. 6.
Cited2 deletion leads to increased phosphorylation of FoxOs in HSCs. (A) Representative plot of p-FoxO1 (Ser256) staining. (B) Quantitative analysis of p-FoxO1 MFI on cells. (C) Representative plot of p-FoxO3a (Ser253) staining. (D) Quantitative analysis of p-FoxO3a MFI on cells. (E) Relative expression of genes determined by real-time quantitative PCR. Fresh bone marrow cells were cultured at 37°C under normoxia for 90 min in DMEM supplemented with 15% FBS, 50 ng/mL rmSCF, 20 ng/mL rmIL-3, and 50 ng/mL rhIL-6, with or without 10 μM PI3K/Akt inhibitor LY294002. LT-HSCs were then sorted for RNA extraction and gene expression analysis. The results are expressed as mean±SD (n=4–6). Asterisks are explained in Materials and Methods Statistics section.
Cited2/HIF-1α double KO HSCs display altered metabolism
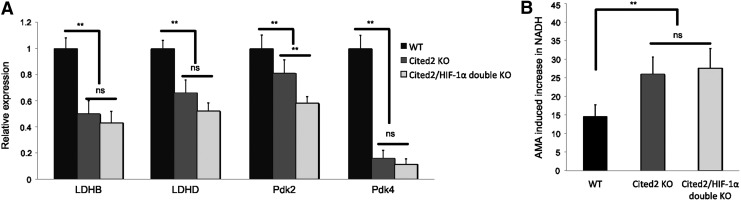
We previously showed that deletion of Cited2 results in loss of HSC quiescence that can be partially rescued by additional deletion of HIF-1α [40]. Thus, we further determined whether deletion of HIF-1α could partially rescue the dysregulation of LDHB, LDHD, Pdk2, and Pdk4 observed in Cited2 KO HSCs. As shown in Fig. 7A, expression of none of these genes was rescued in Cited2/HIF-1α double KO LT-HSCs. In addition, AMA treatment led to comparable accumulation of NADH in double KO LT-HSCs as seen in Cited2 KO LT-HSCs (Fig. 7B), suggesting that deletion of HIF-1α could not rescue dysregulated glucose metabolism caused by the Cited2 deficiency.
FIG. 7.
Cited2/HIF-1α double KO LT-HSCs display reduced glycolytic metabolism. (A) Gene expression analysis of LT-HSCs by real-time quantitative PCR (mean±SD, n=4). (B) Percentage of LT-HSCs that display increased NADH fluorescence in response to AMA treatment (mean±SD, n=4). Asterisks are explained in Materials and Methods Statistics section.
Discussion
Compared with lineage-specific progenitors, mammalian adult HSCs exhibit distinct metabolic features, including more utilization of glycolysis, less mitochondrial activity, lower mitochondrial membrane potential, and lower ATP levels [3,5,11,13,29,56]. In this study, we revealed for the first time an essential role of Cited2 in the metabolic regulation of HSCs. Deletion of Cited2 in HSCs resulted in increased mitochondrial mass, elevated mitochondrial membrane potential, elongation of mitochondrial shape, decreased cellular GSH content, and elevated ROS production (Figs. 1 and 2). Mitochondrial copy number remained unchanged, which is similar to PTPMT1-depleted HSCs [29]. Interestingly, an increase in mitochondrial mass but not copy number has been observed previously in other cell types. Apostolova et al. reported that human hepatic cells display increased mitochondrial mass during Efavirenz-induced apoptosis, which is not paralleled by an increase in the ratio of mitochondrial DNA/nuclear DNA copy number [57].
We found that, in comparison with WT HSCs, Cited2-deficient HSCs display elongated and brighter mitochondria, resembling those in older WT HSCs shown previously [48]. In fact, dysfunction of mitochondria and abnormal levels of ROS have been implicated in aging HSCs [58]. It will be interesting to investigate the relationship between Cited2 and HSC aging in the future. On the other hand, increasing evidence has indicated that tight regulation of mitochondrial homeostasis is critical for HSC integrity. Indeed, Lkb1-deficient HSCs display defective mitochondrial function and are prone to exhaustion [25–27]. Conversely, abnormally increased mitochondrial biogenesis in Tsc1 KO HSCs leads to elevated levels of ROS and impaired self-renewal capacity [16]. These findings underscore the need of precisely controlling mitochondrial homeostasis to ensure HSC maintenance. Moreover, the mitochondrial fusion/fission dynamics, which regulates mitochondrial morphology, is closely related to the survival and differentiation of stem cells. For instance, overexpression of Gfer (growth factor erv1-like) in mouse embryonic stem cells results in decreased levels of Drp1 (the mitochondrial fission GTPase dynamin-related protein 1), elongated mitochondria, and protection from apoptosis [59]. Elongated mitochondria are also found during differentiation of mouse embryonic stem cells to epiblast stem cells [49]. Therefore, mitochondrial elongation observed in Cited2 KO HSCs could result from increased fusion and/or decreased fission of mitochondria. Although no significant changes were observed in the mRNA levels of mitochondrial fusion proteins Mfn1 and Mfn2, and fission protein Drp1 (data not shown), their roles could not be excluded in this context because their activities are largely modulated at the post-translational level.
Glycolysis in Cited2 KO HSCs was markedly reduced as indicated by decreased 13C-lactate production and increased NADH accumulation upon AMA treatment (Fig. 3D–F). Interestingly, the steady-state cellular ATP level was maintained in Cited2 KO LT-HSCs (Fig. 3C). It has been shown that in PTPMT1-depleted LT-HSCs where mitochondrial metabolism is altered, the steady-state level of ATP is maintained [29]. In HIF-1α-deficient LT-HSCs, mitochondrial activity is increased while the overall ATP level is decreased [13]. Given that about 18-fold more ATP is generated by mitochondrial OXPHOS than by glycolysis (36–38 vs. 2 ATP), the finding that LT-HSCs fail to produce more ATP upon switch from glycolysis to OXPHOS suggests a limited ability of LT-HSCs to utilize OXPHOS, a hallmark of quiescent HSCs, and highlights an essential role for anaerobic glycolysis in energy production by LT-HSCs [60].
Akt-mTOR signaling was elevated in Cited2 KO HSCs (Fig. 5A–D). We previously showed that the expression of Egr1, an early response transcription factor, is significantly decreased in Cited2 KO HSCs [40]. In addition, Egr1 has been shown to block the PI3K/Akt signaling pathway [61]. Therefore, the elevated Akt activity in Cited2 KO HSCs may be, at least in part, due to the reduced level of Egr1. Loss of Cited2 significantly decreased the expression of LDHB, LDHD, Pdk2, and Pdk4, which encode critical enzymes for the maintenance of glycolysis (Fig. 4A). Although mTORC1 plays important roles in the regulation of mitochondrial activity and the maintenance of HSCs, inhibition of mTORC1 activity by rapamycin did not rescue the expression of LDHB, LDHD, Pdk2, and Pdk4 in Cited2 KO HSCs (Fig. 4E), which is similar to the observations in Lkb1 KO HSCs. HSCs with deletion of Lkb1 lose quiescence and metabolic homeostasis in an mTOR-independent manner [25–27].
FoxO proteins play essential roles in HSCs in response to physiologic oxidative stress [23,24]. In addition to the elevated activity of mTORC1, Cited2 KO HSCs displayed increased phosphorylation of FoxO1 and FoxO3a (Fig. 6A–D), which are known to regulate transcription of the Pdk4 gene. More importantly, inhibition of PI3/Akt signaling partially rescued the expression of Pdk4 (Fig. 6E), thus suggesting that inactivation of FoxO1 and FoxO3a may partially contribute to the reduced expression of Pdk4 in Cited2 KO HSCs. On the other hand, loss of Cited2 has been shown to increase the p53 protein level and the expression of its target gene p21 [39]. Interestingly, p53 negatively regulates the expression of Pdk2 in a p21-dependent manner [62]. Therefore, the downregulation of Pdk2 in Cited2 KO HSCs could be mediated through the increased p53 activity. The expression of numerous metabolism-related genes was not significantly affected in Cited2 KO HSCs, such as PGC-1, Lkb1, PTPMT1, Esrrb, Pfkm, Pklr, Tpi1, and Tfam (data not shown). Since many of these molecules are regulated by post-translational modifications, their roles in Cited2 KO HSCs cannot be excluded.
Cited2 KO HSCs exhibit loss of quiescence that can be partially rescued by additional deletion of HIF-1α [40]. An important question is whether the altered metabolism observed in Cited2 KO HSCs is the cause or the consequence of loss in quiescence? Interestingly, deletion of HIF-1α did not rescue the metabolic dysregulation caused by Cited2 deficiency (Fig. 7), indicating that metabolic alterations in Cited2 KO HSCs are not likely consequent to loss of quiescence, which is consistent with our recent finding that deletion of Cited2 in mouse embryonic stem cells results in changes in metabolic activities (unpublished data). Because deletion of HIF-1α only partially rescues the defects caused by Cited2 deficiency [40], suggesting the involvement of both HIF-1α-dependent and -independent pathways in regulating quiescence of HSCs by Cited2, we cannot rule out the possibility that the increased mitochondrial activity in Cited2 KO HSCs is triggered to support increased proliferation and/or differentiation in these cells. On the other hand, Takubo et al. [13] showed that Pdk4, a target of HIF-1, functions as a metabolic checkpoint in HSCs. In this study, we found that Pdk4 expression was markedly reduced in Cited2 KO HSCs, which was partially rescued by PI3/Akt inhibitor under normoxia (Fig. 6E). In Cited2 KO HSCs, we also observed increased phosphorylation of FoxO1 and FoxO3, which are known regulators of Pdk4 expression [51–55]. Although HIF-1 has been shown to induce the expression of FoxO3 [63], the FoxO3 mRNA level remains unchanged in Cited2 KO HSCs, suggesting that Cited2 controls the level of Pdk4 independent of HIF-1. Collectively, these results favor the notion that Cited2 has a direct role on metabolic regulation of HSCs, and further studies are hence required to thoroughly address this issue.
Together, our results suggest a critical role of Cited2 in the maintenance of glycolytic metabolism in murine adult HSCs. This finding has significant implications for clinical medicine. Emerging evidence has shown that quiescent leukemic stem cells (LSCs) display low glycolysis rate and depend more on mitochondrial respiration to meet their energy demands and maintain their survival, in contrast to proliferating leukemia cells that utilize anaerobic glycolysis (referred to as “Warburg effect”) [64]. Meanwhile, downregulation of Cited2 has been implicated in various types of leukemia [65]. Therefore, it will be interesting to explore whether downregulation of Cited2 favors the survival of LSCs through facilitating mitochondrial metabolism and whether manipulation of Cited2 activities could be a potential therapeutic alternative that targets metabolism of LSCs.
Acknowledgments
The authors thank Dr. Henri Brunengraber at Mouse Metabolic Phenotyping Center (MMPC) and Department of Nutrition (CWRU) for help with experimental design and discussion of data; Michael Sramkoski at Cytometry & Imaging Microscopy Core Facility (CWRU) for help with flow cytometric analysis; and Dr. Diana Ramirez at Case Cardiovascular Research Institute (CWRU) for help with cell culture under hypoxia.
This study was supported by the National Institutes of Health (R01HL091896 to Y.-C.Y.), CTSC Core Utilization Pilot Grant (to Y.-C.Y.), the National Health and Medical Research Council (SRF 1042002 to S.L.D.), and National Institutes of Health (U24 DK76174 to Mouse Metabolic Phenotyping Center, CWRU). This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the NIH and NIH road map for Medical Research.
Author Disclosure Statement
The authors declare no conflict of financial interests.
References
- 1.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, et al. (2003). Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425:836–841 [DOI] [PubMed] [Google Scholar]
- 2.Eliasson P. and Jonsson JI. (2010). The hematopoietic stem cell niche: low in oxygen but a nice place to be. J Cell Physiol 222:17–22 [DOI] [PubMed] [Google Scholar]
- 3.Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC. and Sadek HA. (2010). The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7:380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu S, Broxmeyer HE. and Hangoc G. (2013). Peroxisome proliferator-activated-gamma coactivator-1alpha-mediated mitochondrial biogenesis is important for hematopoietic recovery in response to stress. Stem cells Dev 22:1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norddahl GL,, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, Sigvardsson M. and Bryder D. (2011). Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell 8:499–510 [DOI] [PubMed] [Google Scholar]
- 6.Romero-Moya D, Bueno C, Montes R, Navarro-Montero O, Iborra FJ, Lopez LC, Martin M. and Menendez P. (2013). Cord blood-derived CD34+ hematopoietic cells with low mitochondrial mass are enriched in hematopoietic repopulating stem cell function. Haematologica 98:1022–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y. and Suda T. (2006). Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med 12:446–451 [DOI] [PubMed] [Google Scholar]
- 8.Jang YY. and Sharkis SJ. (2007). A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 110:3056–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M. and Suda T. (2010). Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7:391–402 [DOI] [PubMed] [Google Scholar]
- 10.Miharada K, Karlsson G, Rehn M, Rorby E, Siva K, Cammenga J. and Karlsson S. (2011). Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell 9:330–344 [DOI] [PubMed] [Google Scholar]
- 11.Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, Copeland NG, Jenkins NA, Grimes HL. and Kumar AR. (2012). Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood 120:4973–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kocabas F, Zheng J, Thet S, Copeland NG, Jenkins NA, DeBerardinis RJ, Zhang C. and Sadek HA. (2012). Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 120:4963–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, et al. (2013). Regulation of glycolysis by pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12:49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yilmaz OH,, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H. and Morrison SJ. (2006). Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441:475–482 [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, et al. (2006). PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441:518–522 [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Liu Y, Liu R, Ikenoue T, Guan KL. and Zheng P. (2008). TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med 205:2397–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ. and DePinho RA. (2008). mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci U S A 105:19384–19389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell TB,, Basu S, Hangoc G, Tao W. and Broxmeyer HE. (2009). Overexpression of Rheb2 enhances mouse hematopoietic progenitor cell growth while impairing stem cell repopulation. Blood 114:3392–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Nguyen-McCarty M, Hexner EO, Danet-Desnoyers G. and Klein PS. (2012). Maintenance of hematopoietic stem cells through regulation of Wnt and mTOR pathways. Nat Med 18:1778–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, Sinha AU, Lane SW, Souza AL, et al. (2012). mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell 11:429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juntilla MM,, Patil VD, Calamito M, Joshi RP, Birnbaum MJ. and Koretzky GA. (2010). AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood 115:4030–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kharas MG,, Okabe R, Ganis JJ, Gozo M, Khandan T, Paktinat M, Gilliland DG. and Gritsman K. (2010). Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood 115:1406–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, et al. (2007). Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1:101–112 [DOI] [PubMed] [Google Scholar]
- 24.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, et al. (2007). FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325–339 [DOI] [PubMed] [Google Scholar]
- 25.Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, et al. (2010). The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468:659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakada D, Saunders TL. and Morrison SJ. (2010). Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468:653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, Fletcher-Sananikone E, Colla S, Wang YA, Chin L. and Depinho RA. (2010). Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468:701–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH. and Pandolfi PP. (2012). A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med 18:1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu WM,, Liu X, Shen J, Jovanovic O, Pohl EE, Gerson SL, Finkel T, Broxmeyer HE. and Qu CK. (2013). Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell 12:62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun HB,, Zhu YX, Yin T, Sledge G. and Yang YC. (1998). MRG1, the product of a melanocyte-specific gene related gene, is a cytokine-inducible transcription factor with transformation activity. Proc Natl Acad Sci U S A 95:13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharya S, Michels CL, Leung MK, Arany ZP, Kung AL. and Livingston DM. (1999). Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev 13:64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bamforth SD,, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, Kranc KR, Farza H, Henderson DJ, Hurst HC. and Bhattacharya S. (2001). Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet 29:469–474 [DOI] [PubMed] [Google Scholar]
- 33.Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, Yuan S, Prabhakar NR, Herrup K, et al. (2002). The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci U S A 99:10488–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bamforth SD,, Braganca J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, et al. (2004). Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet 36:1189–1196 [DOI] [PubMed] [Google Scholar]
- 35.Weninger WJ,, Lopes Floro K, Bennett MB, Withington SL, Preis JI, Barbera JP, Mohun TJ. and Dunwoodie SL. (2005). Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development 132:1337–1348 [DOI] [PubMed] [Google Scholar]
- 36.Qu X, Lam E, Doughman YQ, Chen Y, Chou YT, Lam M, Turakhia M, Dunwoodie SL, Watanabe M, et al. (2007). Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J 26:4445–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glenn DJ. and Maurer RA. (1999). MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J Biol Chem 274:36159–36167 [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Haviernik P, Bunting KD. and Yang YC. (2007). Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood 110:2889–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kranc KR,, Schepers H, Rodrigues NP, Bamforth S, Villadsen E, Ferry H, Bouriez-Jones T, Sigvardsson M, Bhattacharya S, Jacobsen SE. and Enver T. (2009). Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell 5:659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du J, Chen Y, Li Q, Han X, Cheng C, Wang Z, Danielpour D, Dunwoodie SL, Bunting KD. and Yang YC. (2012). HIF-1alpha deletion partially rescues defects of hematopoietic stem cell quiescence caused by Cited2 deficiency. Blood 119:2789–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Ramirez-Bergeron DL, Dunwoodie SL. and Yang YC. (2012). Cited2 controls pluripotency and cardiomyocyte differentiation of murine embryonic stem cells through Oct4. J Biol Chem 287:29088–29100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong JF,, Zhao Y, Sutton S, Su A, Zhan Y, Zhu L, Yan C, Gallaher T, Johnston PB, Anderson WF. and Cooke MP. (2005). Gene expression profile of murine long-term reconstituting vs. short-term reconstituting hematopoietic stem cells. Proc Natl Acad Sci U S A 102:2448–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pendergrass W, Wolf N. and Poot M. (2004). Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A 61:162–169 [DOI] [PubMed] [Google Scholar]
- 44.Rice GC,, Bump EA, Shrieve DC, Lee W. and Kovacs M. (1986). Quantitative analysis of cellular glutathione by flow cytometry utilizing monochlorobimane: some applications to radiation and drug resistance in vitro and in vivo. Cancer Res 46:6105–6110 [PubMed] [Google Scholar]
- 45.Hedley DW. and Chow S. (1994). Evaluation of methods for measuring cellular glutathione content using flow cytometry. Cytometry 15:349–358 [DOI] [PubMed] [Google Scholar]
- 46.Yang L, Kasumov T, Kombu RS, Zhu SH, Cendrowski AV, David F, Anderson VE, Kelleher JK. and Brunengraber H. (2008). Metabolomic and mass isotopomer analysis of liver gluconeogenesis and citric acid cycle: II. Heterogeneity of metabolite labeling pattern. J Biol Chem 283:21988–21996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye F, Lemieux H, Hoppel CL, Hanson RW, Hakimi P, Croniger CM, Puchowicz M, Anderson VE, Fujioka H. and Stavnezer E. (2011). Peroxisome proliferator-activated receptor gamma (PPARgamma) mediates a Ski oncogene-induced shift from glycolysis to oxidative energy metabolism. J Biol Chem 286:40013–40024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warr MR,, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J. and Passegue E. (2013). FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494:323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C. and Ruohola-Baker H. (2012). HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J 31:2103–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magnuson B, Ekim B. and Fingar DC. (2012). Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J 441:1–21 [DOI] [PubMed] [Google Scholar]
- 51.Furuyama T, Kitayama K, Yamashita H. and Mori N. (2003). Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J 375:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim YI,, Lee FN, Choi WS, Lee S. and Youn JH. (2006). Insulin regulation of skeletal muscle PDK4 mRNA expression is impaired in acute insulin-resistant states. Diabetes 55:2311–2317 [DOI] [PubMed] [Google Scholar]
- 53.Nahle Z, Hsieh M, Pietka T, Coburn CT, Grimaldi PA, Zhang MQ, Das D. and Abumrad NA. (2008). CD36-dependent regulation of muscle FoxO1 and PDK4 in the PPAR delta/beta-mediated adaptation to metabolic stress. J Biol Chem 283:14317–14326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long YC,, Cheng Z, Copps KD. and White MF. (2011). Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Mol Cell Biol 31:430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo L, Xie B. and Mao Z. (2012). Autophagy in premature senescent cells is activated via AMPK pathway. Int J Mol Sci 13:3563–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue S, Noda S, Kashima K, Nakada K, Hayashi J. and Miyoshi H. (2010). Mitochondrial respiration defects modulate differentiation but not proliferation of hematopoietic stem and progenitor cells. FEBS Lett 584:3402–3409 [DOI] [PubMed] [Google Scholar]
- 57.Apostolova N, Gomez-Sucerquia LJ, Moran A, Alvarez A, Blas-Garcia A. and Esplugues JV. (2010). Enhanced oxidative stress and increased mitochondrial mass during efavirenz-induced apoptosis in human hepatic cells. Br J Pharmacol 160:2069–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geiger H, de Haan G. and Florian MC. (2013). The ageing haematopoietic stem cell compartment. Nat Rev Immunol 13:376–389 [DOI] [PubMed] [Google Scholar]
- 59.Todd LR,, Damin MN, Gomathinayagam R, Horn SR, Means AR. and Sankar U. (2010). Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell 21:1225–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Folmes CD,, Dzeja PP, Nelson TJ. and Terzic A. (2012). Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11:596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X, Shen N, Zhang ML, Pan FY, Wang C, Jia WP, Liu C, Gao Q, Gao X, Xue B. and Li CJ. (2011). Egr-1 decreases adipocyte insulin sensitivity by tilting PI3K/Akt and MAPK signal balance in mice. EMBO J 30:3754–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Contractor T. and Harris CR. (2012). p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res 72:560–567 [DOI] [PubMed] [Google Scholar]
- 63.Bakker WJ,, Harris IS. and Mak TW. (2007). FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell 28:941–953 [DOI] [PubMed] [Google Scholar]
- 64.Lagadinou ED,, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, et al. (2013). BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, De Vos J, Hernandez JM, Hofmann WK, et al. (2010). Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol 28:2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]