Abstract
Hepatitis C virus (HCV) establishes chronic infection in a large number of infected individuals. We have previously shown that HCV infection in hepatocytes blocks poly (I-C) or interferon (IFN)-α-mediated IRF-7 nuclear translocation (Raychoudhuri and others 2010). However, the mechanism of IRF-7 regulation by HCV remained unknown. In this study, we have observed that HCV NS5A physically associates with IRF-7. A subsequent study suggested that the HCV NS5A protein blocks IRF-7-mediated IFN-α14 promoter activation. Further analyses demonstrated that site-specific mutagenesis of the 2 basic arginine residues (amino acids Arg216 and Arg217) in the NS5A is critical for IRF-7-mediated IFN-α14 promoter regulation. Together, our results suggested that the HCV NS5A protein limits the IFN-α-signaling pathway in association with IRF-7, and may, in part, be responsible for the establishment of chronic infection.
Introduction
Hepatitis C virus (HCV) infection affects approximately 3.2 million people in the United States and a large number in infected individuals develop chronicity (Armstrong and others 2006). The host response is triggered when a pathogen-associated molecular pattern (PAMP) presented by the infecting virus is recognized and engaged by specific PAMP receptor factors expressed in the host cell, initiating signals that ultimately induce the expression of antiviral effector genes (Ramos and Gale 2011). Interferon (IFN)-α and IFN-β are rapidly synthesized after virus infection and trigger intracellular signaling events. The subsequent expression of IFN-stimulated genes (ISGs) is central to these antiviral responses. The transcription factors STAT1 and STAT2 are phosphorylated by Janus protein tyrosine kinases Jak1 and Tyk2 following IFN receptor activation, and released from their docking sites on the receptor (Kisseleva and others 2002). STAT1/STAT2 associate with IRF-9 and form the ISGF3 complex, which stimulates IFN-α/β-dependent gene transcription by binding to the IFN-stimulated response element (ISRE) sequences located in the promoter of target genes (Sen 2001). ISRE sequences are also found in the promoter of IRF-7, and ISGF3 has been shown to activate the IRF-7 gene (Lu and others 2000).
In a majority of cell types, including epithelial, fibroblastic, and myeloid dendritic cells, virus-induced IFN-α gene expression is mediated through both IRF-3 and IRF-7 (Schröder and Bowie 2007). IRF-7 undergoes phosphorylation when activated and translocates into the nucleus. IRF-7 amplifies the type I IFN response by inducing expression of IFN-α, which also acts in both autocrine and paracrine manners through the IFN-α/β receptors. IFN and ISGs are amplified during chronic HCV infection (MacQuillan and others 2003; Bigger and others 2004; Sarasin-Filipowicz and others 2008), but fail to eliminate virus from the liver in a large number of HCV-infected patients. We have previously observed that IRF-7 remains localized in the cytoplasm of HCV-infected hepatocytes (Raychoudhuri and others 2010), although the underlying mechanism remains unknown. In this study, we have observed that the HCV NS5A protein physically associates with IRF-7 and blocks IFN-α14 promoter activity. Further, we identified amino acid residues, Arg216 and Arg217 of NS5A, critical for blocking IRF-7-mediated IFN-α promoter activation. Taken together, we have determined that HCV NS5A is responsible for impairment of IFN-α synthesis.
Materials and Methods
Cell culture and transfection
Immortalized human hepatocytes (IHH) were generated by transfection of the HCV core gene from genotype 1a into primary human hepatocytes (Ray and others 2000). IHH supported the growth of both the HCV genotype 1a and genotype 2a (Kanda and others 2006). IHH were grown in the Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 100 U/mL of penicillin G, and 100 μg/mL of streptomycin at 37°C in a 5% CO2 atmosphere (Raychoudhuri and others 2010). IRF-7-GFP (kindly provided by Betsy Barnes, NJMSUH Cancer Center) and HCV NS5A or NS5A-mutant (fused with a FLAG epitope and cloned under the control of a CMV promoter) plasmid DNAs were transfected into IHH by using the Lipofectamine reagent (Invitrogen). Cells were incubated for 48–72 h before analysis.
Immunofluorescence
IHH transfected with IRF-7-GFP and HCV NS5A or NS5A-mutant (fused with a FLAG tag). After 48h of transfection, cells were fixed with 3.7% formaldehyde for 20 min at room temperature, permeabilized with 0.2% Triton-X 100 for 5 min, and blocked with 3% bovine serum albumin for 1 h. Fixed cells were incubated with the anti-FLAG M2 (Sigma) mouse monoclonal antibody for 1 h. Cells were washed and incubated with anti-mouse Ig conjugated with Alexa 594 (Invitrogen) for 1 h at room temperature. Finally, cells were washed and mounted for confocal microscopy (Olympus FV1000).
Site directed mutagenesis of NS5A
HCV NS5A-mutant (fused with FLAG tag) was constructed by site-directed mutagenesis at Arg216 and Arg217 to Ala using a QuickChange II E Site-Directed Mutagenesis kit (Agilent Technologies) following the supplier's protocol. The primers used for the cloning of NS5A-mutant were forward primer; 5′-GCCGGGGCAGCGTTGGCGAGAGGGTCA-3′ and reverse primer; 5′-TGACCCTCTCGCCAACGCTGCC CCGGC-3′.
In vitro promoter assay using luciferase reporter gene
IHH were cotransfected with a human IFN-α-luc reporter plasmid DNA and IRF-7 with or without the HCV NS5A plasmid or its mutant using Lipofectamine. In a different experiment, IHH were cotransfected with the IFN-α-luc reporter plasmid and IRF-7 with or without HCV genomic region encoding specific protein(s). Luciferase assay was performed 48 h post-transfection as previously described (Kanda and others 2007). Briefly, cells were treated with the lysis buffer (Promega), and the luciferase activity in the lysates was analyzed by integrating the total light emission over 10 s using a luminometer (Glomax; Promega). The luciferase activity was normalized based on the protein concentration. The results are presented as mean of 3 independent experiments. A separate set of transfected cells was examined for NS5A and its mutant protein expression by Western blot analysis using a FLAG tag-specific antibody (Sigma).
Immunoprecipitation assay
IRF-7-GFP or vector alone was transfected into Huh7 cells harboring HCV subgenomic replicon 1b or 2a. In a different experiment, human embryonic kidney epithelial (293) cells were cotransfected with IRF-7 GFP and HCV NS5A or vector control (pFLAG-CMV). Cells were lysed after 48 h of post-transfection by the lysis buffer supplemented with protease inhibitor cocktail (Roche). Cell lysates were clarified by centrifugation at 14,000 rpm for15 min and incubated with a rabbit antibody to GFP (SantaCruz) overnight at 4°C. Protein G-agarose beads were added and incubated at 4°C for 6 h. Immunoprecipitates were washed 4–5 times with the lysis buffer, boiled with a 1×protein loading buffer, and subjected to Western blot analysis using specific antibodies.
Results
IRF-7-mediated IFN promoter activation is inhibited by HCV NS5A
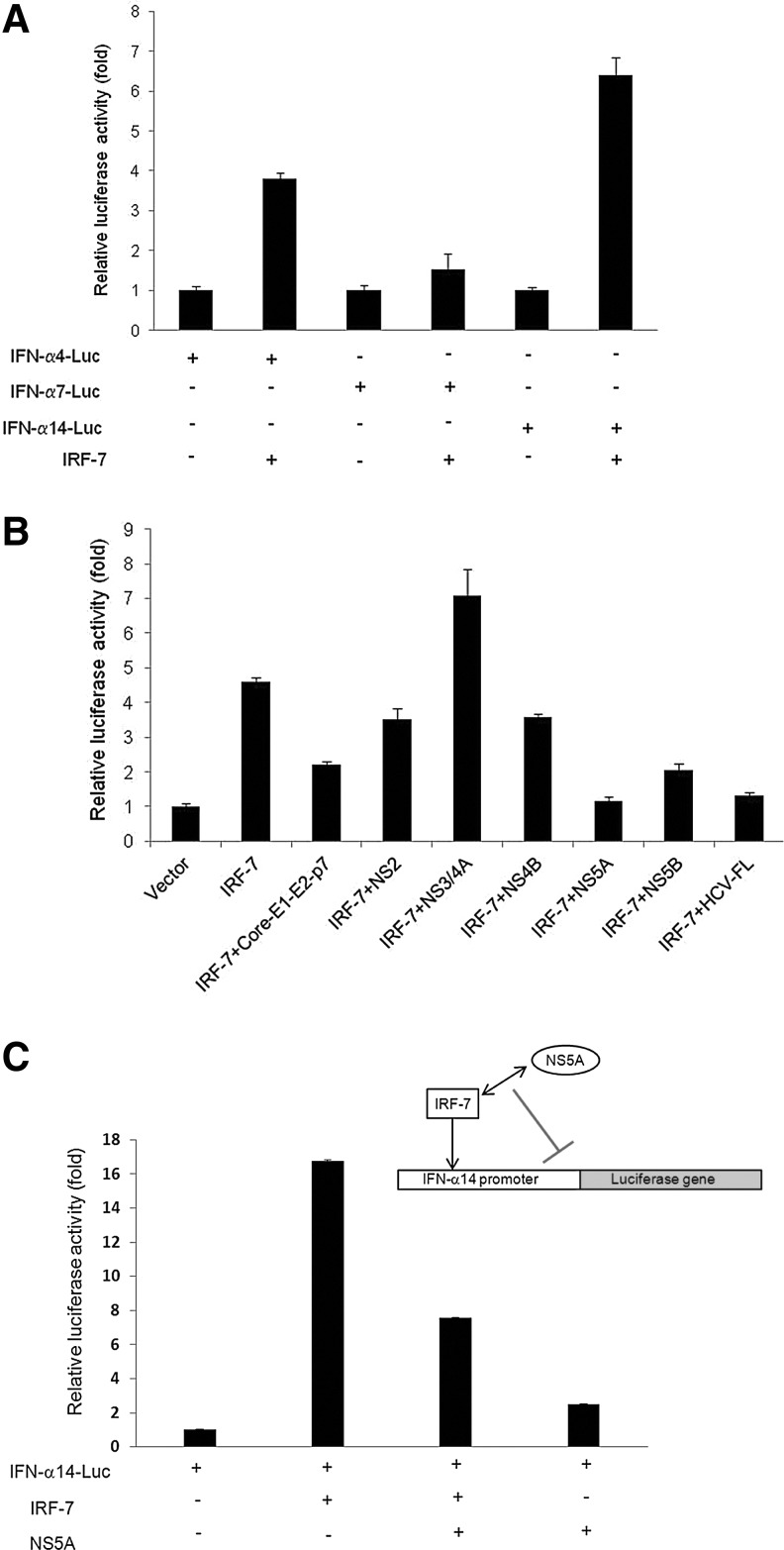
Since IRF-7 is the major inducer for IFN-α synthesis, we initially investigated the role of HCV proteins in IRF-7-mediated activation of IFN-α promoter. IRF-7 can induce IFN-α4, IFN-α7, and IFN-α14 promoter activities (Lin and others 2000). We tested all 3 promoters cloned into a luciferase construct along with IRF-7 in an in vitro assay and observed a strong activation of IFN-α14 promoter by IRF-7 in hepatocytes as compared to IFN-α4 or IFN-α7 promoter (Fig. 1A). Thus, we performed a subsequent study using IFN-α14 promoter.
FIG. 1.
Hepatitis C virus (HCV) NS5A suppresses IRF-7-mediated interferon (IFN)-α14 promoter activity. (A) Immortalized human hepatocytes (IHH) were cotransfected with human IFN-α4, IFN-α7, or IFN-α14 promoter with the Luc reporter and IRF-7 plasmid constructs. Cell extracts were prepared 48 h post-transfection and the luciferase activity was measured. (B) IHH were cotransfected with human IFN-α14-luc reporter plasmid, IRF-7, and HCV genomic region encoding specific protein(s). Cell extracts were prepared for luciferase assay 48 h post-transfection. (C) IHH were cotransfected with human IFN-α14-luc reporter plasmid, IRF-7 construct, and/or HCV NS5A. Cell extracts were prepared for luciferase assay 48 h post-transfection. Results presented are mean with standard error from 3 independent experiments. Empty vector DNA was used as a negative control and the mean basal value was arbitrarily set at 1 for all luciferase assays.
Next, an in vitro reporter assay was performed by transfecting IHH with the human IFN-α14-Luc construct, IRF-7, and/or different HCV genomic regions. Cell lysates were prepared 48 h post-transfection for luciferase assay. We observed that HCV NS5A display the most potent inhibitory effect on the IRF-7-mediated IFN-α14 promoter activity (Fig. 1B). Cells transfected with NS5A or mutants were separately examined and displayed protein expression using a tag antibody by immunofluorescence (not shown). To further verify whether HCV NS5A has an effect on IFN-α14 promoter alone, we cotransfected IHH with human IFN-α14-luc, IRF-7, and HCV NS5A plasmid DNAs for luciferase assay. Our results suggested that HCV NS5A, in the absence of IRF-7, has no regulatory effects upon IFN-α14 promoter (Fig. 1C).
HCV NS5A associates with IRF-7
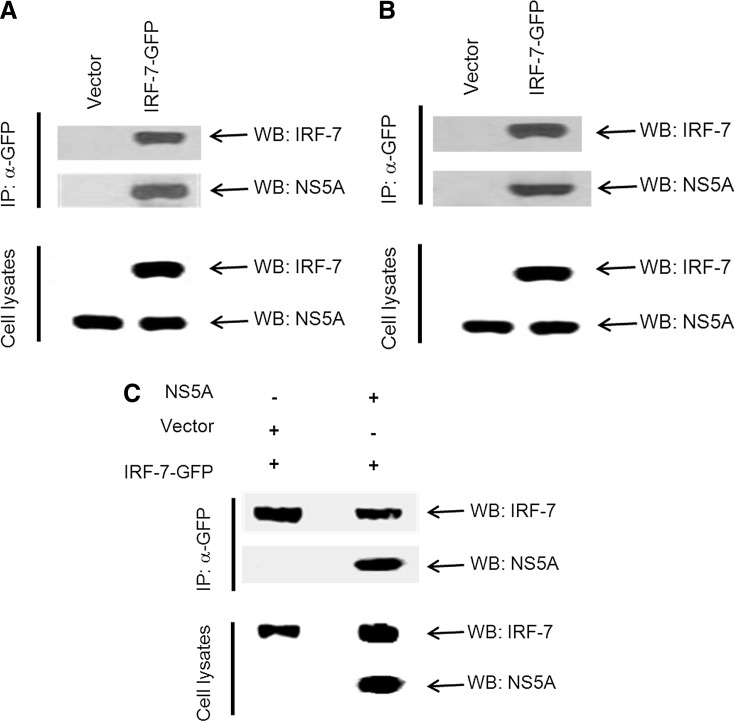
We previously observed that hepatocytes treated with poly (I-C) or IFN-α display nuclear localization of IRF-7 (Raychowdhuri and others 2010). In contrast, IRF-7 was retained in the cytoplasm in HCV-infected IHH after treatment with poly (I-C) or IFN-α. Here we examined whether the HCV NS5A protein is responsible for retaining IRF-7 in the cytoplasm. For this, IRF-7-GFP plasmid DNA was transfected into Huh7 cells harboring the HCV genotype 1b or HCV 2a subgenomic replicon. After 48 h of transfection, cell lysates were used for immunoprecipitation using the anti-GFP antibody. The HCV NS5A protein was co-immunoprecipitated with IRF-7 GFP from both the replicon harboring cell lines (Fig. 2A, B). A portion of the cell lysates was run separately for Western blot analysis to detect the presence of HCV NS5A or IRF-7. To further verify the association, we cotransfected 293 cells with HCV NS5A and IRF-7. After 48 h of transfection, cell lysates were processed for co-immunoprecipitation. An association between HCV NS5A and IRF-7-GFP was observed (Fig. 2C).
FIG. 2.
HCV NS5A associates with IRF-7. (A, B) IRF-7-GFP or control vector was transfected into Huh-7 cells harboring HCV subgenomic replicon from genotype 1b (A) or genotype 2a (B). Cell lysates were prepared 48 h post-transfection, and immunoprecipitated with the anti-GFP antibody. Association of IRF-7 and HCV NS5A was detected by immunoblotting with specific antibodies. The IRF-7 protein was detected with a specific antibody. (C) Human embryonic kidney 293 cells were cotransfected with IRF-7-GFP and control vector or HCV NS5A. Cell lysates were prepared after 48 h of transfection and immunoprecipitated with the anti-GFP antibody. A specific antibody was used for detection of HCV NS5A or IRF-7.
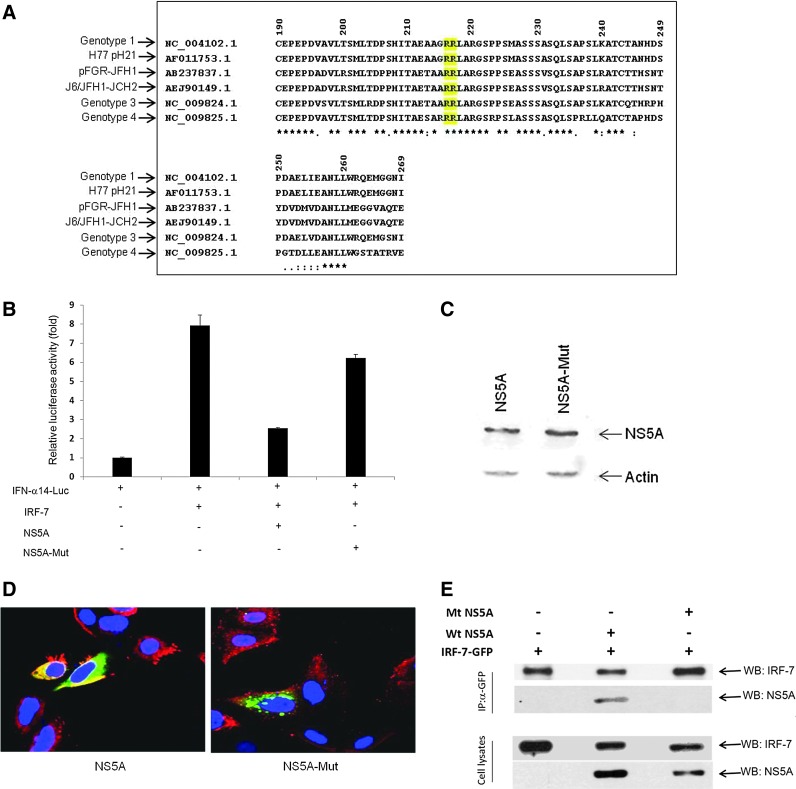
Arg216 and Arg217 act as critical residues of NS5A for suppression of IRF-7 activity
To determine functional consequence of the NS5A, we next analyzed amino acid sequences of the region among different HCV genotypes. For this, the ClustalW program (Eddy 1995) was used to align the amino acid sequences. Alignment of NS5A amino acids suggested 2 Arg residues at 216 and 217 of NS5A are conserved among different HCV genotypes (Fig. 3A). Arg residues are quite frequently found in protein-active or -binding sites. Arg is a positively charged, polar amino acid and frequently plays an important role in the protein structure. Arg residues are also involved in salt bridges, where they pair with negatively charged amino acids to create stabilizing hydrogen bonds for the properties of the protein. On the other hand, alanine is not particularly hydrophobic and is nonpolar. The alanine side chain is very nonreactive and rarely involved in protein functions. Thus, Arg residues are important for both the structural and functional activity of proteins, as mutation of Arg can change the tertiary structure of the protein (Kumar and others 1999). We introduced mutations in both Arg 216 and 217 residues in NS5A by substituting with alanine, and determined NS5A mediated regulation of IRF-7 functional activity. An in vitro reporter luciferase assay demonstrated that Arg to Ala substitutions at 216 and 217 residues of NS5A does not inhibit IRF-7-mediated IFN-α14 promoter activation (Fig. 3B). Thus, our results suggested an important role of Arg216/Arg217 in the NS5A functional activity for blocking IFN signaling by hindering the IRF-7 activity. The expression of mutant HCV NS5A was similar to wild-type NS5A as shown in the Western blot analysis (Fig. 3C).
FIG. 3.
NS5A Arg216 and Arg217 are critical for inhibition of IRF-7-mediated IFN-α14 promoter activation. (A) Alignment of partial NS5A sequences from amino acid residues 190–269 among different genotypes is shown. The sequence analyses displayed conserved arginine residues marked in yellow. (B) IHH were cotransfected with human IFN-α14-luc reporter plasmid, IRF-7, and wild-type or mutant HCV NS5A constructs. The luciferase activity was measured 48 h post-transfection. Results presented are mean with standard error from 4 independent experiments. Empty vector DNA was used as a negative control and mean basal value was arbitrarily set at 1. (C) HCV NS5A (wild-type or mutant) protein expression is shown by immunoblotting with the FLAG antibody for detection of the NS5A protein. (D) IHH were cotransfected with IRF-7-GFP and wild-type or mutant HCV NS5A. Cells were stained after 48 h of transfection for expression of NS5A (red) and IRF-7 (green). Cell nuclei were stained with DAPI (blue). Cytoplasmic localization of IRF-7 and NS5A was observed from images superimposed digitally for fine comparisons in confocal microscopy. (E) IRF-7-GFP plasmid DNA was cotransfected with wild-type NS5A (Wt NS5A) or mutant NS5A (Mt NS5A) into 293 cells. Cell lysates were prepared after 48 h of transfection and immunoprecipitated with the anti-GFP antibody. Association of IRF-7 and wild-type or mutant NS5A was detected by immunoblotting with specific antibodies.
To further verify that these 2 Arg residues are important for association with IRF-7, we performed colocalization experiments using IRF-7 and wild-type or mutant HCV NS5A. Confocal microscopy suggested that mutant HCV NS5A fails to associate with IRF-7, in contrast to wild-type HCV NS5A (Fig. 3D). Co-immunoprecipitation of the wild-type or mutant NS5A with IRF-7 further suggested that the wild-type NS5A protein physically associates with the IRF-7 GFP construct, whereas mutant NS5A fails to associate with IRF-7 (Fig. 3E).
Discussion
We previously reported that HCV infection blocks IFN-α-mediated nuclear translocation of IRF-7 (Raychoudhuri and others 2010). IRF-7 remains localized in the cytoplasm of HCV-infected IHH in the presence or absence of IFN-α. In this study, we have observed that the HCV NS5A protein physically associates with IRF-7 and impairs its function by inhibiting IFN-α14 promoter activation. IRF-7 is a master regular for IFN-α production. IRF-7 undergoes phosphorylation when activated and translocates into the nucleus. IRF-7 amplifies type I IFN response by inducing IFN-α expression, which also acts in both autocrine and paracrine manners through the IFN-α/β receptor. HCV infection enhances the ISRE promoter activity and production of ISGs, but it fails to induce subsequent downstream signaling molecules (Raychoudhuri and others 2010). In fact, several ISGs are amplified during HCV infection, but they are not sufficient to clear virus infection from host (Bigger and others 2004; Sarasin-Filipowicz and others 2008). IRF-7 knockout mice are unable to express type 1 IFN following infection (Honda and others 2005). Overexpression of DNIRF-7 (lacking DNA binding region) in Hus-EZ cells showed higher HCV infectivity (Aly and others 2007) implying that IRF-7-mediated signaling can impair HCV growth.
IRF-7-dependent induction of systemic IFN response for innate antiviral immunity is crucial for the host immune system to combat against viruses. Virus adapts several strategies to counteract the function of IRF-7. Enterovirus 71 is a positive-stranded RNA virus and capable of inhibiting innate immunity. Enterovirus 71 downregulates IRF-7 through the 3C protein (Lei and others 2011). Moreover, 3C protein mediates cleavage at the Q189 and S190 junction within the constitutive activation domain of IRF-7, resulting in 2 cleaved IRF-7 fragments incapable of activating IFN expression.
Structure/function analysis suggested that Arg216 and Arg217 of NS5A are 2 crucial basic amino acids, and changing Arg to Ala on those 2 regions of NS5A renders it inactive to block IRF-7 function. Arg residues are important for both the structural and functional activity of proteins. Mutation of Arg can change the tertiary structure of a protein (Kumar and others 1999), as well as it can alter the catalytic activity without changing protein conformation (Shah and others 1996). In the TGBp1 of bamboo, mosaic potexvirus (BaMV) 2 arginine residues (Arg16 and Arg21) were shown to be essential for RNA binding and required for the ATP-utilizing activity of the soluble TGBp1 (Liou and others 2000). Amino acid residue Arg356 in HCV NS5A plays a critical role for stabilization of the NS5A domain and SH3 interaction by the formation of a salt bridge with an acidic residue. The binding of NS5A to SH3 domain is abolished when Arg356 was substituted with alanine (Macdonald and others 2005). Amino acid residues Arg216 and/or Arg217 are essential to the binding of IRF-7, and each of the 2 Arg-to-Ala substitutions may alter the overall conformation of NS5A and render it inactive from binding with IRF-7. Our results suggest the 2 arginine residues, Arg216 and Arg217, are important for the IRF-7-mediated activity, indicating that a same structure motif is required for the function of HCV NS5A. Further characterization of a replication phenotype of a virus or replicon with the mutations should clarify contributions of specific amino acids in the HCVNS5A/IRF-7 association and the underlying mechanism.
In summary, our results indicate that HCV regulates IFN signaling by impairing IRF-7 nuclear localization through its NS5A protein. PEG-IFN and ribavirin treatment of HCV-infected patients augment the IRF-7 level (Thomas and others 2011). Since IRF-7 is critical in mounting antiviral responses against HCV infection, our study suggests a new therapeutic target for drug development against HCV infection.
Acknowledgments
We thank Dr. Jung San Huang, Department of Biochemistry and Molecular Biology, Saint Louis University for helpful suggestions. This work was supported by research grant U54-AI057160 to the Midwest Regional Center of Excellence (MRCE) for Biodefense and Emerging Infectious Diseases Research and grant DK081817 from the National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- Aly HH, Watashi K, Hijikata M, Kaneko H, Takada Y, Egawa H, Uemoto S, Shimotohno K. 2007. Serum-derived hepatitis C virus infectivity in interferon regulatory factor-7-suppressed human primary hepatocytes. J Hepatol 46:26–36 [DOI] [PubMed] [Google Scholar]
- Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med 144:705–714 [DOI] [PubMed] [Google Scholar]
- Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, Lemon SM, Lanford RE. 2004. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol 78:13779–13792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 1995. Multiple alignment using hidden Markov models. Proc Int Conf Intell Syst Mol Biol 3:114–120 [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- Kanda T, Basu A, Steele R, Wakita T, Ryerse JS, Ray R, Ray RB. 2006. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J Virol 80, 4633–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Steele R, Ray R, Ray RB. 2007. Hepatitis C virus infection induces the beta interferon signaling pathway in immortalized human hepatocytes. J Virol 81:12375–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1–24 [DOI] [PubMed] [Google Scholar]
- Kumar LV, Ramakrishna T, Rao CM. 1999. Structural and functional consequences of the mutation of a conserved arginine residue in alphaA and alphaB crystallins. J Biol Chem 274:24137–24141 [DOI] [PubMed] [Google Scholar]
- Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. 2011. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by Toll-like receptor 3. J Virol 85:8811–8818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Genin P, Mamane Y, Hiscott J. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol Cell Biol 20:6342–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou DY, Hsu YH, Wung CH, Wang WH, Lin NS, Chang BY. 2000. Functional analyses and identification of two arginine residues essential to the ATP-utilizing activity of the triple gene block protein 1 of bamboo mosaic potexvirus. Virology 277:336–344 [DOI] [PubMed] [Google Scholar]
- Lu R, Au WC, Yeow WS, Hageman N, Pitha PM. 2000. Regulation of the promoter activity of interferon regulatory factor-7 gene. Activation by interferon and silencing by hypermethylation. J Biol Chem 275:31805–31812 [DOI] [PubMed] [Google Scholar]
- Macdonald A, Mazaleyrat S, McCormick C, Street A, Burgoyne NJ, Jackson RM, Cazeaux V, Shelton H, Saksela K, Harris M. 2005. Further studies on hepatitis C virus NS5A-SH3 domain interactions: identification of residues critical for binding and implications for viral RNA replication and modulation of cell signalling. J Gen Virol 86:1035–1044 [DOI] [PubMed] [Google Scholar]
- MacQuillan GC, Mamotte C, Reed WD, Jeffrey GP, Allan JE. 2003. Upregulation of endogenous intrahepatic interferon stimulated genes during chronic hepatitis C virus infection. J Med Virol 70:219–227 [DOI] [PubMed] [Google Scholar]
- Ramos HJ, Gale M., Jr 2011. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol 1:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RB, Meyer K, Ray R. 2000. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 271:193–204 [DOI] [PubMed] [Google Scholar]
- Raychoudhuri A, Shrivastava S, Steele R, Dash S, Kanda T, Ray R, Ray RB. 2010. Hepatitis C virus infection impairs IRF-7 translocation and Alpha interferon synthesis in immortalized human hepatocytes. J Virol 84:10991–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. 2008. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A 105:7034–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M, Bowie AG. 2007. An arms race: innate antiviral responses and counteracting viral strategies. Biochem Soc Trans 35:1512–1514 [DOI] [PubMed] [Google Scholar]
- Sen GC. 2001. Viruses and interferons. Annu Rev Microbiol 55:255–281 [DOI] [PubMed] [Google Scholar]
- Shah MA, Tayyab S, Ali R. 1996. Probing structure-activity relationship in diamine oxidase—reactivities of lysine and arginine residues. Int J Biol Macromol 18:77–81 [DOI] [PubMed] [Google Scholar]
- Thomas E, Feldm JJ, Li Q, Hu Z, Fried MW, Liang TJ. 2011. Ribavirin potentiates interferon action by augmenting interferon-stimulated gene induction in hepatitis C virus cell culture models. Hepatology 53:32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]