Abstract
Prior studies demonstrated that resistance to the ERBB1/2 inhibitor Lapatinib in HCT116 cells was mediated by increased MCL-1 expression. We examined whether inhibition of BCL-2 family function could restore Lapatinib toxicity in Lapatinib adapted tumor cells and enhance Lapatinib toxicity in naive cells. The BCL-2 family antagonist Obatoclax (GX15-070), that inhibits BCL-2/BCL-Xl/MCL-1 function, enhanced Lapatinib toxicity in parental HCT116 and Lapatinib adapted HCT116 cells. In breast cancer lines, regardless of elevated ERBB1/2 expression, GX15-070 enhanced Lapatinib toxicity within 3–12 h.The promotion of Lapatinib toxicity neither correlated with cleavage of caspase 3 nor was blocked by inhibition caspases; and was not associated with changes in the activities of ERK1/2, JNK1/2 or p38 MAPK but with reduced AKT, mTOR and S6K1 phosphorylation. The promotion of Lapatinib toxicity by GX15-070 correlated with increased cytosolic levels of apoptosis inducing factor (AIF) and expression of ATG8 (LC3), and the formation of large vesicles that intensely stained for a transfected LC3-GFP construct. Knockdown of the autophagy regulatory proteins ATG5 or Beclin1 suppressed the induction of LC3-GFP vesicularization and significantly reduced cell killing, whereas knock down of MCL-1 and BCL-Xl enhanced the induction of LC3-GFP vesicularization and significantly enhanced cell killing. Knockdown of Beclin1 and AIF abolished cell killing. Collectively, our data demonstrate that Obatoclax mediated inhibition of MCL-1 rapidly enhances Lapatinib toxicity in tumor cells via a toxic form of autophagy and via AIF release from the mitochondrion.
Keywords: lapatinib, obatoclax, autophagy, cell death, resistance
Introduction
Inhibitors of receptor tyrosine kinases, particularly of ERBB1 and ERBB2, have been under pre-clinical and clinical development for over 10 y e.g., Gefitinib, Erlotinib and Lapatinib.1 In vitro, numerous tumor cell types have been shown to exhibit growth reduction following inhibition of growth factor receptors, e.g., ERBB1, or inhibition of signaling pathways, e.g., MEK1/2.2 However, in many such studies the primary effect of a single kinase inhibitory agent at low “target specific” doses on tumor cells was cyto-static, rather than cyto-toxic.3 And, in contrast to the relatively encouraging findings from pre-clinical in vitro work, clinical studies using many ERBB1/ERBB2 inhibitors as single agents frequently did not demonstrate any form of tumor growth control.4
Where single agent drug-induced toxic effects are particularly pronounced in patients, such as using Imatinib in treatment of BCR-ABL+ CML, it was hypothesized and proven that the tumor control effect was due to CML cells being exquisitely addicted to the kinase activity of the BCR-ABL fusion protein for growth and survival.5 On the contrary, in NSCLC, however, despite the tumors of ~70% of patients overexpressing ERBB1, only a small sub-population of individuals (~10%) responded to ERBB1 inhibitors, and these individuals statistically tended to be non-smokers and with an Asian/female genetic background.6 Subsequently it was shown in responsive NSCLC patients that ERBB1 was mutated to become a constitutively active kinase, with such NSCLC cells being addicted to the survival signals emanating from the mutated receptor.7 Thus only a minority of tumor cell types appear to present with a relatively simplistic single oncogene activating mutation/survival signaling addiction that would predict for use of a single kinase inhibitory drug. These findings make clear that the rational development of approaches which simultaneously target multiple signal transduction/cell survival pathways to kill tumor cells will more likely have broad therapeutic usefulness.
Exposure of tumor cells expressing a mutated active form of ERBB1, but generally not an overexpressed wild type ERBB1, to kinase domain inhibitors results in growth arrest, and tumor cell death.8,9 Over the course of many months exposure to kinase inhibitor(s), secondary mutations in the receptor kinase domain develop which render the receptor resistant to the kinase inhibitor. A more rapid mechanism of resistance to ERBB receptor inhibitors as single agents, prior to the development of secondary mutations, is the compensatory activation of growth factor receptors such as c-MET (+c-Src), and the IGF1R which can act in paral to provide survival signaling.10-12 These receptors can provide a survival signal in their own right as receptor tyrosine kinases as well as causing trans-phosphorylation of inhibited ERBB receptors, thereby permitting the ERBB receptors to act as docking sites for e.g., RAS GTP exchange factors. And, combinations of ERBB receptor inhibitors with inhibitors of c-Met or of the IGF1R have proven efficacious in promoting cell death in naïve cells and reverting significantly the ERBB inhibitor resistant phenotype.13,14 Others have noted lower levels of the pro-apoptotic protein BIM in ERBB1 inhibitor resistant cells, and inhibition of BCL-2/BCL-Xl function can enhance ERBB1 inhibitor-induced caspase-dependent toxicity in NSCLCs.15,16 In breast cancer cells, resistance to the ERBB1/ERBB2 inhibitor Lapatinib was reported to be due to re-activation of the estrogen receptor.17 In contrast, we have found that resistance to Lapatinib in colon cancer cells is unrelated to compensatory growth factor receptor signaling, mutation of the ERBB1 kinase domain or re-activation of the estrogen receptor, but is instead mediated by increased expression of mitochondrial and endoplasmic reticulum protective MCL-1 and BCL-Xl proteins with reduced expression of pro-apoptotic BAX.18
The BCL-2 family of proteins regulates the intrinsic/mitochondrial apoptosis signaling pathway. Protective BCL-2 family proteins (BCL-2, BCL-Xl, MCL-1, A1) associate via BH3 domains with pro-apoptotic family members including BAX and BAK. BAX and BAK, when released from protective BCL-2 proteins, can perturb the mitochondrial membrane forming pores that permit release of cytochrome c, leading to the activation of caspase 9, and ultimately apoptosis. Tumor cells utilize a number of mechanisms to maintain viability, including loss of death receptor expression, e.g., CD95, by losing expression of pro-apoptotic BH3 domain proteins, e.g., BAX, or by increasing expression of anti-apoptotic BCL-2 family members, e.g., MCL-1.19,20 In the case of protective BCL-2 family proteins, several clinically relevant small molecule inhibitors have been developed that specifically bind to the BCL-2 family protein, without altering expression of the protein, and that block the binding of pro-apoptotic BH3 domain proteins, e.g., ABT-737; Gossypol; GX15-070.21,22 The drug-induced dissociation of BCL-2 protein from toxic BH3 domain protein results in greater levels of free BH3 domain protein that will facilitate mitochondrial dysfunction and promote the toxicity of other therapeutic agents.23,24 The present studies determined: whether inhibition of BCL-2 family function could restore Lapatinib toxicity in Lapatinib adapted tumor cells and enhance Lapatinib toxicity in naive cells. Based on our initial findings, we then determined the molecular mechanisms by which GX15-070 (Obatoclax) enhanced Lapatinib toxicity in tumor cells.
Results
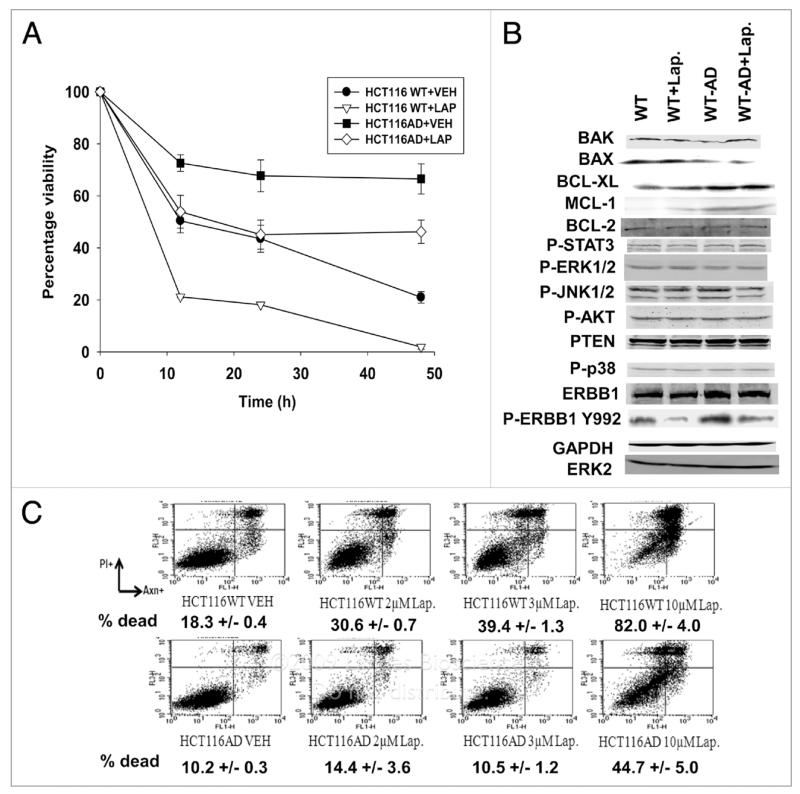
We incubated HCT116 cells in the presence of Lapatinib for >3 mon until a homogeneous population of cells grew out that we determined were resistant to Lapatinib (termed, HCT116-AD) (Fig. 1A). Lapatinib adapted cells did not exhibit significant changes in the activities/phosphorylation of known cyto-protective signaling molecules including ERK1/2, AKT and STAT3. The Lapatinib adapted cells had higher expression of BCL-Xl and MCL-1 and reduced expression of BAX compared to wild-type parental cells; BCL-2 expression did not change (Fig. 1B). Lapatinib treatment suppressed ERBB1 Y992 phosphorylation in both parental and adapted cells, arguing that drug resistance was not due to a mutation in the ERBB1 tyrosine kinase ATP binding domain. Lapatinib resistant cells were also resistant to the lethal effects of Lapatinib when cells were grown in the presence of serum (Figure 1C).
Figure 1.
Lapatinib resistance in HC T116 cells. (A) Parental HC T116 cells (WT) and HC T116 Lapatinib adapted cells (WT-AD) cells were plated, and 24 h after plating grown in serum depleted medium in the presence or absence of vehicle (DMSO) or Lapatinib (2 μM). Cells were isolated at the indicated times and cell viability determined by annexin-PI flow cytometric analysis in triplicate ±SEM (n = 3). (B) Parental HCT116 cells (WT) and HCT116 Lapatinib adapted cells (WT-AD) cells were plated, and 24 h after plating treated with Lapatinib (2 μM). Cells were isolated 30 min after drug treatment and blotting performed to determine the expression levels of proteins and the phosphorylation status of proteins (n = 3). (C) Parental HCT116 cells (WT) and HC T116 Lapatinib adapted cells (WT-AD) cells were plated, and 24h after plating treated with vehicle (DMSO) or Lapatinib (0-10 μM). Cells were isolated 48 h Lapatinib addition and cell viability determined in triplicate by annexin-PI flow cytometry assay ± SEM. The data shown are a representative study from two separate studies.
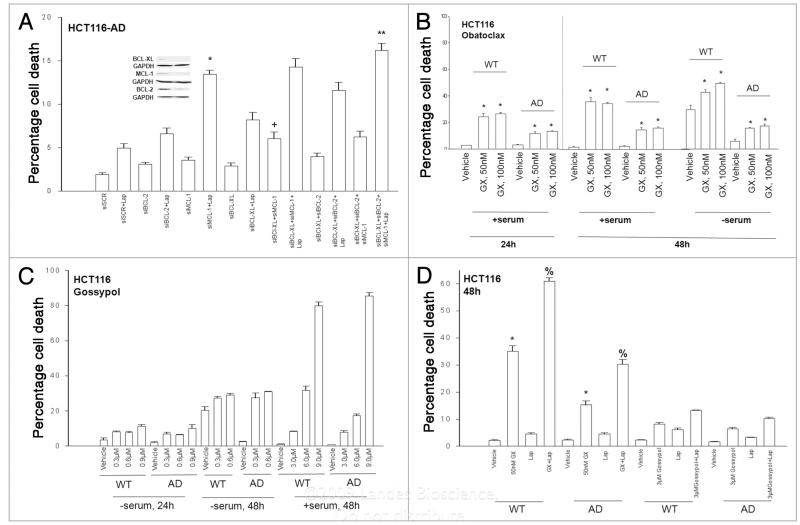
We next determined the relative importance of BCL-2, and elevated BCL-Xl and MCL-1 expression in maintaining the viability of HCT116-AD cells treated with Lapatinib. Knockdown of MCL-1 expression promoted a greater restoration of Lapatinib toxicity in HCT116-AD cells than knock down of either BCL-2 or BCL-Xl (Fig. 2A). Loss of BCL-Xl and MCL-1 expression enhanced basal levels of death, which was not significantly further enhanced by loss of BCL-2. Combined knock down of BCL-2 and BCL-Xl caused approximately the same level of cell killing as did knock down of MCL-1 alone, and combined knock down of BCL-2, BCL-Xl and MCL-1 facilitated more Lapatinibinduced cell killing than knock down of MCL-1 alone.
Figure 2.
Obatoclax enhances Lapatinib toxicity and reverts Lapatinib resistance in HCT116 cells. (a) HCT116 Lapatinib adapted cells (WT-AD) cells were plated, and 12 h later as indicated transfected with siRNA molecules to reduce the expression of; nothing/control (siSCR); BCL-2 (siBCL-2); BCL-Xl(siBCL-Xl); or MCL-1 (siMCL). Forty eight hours after transfection, cells were grown in serum depleted medium in the presence or absence of vehicle (DMSO) or Lapatinib (2 μM), as indicated. Cells were isolated 36 h after serum starvation and cell viability determined in triplicate by trypan blue exclusion assay ±SEM. The data shown are the mean of four separate studies. (+p < 0.05 greater than corresponding HCT116 siscR cell value; *p < 0.05 greater than siMCL-1 HCT116 cell value; **p < 0.05 greater than siMCL-1 + Lap HCT116 cell value). (B) HCT116 wild-type (WT) and HCT116 Lapatinib adapted cells (WT-AD) cells 24 h after plating were grown as indicated in serum depleted medium in the presence or absence of vehicle (DMSO) or Obatoclax (GX, 50 nM; 100 nM). Cells were isolated 48 h after serum starvation and cell viability determined in triplicate by trypan blue exclusion assay ±SEM. The data shown are the mean from three separate studies. (*p < 0.05 greater than corresponding VEH value). (C) HCT116 wild-type (WT) and HCT116 Lapatinib adapted cells (WT-AD) cells 24 h after plating were grown as indicated in serum depleted medium in the presence or absence of vehicle (DMSO) or Gossypol (0.3–9.0 μM). Cells were isolated 48 h after serum starvation and cell viability determined in triplicate by trypan blue exclusion assay ±SEM. The data shown are the mean from three separate studies. (D) HCT116 wild-type (WT) and HCT116 Lapatinib adapted cells (WT-AD) cells 24 h after plating were grown in the presence or absence of vehicle (DMSO), Lapatinib (Lap, 2 μM), Obatoclax (GX, 50 nM) or Gossypol (3.0 μM), as indicated. Cells were isolated 48 h after drug treatment and cell viability determined in triplicate by trypan blue exclusion assay ±SEM. The data shown are the mean from three separate studies. (*p < 0.05 greater than VEH treated value; %p < 0.05 greater than GX treated cell value).
Based on these findings, we explored whether pharmacologic inhibition of protective BCL-2 family protein function could revert the Lapatinib resistant phenotype in the HCT116-AD cells. For these studies we employed the clinically relevant BCL-2 family antagonists Gossypol (e.g., AT-101) and Obatoclax (GX15-070, GX).31,32 Based on published Phase I trial data, the sustainable free plasma concentrations of Gossypol and Obatoclax are ~600 nM and ~200 nM, respectively; the Cmax for Gossypol is ~3 μM and Gossypol has a very short half life in serum.33,34 Obatoclax (50–100 nM) significantly enhanced the toxicity of serum starvation in wild-type and adapted HCT116 cells (Fig. 2B). The inclusion of serum in the growth media did not reduce Obatoclax toxicity in these cells. At clinically relevant drug concentrations (<3 μM) Gossypol promoted the toxicity of serum starvation in wild type and adapted HCT116 cells (Fig. 2C). The inclusion of serum in the growth media profoundly reduced Gossypol toxicity at clinically relevant drug concentrations, although supraphysiologic levels of Gossypol were competent to cause cell death (Fig. 2C, data not shown). We then examined whether Lapatinib exposure could mimic the effect of serum withdrawal, and could enhance either Obatoclax or Gossypol toxicity. In the presence of serum Obatoclax enhanced Lapatinib toxicity in both wildtype and adapted HCT116 cells whereas Gossypol had no effect on reducing cell survival or enhancing Lapatinib toxicity (Fig. 2D). The toxic interaction between Lapatinib and Obatoclax in HCT116 wild-type cells and in HCT116-AD cells was noted to be synergistic in median dose effect isobologram colony formation assays (Table 1).
Table 1.
Lapatinib and Obatoclax synergize to kill hcT116 WT and hcT116 AD cells
| HCT116 WT | |||
|---|---|---|---|
| GX15-070 (nM) | Lapatinib (μM) | Fa | Cl |
| 33.0 | 0.5 | 0.60 | 0.66 |
| 66.0 | 1.0 | 0.94 | 0.37 |
| 99.0 | 1.5 | 0.99 | 0.18 |
| HCT116 AD | |||
| GX15-070 (nM) | Lapatinib ( μ M) | Fa | Cl |
| 33.0 | 0.5 | 0.15 | 0.67 |
| 66.0 | 1.0 | 0.34 | 0.59 |
| 99.0 | 1.5 | 0.46 | 0.58 |
HCT116 WT and HCT116 AD cells were plated as single cells (250–1500 cells/well) in sextuplicate and 12 h after this plating the cells were treated with vehicle (VEH, DMSO), Lapatinib (0.5–1.5 μM) or Obatoclax (GX15-070, 33–99 nM), or with both drugs combined, as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. after drug exposure (24 h), the media was changed and cells cultured in drug free media for an additional 10–14 d. cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. colony formation data were entered into the calcusyn program and combination index (CI) and Fraction affected (Fa) values determined. A CI value of less than 1.00 indicates synergy.
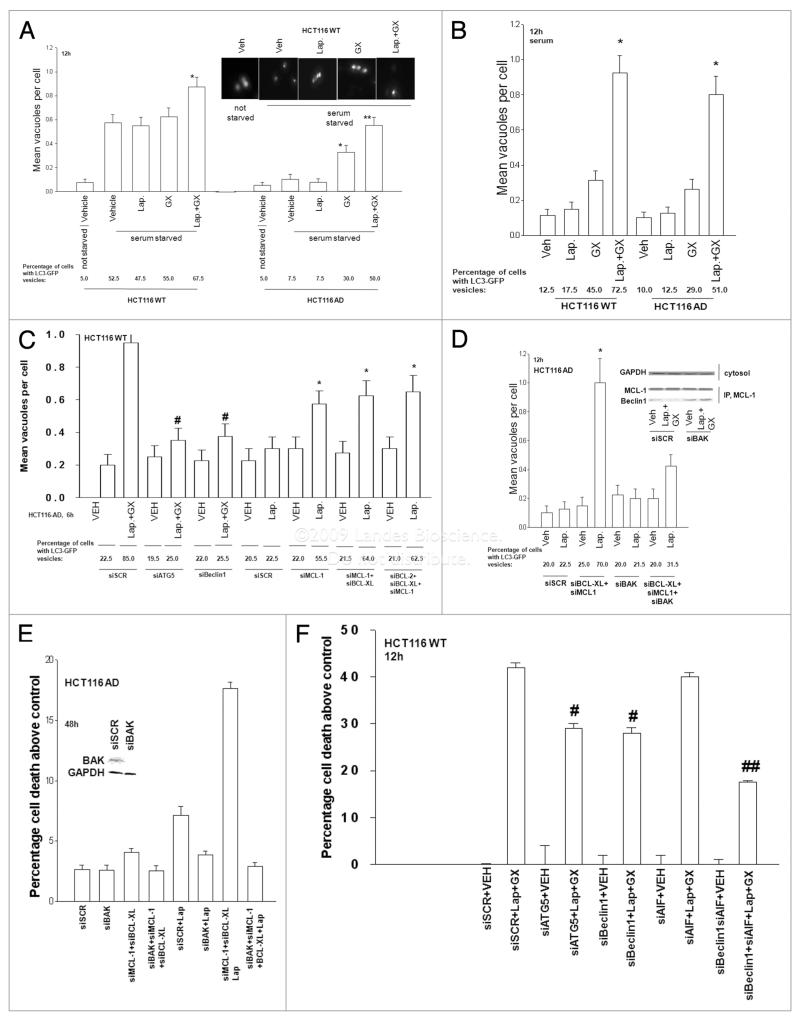
We next examined the mechanism by which Obatoclax enhanced Lapatinib lethality in HCT116 cells. In preliminary studies we found that the toxicity of Obatoclax +/− Lapatinib was suppressed by overexpression of BCL-Xl and was surprisingly modestly enhanced by treatment with the pan-caspase inhibitor zVAD (data not shown). As drug lethality was being enhanced by caspase inhibition, and because of studies by other laboratories that have linked inhibition of caspase function to increased toxic forms of autophagy, we determined whether Obatoclax +/− Lapatinib treatment was causing autophagy in HCT116 cells.35,36 We transfected HCT116 cells with a plasmid to express a GFP tagged version of LC3 (ATG8), an autophagy regulatory protein, and 24 h later serum starved and treated cells with Lapatinib and Obatoclax. Serum starvation increased the number of intense LC3-GFP staining vesicles in parental but not in adapted HCT116 cells (Fig. 3A). Thus HCT116-AD cells are inherently autophagy resistant. In adapted cells Obatoclax enhanced LC3-GFP vesicularization that was further enhanced by Lapatinib; in parental cells only the combination of Obatoclax + Lapatinib treatment further enhanced autophagy. The Lapatinib + Obatoclax drug combination increased LC3-GFP vesicularization in both parental and adapted HCT116 cells the presence of serum (Fig. 3B). Thus Obatoclax and Lapatinib interact to promote autophagy in HCT116 cells, regardless of whether the tumor cell has any inherent Lapatinib resistance.
Figure 3.
Lapatinib enhances Obatoclax-induced autophagy: inhibition of autophagy protects cells from Lapatinib + Obatoclax toxicity. (A) HCT116 WT and AD cells 24 h after plating in glass chambered slides were transfected with a plasmid to express LC3-GFP. Twenty-four hours after transfection cells were grown in serum depleted medium, as indicated and 30 min later treated with Vehicle (VEH, DMSO), Lapatinib (Lap., 2 μM); Obatoclax (GX, 50 nM); or Lap. + GX. Six hours after drug treatment cells were examined under a fluorescent microscope and the number of intense LC3-GFP staining vesicles per cell determined in 40 cells (n = 2; ±SEM). (B) HCT116 WT and AD cells 24 h after plating in glass chambered slides were transfected with a plasmid to express LC3-GFP. Twenty-four hours after transfection cells were treated in the presence of serum with Vehicle (VEH, DMSO), Lapatinib (Lap., 2 μM); Obatoclax (GX, 50 nM); or Lap. + GX. Six hours after drug treatment cells were examined under a fluorescent microscope and the number of intense LC3-GFP staining vesicles per cell determined in 40 cells (n = 2; ±SEM). (C) HCT116 cells 24 h after plating in glass chambered slides were transfected with a plasmid to express LC3-GFP and co-transfected with scrambled siRNA (siSCR); and siRNA molecules to knock down ATG5 (siATG5) and Beclin1 (siBeclin1); in parallel cells were also transfected with siRNA molecules to knock down expression of BCL-2, BCL-Xl and MCL-1, as indicated. Twenty-four hours after transfection cells were treated with Vehicle (VEH, DMSO) or with Lapatinib (Lap., 2 μM) + Obatoclax (GX, 50 nM). Six hours after drug treatment cells were examined under a fluorescent microscope and the number of intense LC3-GFP staining vesicles per cell determined in 40 cells (n = 2; ±SEM). (#p < 0.05 less than corresponding siSCR value; *p < 0.05 greater than corresponding Vehicle treated cell value). (D) HCT116 AD cells 24 h after plating in glass chambered slides or in 60 mm dishes were transfected, as indicated in the panel, with scrambled siRNa (siscR); and siRNA molecules to knock down BCL-Xl (siBCL-Xl), MCL-1 (siMCL-1) and BAK (siBAK). Cells were co-transfected to express LC3-GFP. Twenty-four hours after transfection cells were treated with Vehicle (VEH, DMSO) or with Lapatinib (Lap, 2 μM). Twelve hours after drug treatment cells in chamber slides were examined under a fluorescent microscope and the number of intense LC3-GFP staining vesicles per cell determined in 40 cells (n = 2; ±SEM). (Inset Panel) Transfected cells were treated with Vehicle (VEH, DMSO) or with Lapatinib (Lap, 2 μM) and 6 h after exposure cells were lysed and the association of Beclin1 with MCL-1 determined after immunoprecipitation and SDS PAGE/blotting. (E) HCT116 AD cells 24 h after plating in glass chambered slides or in 60 mm dishes were transfected, as indicated in the panel, with scrambled siRNA (siSCR); and siRNA molecules to knock down BCL-Xl (siBCL-Xl), MCL-1 (siMCL-1) and BAK (siBAK). Twenty-four hours after transfection cells were treated with Vehicle (Veh, DMSO) or with Lapatinib (Lap, 2 μM). Forty eight hours after drug treatment cells in 60 mm dishes were isolated cell viability determined in triplicate by trypan blue exclusion assay (n = 2, ±SEM). (F) HCT116 cells 24 h after plating were transfected with scrambled siRNA (siSCR); and siRNA molecules to knock down ATG5 (siATG5), Beclin1 (siBeclin1) and AIF (siAIF), as indicated. Twenty-four hours after transfection cells were treated with Vehicle (VEH, DMSO) or with Lapatinib (Lap., 2 μM) + Obatoclax (GX, 50 nM). Cells were isolated 12 h after drug exposure and cell viability determined in triplicate by flow cytometry (n = 2, ±SEM). (#p < 0.05 less than corresponding siSCR value; ##p < 0.05 less than corresponding siATG5 or siBeclin1 treated cell value).
Autophagy is regulated by a group of evolutionarily conserved genes (ATG genes) that control the various steps of autophago-some formation, lysosomal fusion and vesicle recycling. The protein Beclin1 (ATG6) contains a BH3 domain and is part of a type III PI3K complex which is required for the formation of autophagic vesicles. Knockdown of ATG5 or Beclin1 expression in parental HCT116 cells blocked Lapatinib + Obatoclax-induced LC3-GFP vesicularization and loss of Beclin1 or ATG5 expression reduced Lapatinib + Obatoclax-induced cell killing (Fig. 3C and D). Previously we have noted that knockdown of apoptosis inducing factor (AIF) significantly reduced Lapatinib toxicity in HCT116 cells.18 Knockdown of AIF did not alter the toxicity of Lapatinib + Obatoclax treatment, however, knock down of both AIF and Beclin1 significantly reduced Lapatinib + Obatoclax-induced cell death (Fig. 3D).
Obatoclax is an inhibitor of BCL-2, BCL-Xl and MCL-1 by interfering with their ability to associate with toxic BH3 domain proteins, and MCL-1 and BCL-Xl are overexpressed in Lapatinib adapted cells.18 MCL-1 and BCL-Xl are also known to be inhibitors of autophagy, in part by binding Beclin1 through Beclin1’s BH3 domain.37,38 Knockdown of MCL-1, MCL-1 and BCL-Xl, or BCL-2 + BCL-Xl + MCL-1 enhanced Lapatinib-induced LC3-GFP vesicularization in parental HCT116 cells (Fig. 3C). In Lapatinib adapted HCT116 cells knock down of BCL-Xl and MCL-1 expression enhanced LC3-GFP vesicularization to a level comparable to that observed in parental cells (Fig. 3D; cf C); this was associated with increased cell death (Fig. 3E). Knockdown of BAK suppressed drug-induced autophagy and reduced drug-induced cell killing. Treatment of adapted HCT116 cells with Lapatinib and Obatoclax reduced the amount of Beclin1 associated with MCL-1 (Fig. 3D, inset panel); autophagy was inhibited by knock down of Beclin1 (Fig. 3C). Combined inhibition of AIF and autophagy was required to strongly suppress Lapatinib + Obatoclax cell killing in parental HCT116 cells (Fig. 3F). Collectively the data in Figures 2 and 3 and Table 1 demonstrate that Obatoclax enhances Lapatinib toxicity and reverted Lapatinib resistance in HCT116 cells through a mechanism that involves inhibition of BCL-Xl and MCL-1, the actions of BAK, and the release of Beclin1 from BCL-2 family proteins, which leads to increased levels of toxic autophagy.
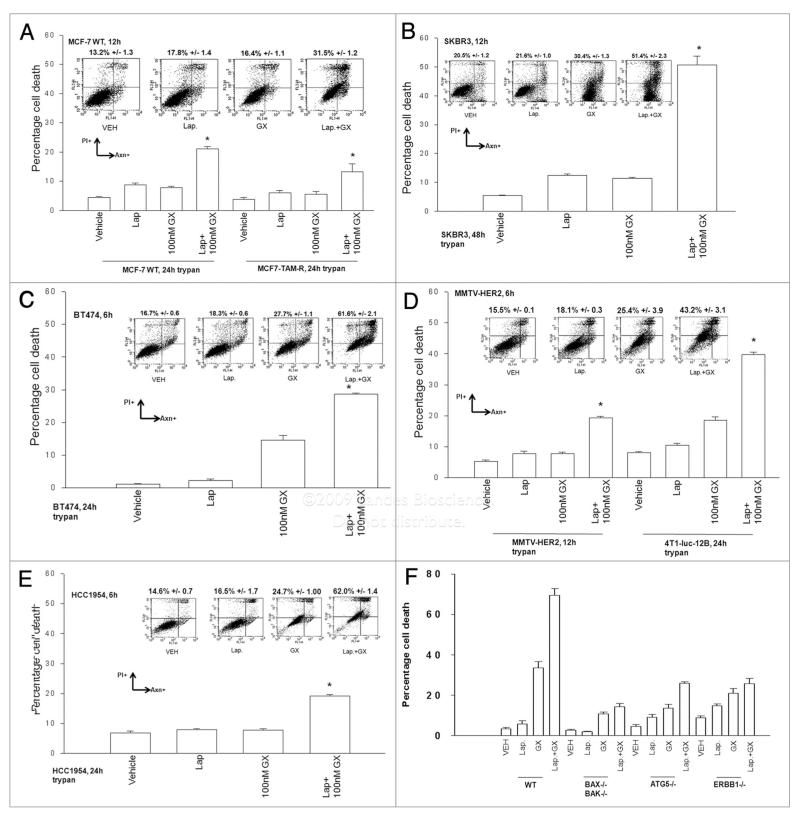
As Lapatinib is FDA approved for the treatment of breast cancer, we then went on to determine whether Obatoclax enhanced Lapatinib toxicity in breast cancer cells. In breast cancer cells with low, e.g., MCF7, or high, e.g., BT474, expression of ERBB1/ERBB2, Lapatinib toxicity was enhanced by Obatoclax (Fig. 4A–E). These effects were noted to be synergistic in median dose effect isobologram colony formation assays in both human and rodent mammary carcinoma cells (Tables 2 and 3). Using SV40 Large T antigen transformed mouse embryonic fibroblasts that had been deleted for specific genes we noted that loss of BAX and BAK function or ATG5 function abolished Lapatinib + Obatoclax toxicity (Fig. 4F). Knock out of ERBB1 also significantly reduced the toxic interaction between Lapatinib and Obatoclax in MEFs.
Figure 4.
Obatoclax enhances Lapatinib toxicity in multiple breast cancer cell lines. (A) human MCF7 parental and MCF7 tamoxifen resistant; (B) human SKBR3; (C) human BT474; (D) rodent MMTV-HE R2 and 4T1-luc-12B; (E) human HCC 1954; mammary carcinoma cells; 24 h after plating were treated with vehicle (DMSO), Lapatinib (Lap, 2 μM), Obatoclax (GX, 50 nM) or Lapatinib + Obatoclax. Floating and attached cells were isolated 24 h after drug exposure and cell viability determined using: lower graph, trypan blue exclusion assay; upper plots, Annexin-PI flow cytometry assay (±SEM, n = 3). Upper inset immunoblots: Cells were isolated 6 h after drug exposure and immunoblotting performed to determine the expression and phosphorylation of ERBB1, ERBB2 and GAPDH (a representative set of blots from n = 2–4 is shown). Twenty-four hours after plating were treated with vehicle (DMSO), Lapatinib (Lap, 2 μM), Obatoclax (GX, 50 nM) or Lapatinib + Obatoclax. In (F) transformed mouse embryonic fibroblasts, either wild-type (WT) or genetically deleted for the indicated proteins, were treated with vehicle (DMSO), Lapatinib (Lap, 1 μM), Obatoclax (GX, 50 nM) or Lapatinib + Obatoclax. Floating and attached cells were isolated 24 h after drug exposure and cell viability determined using: lower graph, trypan blue exclusion assay; upper plots, Annexin-PI flow cytometry assay (±SEM, n = 3).
Table 2.
Lapatinib and Obatoclax synergize to kill human mammary carcinoma cells
| MCF7 | |||
|---|---|---|---|
| GX15-070 (nM) | Lapatinib (μM) | Fa | Cl |
| 33.0 | 0.5 | 0.08 | 0.73 |
| 66.0 | 1.0 | 0.29 | 0.63 |
| 99.0 | 1.5 | 0.46 | 0.62 |
| SKBR3 | |||
| GX15-070 (nM) | Lapatinib (μM) | Fa | Cl |
| 33.0 | 0.5 | 0.38 | 0.61 |
| 66.0 | 1.0 | 0.48 | 0.58 |
| 99.0 | 1.5 | 0.59 | 0.45 |
| BT474 | |||
| GX15-070 (nM) | Lapatinib (μM) | Fa | Cl |
| 15.0 | 0.5 | 0.28 | 0.67 |
| 30.0 | 1.0 | 0.54 | 0.57 |
| 45.0 | 1.5 | 0.82 | 0.50 |
McF7, sKBR3 and BT474 cells were plated as single cells (250–1500 cells/ well) in sextuplicate and 12 h after this plating the cells were treated with vehicle (Veh, DMsO), Lapatinib (0.5–1.5 μM) or Obatoclax (GX15-070, 33–99 nM), or with both drugs combined, as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. after drug exposure (24 h), the media was changed and cells cultured in drug free media for an additional 10–14 d. cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. colony formation data were entered into the calcusyn program and combination index (CI) and Fraction affected (Fa) values determined. A CI value of less than 1.00 indicates synergy.
Table 3.
Lapatinib and Obatoclax synergize to kill rodent mammary carcinoma cells
| Lapatinib (μM) | GX15-070 (nM) | Fa | Cl |
|---|---|---|---|
| 0.5 | 33.0 | 0.44 | 0.45 |
| 1.0 | 66.0 | 0.54 | 0.47 |
| 1.5 | 99.0 | 0.66 | 0.35 |
The rodent 4T1-1uc-12B mammary carcinoma cell line derived from a spontaneous BaLB/c mammary tumor was cultured in vitro and were plated as single cells (250–1500 cells/well) in sextuplicate and 12 h after this plating the cells were treated with vehicle (VEH, DMSO), Lapatinib (0.5–1.5 μM) or Obatoclax (GX15-070, 33–99 nM), or with both drugs combined, as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. after drug exposure (24 h), the media was changed and cells cultured in drug free media for an additional 10–14 d. cells were fixed, stained with crystal violet and colonies of >50 cells/colony counted. colony formation data were entered into the calcusyn program and combination index (CI) and Fraction affected (Fa) values determined.A CI value of less than 1.00 indicates synergy.
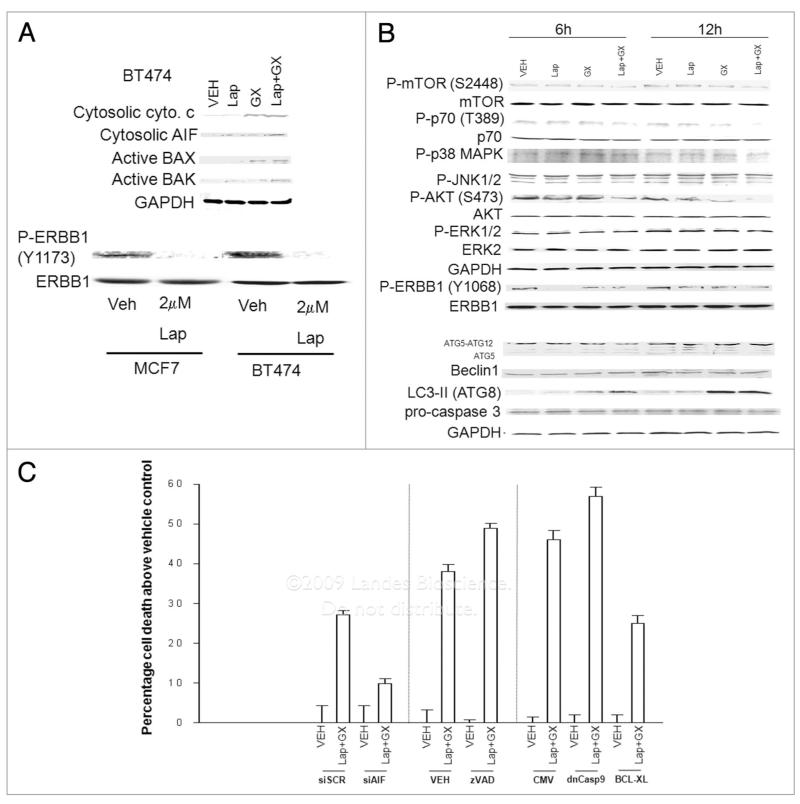
Treatment of BT474 breast cancer cells with Obatoclax promoted activation of BAK and release of cytochrome c into the cytosol (Fig. 5A). Treatment of these cells with both Lapatinib + Obatoclax, however, was required to promote cytosolic release of AIF. Treatment of BT474 cells with Lapatinib ± Obatoclax did not cause cleavage of pro-caspase 3 despite causing profound levels of cell killing over this time period, and this treatment rapidly reduced ERBB1 Y1173/Y1068 phosphorylation but did not alter the activities of ERK1/2 or JNK1/2; the activity of p38 MAPK was weakly enhanced by Lapatinib + Obatoclax (Figs. 4C and 5B). Lapatinib + Obatoclax treatment, but not treatment with either agent individually, reduced the phosphorylation of AKT (S473), S6K1 (T389) and of mTOR (S2448); the mobility of mTOR also increased on SDS PAGE, indicative of a general dephosphorylation of this protein. Also of note, Obatoclax increased the expression of ATG8 (LC3), which was enhanced by Lapatinib. Based on our findings in HCT116 cells as well as the above data, we explored the mechanisms by which Lapatinib and Obatoclax enhanced breast cancer cell death. Knockdown of AIF or overexpression of BCL-Xl suppressed Lapatinib + Obatoclax lethality whereas inhibition of caspase function enhanced drug toxicity (Fig. 5C).
Figure 5.
Obatoclax enhances Lapatinib toxicity in a manner that is dependent on AIF and inhibited by BCL-Xl. (A) BT474 cells 24 h after plating were treated with vehicle (DMSO) or GX15-070 (Obatoclax; GX, 50 nM) in the presence or absence of Lapatinib (2 μM). Cells were isolated 6 h after drug exposure and portions of the cells were lysed and the crude granular and cytosolic fractions isolated. In parallel, portions of the cells were lysed and the activated form of BAX immunoprecipitated. The levels of cytochrome c, AIF and activated BAX were determined after SDS PAGE Data are representative of 2–3 independent studies. (B) BT474 cells 24 h after plating were treated with vehicle (DMSO), GX15-070 (Obatoclax; GX, 50 nM) or Lapatinib (Lap, 2 μM), as indicated. Cells were isolated 6 and 12 h after drug exposure and immunoblotting performed after SDS PAGE on cell lysates to determine the expression/phosphorylation of the indicated proteins. (C) BT474 cells 24 h after plating were: (A) transfected with scrambled siRNA (siscR) or siRNA to knock down AIF (siAIF); (b) cells were infected with empty vector control adenovirus (CMV), or viruses to express BCL-Xl or dominant negative caspase 9; (c) not manipulated. Twenty-four hours after manipulation, cells are treated with vehicle (DMSO) or Obatoclax (GX, 50 nM) + Lapatinib (Lap, 2 μM). Cells were isolated 3 h after drug exposure and viability determined using Annexin-PI flow cytometry (±SEM, n = 3).
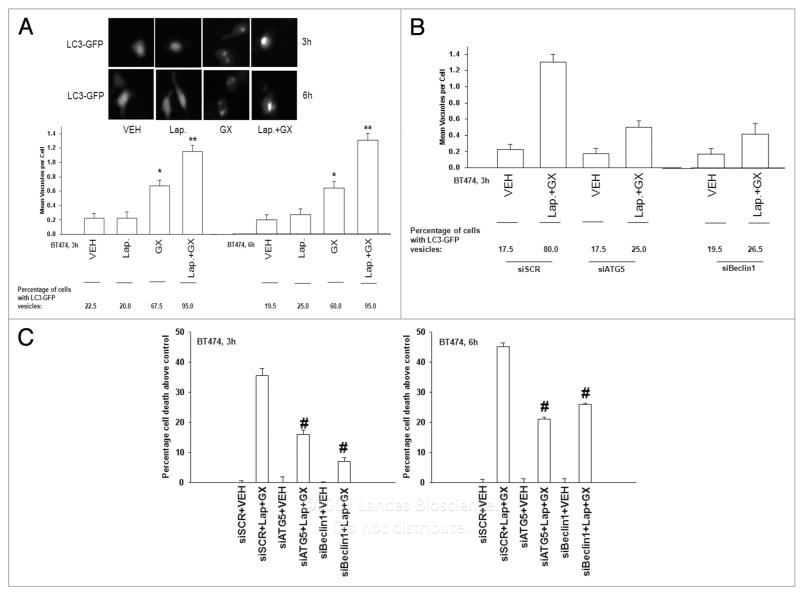
Based on our findings with increased LC3 expression in BT474 cells, and our prior data in HCT116 cells, we determined whether autophagy was playing any role in Lapatinib and Obatoclax toxicity. In BT474 cells Obatoclax, but not Lapatinib, promoted formation of vesicles that included a transfected LC3-GFP construct (Fig. 6A). Knockdown of either ATG5 or Beclin1 suppressed drug-induced LC3-GFP vesicularization and knockdown of either ATG5 or Beclin1 significantly reduced drug-induced cell death (Fig. 6B and C). Collectively, the data in Figures 4–6, as well as Tables 2 and 3 demonstrates that Lapatinib and Obatoclax interact in a synergistic fashion to kill breast cancer cells through a toxic form of autophagy.
Figure 6.
Lapatinib and Obatoclax cause a toxic form of autophagy in breast cancer cells. (A) BT474 cells 24 h after plating were transfected with a construct to express LC3-GFP, and 24 h after transfection cells were treated as indicated with vehicle (VEH , DMSO), Lapatinib (Lap, 2 μM); or Obatoclax (GX, 50 nM). Three and 6 h after exposure, cells were examined in a fluorescent microscope for the formation of vesicles with intense LC3-GFP staining (±SEM, n = 3). *p < 0.05 greater than corresponding value in VEH vector cells. Upper inset: Pictures of cells were treated with drugs 3 and 6 h after exposure. (B) BT474 cells 24 h after plating were transfected with scrambled siRNA (siSC R, 20 nM) or transfected with siRNAs to knockdown ATG5 (siATG5) or Beclin1 (siBeclin1). In parallel, cells were transfected with a construct to express LC3-GFP. Twenty four h after transfection, cells were treated with vehicle (VEH , DMSO) or Lapatinib (Lap, 2 μM) and Obatoclax (GX, 50 nM). Three hours after drug exposure, cells were examined in a fluorescent microscope for the formation of vesicles with intense LC3-GFP staining (±SEM, n = 3). (C) BT474 cells 24 h after plating were transfected with scrambled siRNA (siSC R, 20 nM) or transfected with siRNAs to knock down ATG5 (siATG5) or Beclin1 (siBeclin1). Twenty four hours after transfection, cells were treated with vehicle (VEH , DMSO) or Lapatinib (Lap, 2 μM) and Obatoclax (GX, 50 nM). Three hours and 6 h after drug exposure cells were isolated and viability determined by Annexin-PI staining and flow cytometry (±SEM, n = 2). #p < 0.05 less than corresponding value in siSCR cells.
Discussion
In the present manuscript we have attempted to determine in tumor cells whether inhibition of MCL-1 function represents a valid approach for enhancing the toxicity of the ERBB1/2 inhibitor Lapatinib. This hypothesis was based on previous studies that demonstrated Lapatinib resistant HCT116 colon cancer cells overexpressed MCL-1 and that knock down of MCL-1 partially restored Lapatinib sensitivity.18
In the present series of studies in HCT116 cells, we discovered that knock down of MCL-1 enhanced Lapatinib toxicity to a greater extent than knockdown of either BCL-Xl or BCL-2 expression. Inhibition of BCL-2 family protein function using the small molecule BH3 domain antagonist Obatoclax, a drug that is entering Phase II trials, enhanced Lapatinib toxicity in both parental and in Lapatinib adapted HCT116 cells. Several drugs designed to inhibit protective BCL-2 family function are presently undergoing clinical evaluation including ABT-263 and AT-101 (R+ gossypol).39,40 ABT-263 inhibits only BCL-2 and BCL-Xl, whereas AT-101 is claimed, like Obatoclax, to inhibit BCL-2, BCL-Xl and MCL-1. In many studies examining resistance to ERBB1 inhibitors, the mechanisms of resistance have been ascribed to: mutation within the kinase domain; activation of compensatory survival signaling growth factor receptors; and reactivation of the estrogen receptor (reviewed in ref. 18). The selection for tumor cells with enhanced expression of mitochondrial protective proteins after chemotherapy has been established for many years (reviewed in ref. 41), though surprisingly few studies have suggested that resistance to ERBB receptor inhibitors could be due to alterations in the expression of mitochondrial protective/toxic proteins.15,16,18 The lack of studies describing this mechanism of ERBB1 inhibitor resistance is also unexpected as it has been shown in lung cancer cells addicted for survival to mutant active ERBB1 signaling that inhibition of BCL-2/BCL-Xl using ABT-737 enhances Gefitinib toxicity and that in other tumor cell types ERBB1 inhibitor toxicity is mediated via mitochondrial dysfunction.42,43 Collectively, our findings in colon and breast cancer cells demonstrate that one useful approach to both sensitize and reverted resistance to ERBB1/ERBB2 inhibitors is to inhibit the function of protective BCL-2 family proteins.
Many of the findings made in this manuscript center on the induction of autophagy in Lapatinib + Obatoclax-exposed tumor cells. Autophagy is an evolutionarily conserved process from yeast to mammals and simplistically is a catabolic process in which a cell lacking nutrients begins to degrade its own components, e.g., mitochondria, through lysosomal fusion and the subsequent recycling of nutrients to sustain cell viability. For obvious reasons the process is tightly regulated as it plays a key homeostatic survival mechanism. In cancer, however, the role of autophagy in cell survival processes appears to be more complicated. For example, autophagy is thought to be a tumor suppression mechanism because genetic haplo-deficiency in autophagy regulatory proteins, e.g., Beclin1, in breast cancer cells leads to increased cancer incidence; and activated oncogenes in general have been shown to inhibit autophagy and tumor suppressors to promote autophagy.44 We noted that H-RAS V12, in protecting HCT116 cells from serum withdrawal + Lapatinib toxicity also suppressed increased levels of autophagy (Martin A.P. and Dent P., unpublished observations).
Autophagy is also important in the response of tumor cells to cancer therapy, with the toxicity of agents as diverse as Temozolomide and Arsenic Trioxide having been linked to the process.44 Prior studies from this laboratory using the drug OSU-03012 or the toxic cytokine MDA-7/IL24 have shown that increased levels of autophagy play a key role in promoting agent toxicity.26,27 In contrast, treatment of tumor cells with sorafenib + vorinostat or with bile acids have shown that death receptor-induced autophagy in these systems was a protective event against apoptosis.28,29 With regard to the present studies, elevated autophagy levels have been shown to make breast cancer cells resistant to anti-estrogen therapy.17
Obatoclax promoted Lapatinib toxicity by rapidly enhancing the formation of large autophagic vesicles and inhibition of autophagy, combined with knock down of AIF activity, abolished HCT116 cell killing by the drug combination. In breast cancer cells, regardless of high levels of ERBB1/2 expression, Obatoclax and Lapatinib again interacted to rapidly kill cells in a manner consistent with induction of a toxic form of autophagy. In our prior studies examining autophagy, we noted that drug exposure caused LC3-GFP vesicle formation with the development of multiple small intensely staining vesicles that included LC3-GFP.26-29 In the present studies we found that Obatoclax and Obatoclax + Lapatinib treatment rapidly caused the formation of one or two very large intensely staining LC3-GFP positive vesicles per cell. The precise molecular reasons for the differences in size and number of autophagic vesicles per cell between our different studies using different autophagy-inducing drugs will require a large amount of study beyond the scope of the present manuscript.
Many laboratories have presented evidence supporting the concept that apoptosis and autophagy are controlled by a number of identical proteins and that this inter-connection is central to the cross-talk between these processes. Beclin1 (ATG6) is a component of a type III PI3K complex that is required for the formation of autophagic vesicles, and was first identified as a BCL-2 interacting protein.37,38 Association of Beclin1 with BCL-2 family proteins blocks the autophagy-promoting function Beclin1. Thus BCL-2 and BCL-Xl regulate not only apoptosis but also autophagy. The use of a drug such as Obatoclax may thus serve to promote apoptosis by facilitating BAX and BAK activation or may serve to promote a toxic form of autophagy by promoting Beclin1 release from BCL-2 and BCL-Xl. Further studies, beyond the scope of this manuscript, will be required to determine whether Obatoclax and Lapatinib interact to suppress tumor viability in vivo.
Materials and Methods
Materials
Obatoclax (GX-15-070) was generously provided by Gemin X Pharmaceuticals. Lapatinib was purchased from LC Laboratories (Woburn, MA). Phospho-/total-(mTOR, ERK1/2; JNK1/2; p38 MAPK, ERBB1 Y992 and Y1068 and Y1173) antibodies, phospho-/total-AKT (T308; S473); anti-ATG5 and Beclin1, and the total and cleaved caspase 3 antibodies were purchased from Cell Signaling Technologies (Worcester, MA). All the secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). TUNEL kits were purchased from NEN Life Science Products (NEN Life Science Products, Boston, MA) and Boehringer Mannheim (Manheim, Germany), respectively. Trypsin-EDTA, DMEM, RPMI, penicillin-streptomycin were purchased from GIBCOBRL (GIBCOBRL Life Technologies, Grand Island, NY). Commercially available validated short hairpin RNA molecules to knockdown RNA/protein levels were from Qiagen (Valencia, CA): AIF (SI02654463; SI03118255); ATG5 (SI02655310); Beclin 1 (SI00055573, SI00055587). We also made use for confirmatory purposes of the short hairpin RNA construct targeting ATG5 (pLVTHM/Atg5) that was a gift from Dr. Yousefi, Department of Pharmacology, University of Bern, Switzerland. The plasmids to express green fluorescent protein (GFP)-tagged human LC3 was kindly provided by Dr. S. Spiegel, VCU. MEFs lacking expression of BAX, BAK and BIM were from the laboratory of Dr. S. Korsmeyer (Harvard University). ERBB1−/− MEFs were supplied by Dr. J. Grandis (University of Pitsburgh). ATG5−/− MEFs were supplied by Dr. M. Czaja (Yeshiva University, New York). Mammary carcinoma cells were from the ATCC and from Dr. Nephew and Dr. Larner. Reagents and the detailed performance of all experimental procedures were as described refs. 18; 25–30.
Methods
Culture and in vitro exposure of cells to drugs
All established cell lines were cultured at 37%C (5% (v/v CO2) in vitro using RPMI supplemented with 5% (v/v) fetal calf serum and 10% (v/v) Non-essential amino acids. For short term cell killing assays, immunoblotting and cytochrome c release/BH3 domain protein activation studies, cells were plated at a density of 3 × 103 per cm2 (~2 × 105 cells per well of a 12-well plate) and 48 h after plating treated with various drugs, as indicated. In vitro “drug” treatments were from 100 mM stock solutions of each drug and the maximal concentration of Vehicle (DMSO) in media was 0.02% (v/v).
In vitro cell treatments, microscopy, SDS-PAGE and western blot analysis
For in vitro analyses of short-term cell death effects, cells were treated with vehicle or drugs for the indicated times in the figure legends. For apoptosis assays where indicated, cells were pre-treated with vehicle (VEH, DMSO) and therapeutic drugs; cells were isolated at the indicated times, and either subjected to trypan blue cell viability assay by counting in a light microscope or fixed to slides, and stained using a commercially available Diff Quick (Geimsa) assay kit. Alternatively, the Annexin V/propidium iodide assay was carried to determine cell viability out as per the manufacturer’s instructions (BD PharMingen) using a Becton Dickinson FACScan flow cytometer (Mansfield, MA).
For SDS PAGE and immunoblotting, cells were plated at 5 × 105 cells/cm2 and treated with drugs at the indicated concentrations and after the indicated time of treatment, lysed in wholecell lysis buffer (0.5 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 0.02% bromophenol blue), and the samples were boiled for 30 min. The boiled samples were loaded onto 10–14% SDS-PAGE and electrophoresis was run overnight. Proteins were electrophoretically transferred onto 0.22 μm nitrocellulose, and immunoblotted with various primary antibodies against different proteins. Immunoblots were visualized by an Odyssey infra red imaging system. For presentation, immunoblots after scanning were processed using Adobe PhotoShop CS2 and Figures generated in MicroSoft PowerPoint.
Infection of cells with recombinant adenoviruses
Cells were plated at 3 × 103 per cm2 in each well of a 12-well, 6-well or 60 mm plate. After plating (24 h), cells were infected (at a multiplicity of infection of 50) with a control empty vector virus (CMV) and adenoviruses to express CRM A, c-FLIP-s, BCL-Xl, dominant negative caspase 9 or XIAP (Vector Biolabs, Philadelphia, PA). After infection (24 h) cells were treated with the indicated concentrations of drugs, and cell survival or changes in expression/phosphorylation determined 0–96 h after drug treatment by trypan blue/TUNEL/flow cytometry assays and immunoblotting, respectively.
Transfection of cells with siRNA or with plasmids
For plasmids
Cells were plated as described above and 24 h after plating, transfected. Cells were transfected with (0.5 μg) plasmids expressing a specific mRNA (or siRNA) or appropriate vector control plasmid DNA was diluted in 50 μl serum-free and antibiotic-free medium (one portion for each sample). Concurrently, 2 μl Lipofectamine 2000 (Invitrogen), was diluted into 50 μl of serum-free and antibiotic-free medium (one portion for each sample). Diluted DNA was added to the diluted Lipofectamine 2000 for each sample and incubated at room temperature for 30 min. This mixture was added to each well/dish of cells containing 200 μl serum-free and antibiotic-free medium for a total volume of 300 μl, and the cells were incubated for 4 h at 37°C. An equal volume of ×2 medium was then added to each well. Cells were incubated for 24 h, and then treated with vorinostat/sorafenib.
Transfection of cells with siRNA or with plasmids
For plasmids. Cells were plated as described above and 24 h after plating, transfected. Cells were transfected with (0.5 μg) plasmids expressing a specific mRNA (or siRNA) or appropriate vector control plasmid DNA was diluted in 50 μl serum-free and antibiotic-free medium (one portion for each sample). Concurrently, 2 μl Lipofectamine 2000 (Invitrogen), was diluted into 50 μl of serum-free and antibiotic-free medium (one portion for each sample). Diluted DNA was added to the diluted Lipofectamine 2000 for each sample and incubated at room temperature for 30 min. This mixture was added to each well/dish of cells containing 200 μl serum-free and antibiotic-free medium for a total volume of 300 μl, and the cells were incubated for 4 h at 37°C. An equal volume of ×2 medium was then added to each well. Cells were incubated for 24 h, and then treated with vorinostat/sorafenib.
Transfection with siRNA. Cells were plated in 60 mm dishes from a fresh culture growing in log phase as described above, and 24 h after plating transfected. Prior to transfection, the medium was aspirated and 1 ml serum-free medium was added to each plate. For transfection, 10 nM of the annealed siRNA, the positive sense control doubled stranded siRNA targeting GAPDH or the negative control (a “scrambled” sequence with no significant homology to any known gene sequences from mouse, rat or human cell lines) were used. 10 nM siRNA (scrambled or experimental) was diluted in serum-free media. 4 μl Hiperfect (Qiagen) was added to this mixture and the solution was mixed by pipetting up and down several times. This solution was incubated at room temp for 10 min, then added dropwise to each dish. The medium in each dish was swirled gently to mix, then incubated at 37°C for 2 h. One ml of 10% (v/v) serum-containing medium was added to each plate, and cells were incubated at 37°C for 48 h before re-plating (50 × 103 cells each) onto 12-well plates. Cells were allowed to attach overnight, then treated with drugs (0–48–96 h). Trypan blue/TUNEL/flow cytometry assays and SDSPAGE/immunoblotting analyses were performed at the indicated time points in each figure.
Isolation of a crude cytosolic fraction
A crude membrane fraction was prepared from treated cells. Briefly, cells were washed twice in ice cold isotonic HEPES buffer (10 mM HEPES pH 7.5, 200 mM mannitol, 70 mM sucrose, 1 μM EGTA, 10 μM protease inhibitor cocktail (Sigma, St. Louis, MO). Cells on ice were scraped into isotonic HEPES buffer and lysed by passing 20 times through a 25 gauge needle. Large membrane pieces, organelles and unlysed cells were removed from the suspension by centrifugation for 5 min at 120 xg. The crude granular fraction and cytosolic fraction was obtained from by centrifugation for 30 min at 10,000 xg, leaving the cytosol as supernatant.
Microscopic assessment of autophagy
For autophagy studies, cells transfected with LC3-GFP in glass chambered slides were applied after drug exposure to high powered light/confocal microscopes (Zeiss LSM 510 Meta-confocal scanning microscope; Zeiss HBO 100 microscope with Axio Cam MRm camera) at the indicated magnification in the figure/figure legend.
Data analysis
Comparison of the effects of various treatments was performed using ANOVA and the Student’s t test. Differences with a p-value of <0.05 were considered statistically significant. Experiments shown are the means of multiple individual points (±SEM). Median dose effect isobologram analyses to determine synergism of drug interaction were performed according to the Methods of T.-C. Chou and P. Talalay using the Calcusyn program for Windows (BIOSOFT, Cambridge, UK). Cells are treated with agents at a fixed concentration dose. A combination index (CI) value of less than 1.00 indicates synergy of interaction between the drugs; a value of 1.00 indicates additivity; a value of >1.00 equates to antagonism of action between the agents.
Acknowledgements
This work was funded by The Goodwin Foundation for Cancer Research (to Massey Cancer Center). P.D. is the holder of the Universal Inc., Professorship in Signal Transduction Research. Dr. Nephew was funded by NIH grant U54CA113001.
Abbreviations
- Lap
lapatinib
- GX
GX15-070 (Obatoclax)
- FP
flavopiridol
- Rosc
roscovitine
- ERK
extracellular regulated kinase
- MEK
mitogen activated extracellular regulated kinase
- EGF
epidermal growth factor
- PARP
poly ADP ribosyl polymerase
- PI3K
phosphatidyl inositol 3 kinase
- −/−
null/gene deleted
- ERK
extracellular regulated kinase
- MAPK
mitogen activated protein kinase
- MEK
mitogen activated extracellular regulated kinase
- R
receptor
- JNK
c-Jun NH2-terminal kinase
- dn
dominant negative
- P
phospho-
- ca
constitutively active
- WT
wild-type
References
- 1.Pytel D, Sliwinski T, Poplawski T, Ferriola D, Majsterek I. Tyrosine kinase blockers: new hope for successful cancer therapy. Anticancer Agents Med Chem. 2009;9:66–76. doi: 10.2174/187152009787047752. [DOI] [PubMed] [Google Scholar]
- 2.McCubrey JA, Milella M, Tafuri A, Martelli AM, Lunghi P, Bonati A, et al. Targeting the Raf/MEK/ERK pathway with small-molecule inhibitors. Curr Opin Investig Drugs. 2008;9:614–30. [PubMed] [Google Scholar]
- 3.Benavente S, Huang S, Armstrong EA, Chi A, Hsu KT, Wheeler DL, Harari PM. Establishment and characterization of a model of acquired resistance to epidermal growth factor receptor targeting agents in human cancer cells. Clin Cancer Res. 2009;15(5):1585–92. doi: 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hida T, Ogawa S, Park JC, Park JY, Shimizu J, Horio Y, Yoshida K. Gefitinib for the treatment of non-small-cell lung cancer. Expert Rev Anticancer Ther. 2009;9:17–35. doi: 10.1586/14737140.9.1.17. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–17. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 6.Ladanyi M, Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol. 2008;21:16–22. doi: 10.1038/modpathol.3801018. [DOI] [PubMed] [Google Scholar]
- 7.Johnson BE, Jänne PA. Epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Cancer Res. 2005;65:7525–9. doi: 10.1158/0008-5472.CAN-05-1257. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinibsensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 10.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Q, Esteva FJ. Cross-talk between the ErbB/HER family and the type I insulin-like growth factor receptor signaling pathway in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:485–98. doi: 10.1007/s10911-008-9107-3. [DOI] [PubMed] [Google Scholar]
- 12.Mueller KL, Hunter LA, Ethier SP, Boerner JL. Met and c-Src cooperate to compensate for loss of epidermal growth factor receptor kinase activity in breast cancer cells. Cancer Res. 2008;68:3314–22. doi: 10.1158/0008-5472.CAN-08-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arteaga CL. HER3 and mutant EGFR meet MET. Nat Med. 2007;13:675–7. doi: 10.1038/nm0607-675. [DOI] [PubMed] [Google Scholar]
- 14.Huang F, Greer A, Hurlburt W, Han X, Hafezi R, Wittenberg GM, et al. The mechanisms of differential sensitivity to an insulin-like growth factor-1 receptor inhibitor (BMS-536924) and rationale for combining with EGFR/HER2 inhibitors. Cancer Res. 2009;69:161–70. doi: 10.1158/0008-5472.CAN-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, et al. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–75. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- 16.Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4:1681–9. doi: 10.1371/journal.pmed.0040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen FL, Xia W, Spector NL. Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin Cancer Res. 2008;14:6730–4. doi: 10.1158/1078-0432.CCR-08-0581. [DOI] [PubMed] [Google Scholar]
- 18.Martin AP, Miller A, Emad L, Rahmani M, Walker T, Mitchell C, et al. Lapatinib resistance in HCT116 cells is mediated by elevated MCL-1 expression and decreased BAK activation and not by ERBB receptor kinase mutation. Mol Pharmacol. 2008;74:807–22. doi: 10.1124/mol.108.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant S, Dent P. Simultaneous interruption of signal transduction and cell cycle regulatory pathways: implications for new approaches to the treatment of childhood leukemias. Curr Drug Targets. 2007;8:751–9. doi: 10.2174/138945007780830764. [DOI] [PubMed] [Google Scholar]
- 20.Grant S. Is the focus moving toward a combination of targeted drugs? Best Pract Res Clin Haematol. 2008;21:629–37. doi: 10.1016/j.beha.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azmi AS, Mohammad RM. Non-peptidic small molecule inhibitors against Bcl-2 for cancer therapy. J Cell Physiol. 2009;218:13–21. doi: 10.1002/jcp.21567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–32. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cragg MS, Harris C, Strasser A, Scott CL. Unleashing the power of inhibitors of oncogenic kinases through BH3 mimetics. Nat Rev Cancer. 2009;9(5):321–6. doi: 10.1038/nrc2615. [DOI] [PubMed] [Google Scholar]
- 24.Li R, Zang Y, Li C, Patel NS, Grandis JR, Johnson DE. ABT-737 synergizes with chemotherapy to kill head and neck squamous cell carcinoma cells via a NOXAmediated pathway. Mol Pharmacol. 2009;75(5):1231–9. doi: 10.1124/mol.108.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park MA, Zhang G, Mitchell C, Rahmani M, Hamed H, Hagan MP, et al. Mitogen-activated protein kinase kinase 1/2 inhibitors and 17-allylamino-17-demethoxygeldanamycin synergize to kill human gastrointestinal tumor cells in vitro via suppression of c-FLIP-s levels and activation of CD95. Mol Cancer Ther. 2008;7:2633–48. doi: 10.1158/1535-7163.MCT-08-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park MA, Yacoub A, Sarkar D, Emdad L, Rahmani M, Spiegel S, et al. PERK-dependent regulation of MDA-7/IL-24-induced autophagy in primary human glioma cells. Autophagy. 2008;4:513–5. doi: 10.4161/auto.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park MA, Yacoub A, Rahmani M, Zhang G, Hart L, Hagan MP, et al. OSU-03012 stimulates PKR-like endoplasmic reticulum-dependent increases in 70-kDa heat shock protein expression, attenuating its lethal actions in transformed cells. Mol Pharmacol. 2008;73:1168–84. doi: 10.1124/mol.107.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, et al. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramidedependent CD95 and PERK activation. Cancer Biol Ther. 2008;7:1648–62. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G, Park MA, Mitchell C, Walker T, Hamed H, Studer E, et al. Multiple cyclin kinase inhibitors promote bile acid-induced apoptosis and autophagy in primary hepatocytes via p53-CD95-dependent signaling. J Biol Chem. 2008;283:24343–58. doi: 10.1074/jbc.M803444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell C, Park MA, Zhang G, Yacoub A, Curiel DT, Fisher PB, et al. Extrinsic pathway- and cathepsindependent induction of mitochondrial dysfunction are essential for synergistic flavopiridol and vorinostat lethality in breast cancer cells. Mol Cancer Ther. 2007;6:3101–12. doi: 10.1158/1535-7163.MCT-07-0561. [DOI] [PubMed] [Google Scholar]
- 31.Lin TS. Novel agents in chronic lymphocytic leukemia: efficacy and tolerability of new therapies. Clin Lymphoma Myeloma. 2008;4:137–43. doi: 10.3816/CLM.2008.s.009. [DOI] [PubMed] [Google Scholar]
- 32.Balakrishnan K, Burger JA, Wierda WG, Gandhi V. AT-101 induces apoptosis in CLL B cells and overcomes stromal cell-mediated Mcl-1 induction and drug resistance. Blood. 2009;113:149–53. doi: 10.1182/blood-2008-02-138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia L, Coward LC, Kerstner-Wood CD, Cork RL, Gorman GS, Noker PE, et al. Comparison of pharmacokinetic and metabolic profiling among gossypol, apogossypol and apogossypol hexaacetate. Cancer Chemother Pharmacol. 2008;61:63–73. doi: 10.1007/s00280-007-0446-3. [DOI] [PubMed] [Google Scholar]
- 34.Schimmer AD, O’Brien S, Kantarjian H, Brandwein J, Cheson BD, Minden MD, et al. A phase I study of the pan bcl-2 family inhibitor obatoclax mesylate in patients with advanced hematologic malignancies. Clin Cancer Res. 2008;14:8295–301. doi: 10.1158/1078-0432.CCR-08-0999. [DOI] [PubMed] [Google Scholar]
- 35.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 36.Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S, et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007;3:561–8. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 38.Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–6. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 39.Ackler S, Xiao Y, Mitten MJ, Foster K, Oleksijew A, Refici M, et al. ABT-263 and rapamycin act cooperatively to kill lymphoma cells in vitro and in vivo. Mol Cancer Ther. 2008;7:3265–74. doi: 10.1158/1535-7163.MCT-08-0268. [DOI] [PubMed] [Google Scholar]
- 40.Paoluzzi L, Gonen M, Gardner JR, Mastrella J, Yang D, Holmlund J, et al. Targeting Bcl-2 family members with the BH3 mimetic AT-101 markedly enhances the therapeutic effects of chemotherapeutic agents in in vitro and in vivo models of B-cell lymphoma. Blood. 2008;111:5350–8. doi: 10.1182/blood-2007-12-129833. [DOI] [PubMed] [Google Scholar]
- 41.Reed JC, Kitada S, Takayama S, Miyashita T. Regulation of chemoresistance by the bcl-2 oncoprotein in non-Hodgkin’s lymphoma and lymphocytic leukemia cell lines. Ann Oncol. 1994;5:61–5. doi: 10.1093/annonc/5.suppl_1.s61. [DOI] [PubMed] [Google Scholar]
- 42.Okumura K, Huang S, Sinicrope FA. Induction of Noxa sensitizes human colorectal cancer cells expressing Mcl-1 to the small-molecule Bcl-2/Bcl-xl inhibitor, ABT-737. Clin Cancer Res. 2008;14:8132–42. doi: 10.1158/1078-0432.CCR-08-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]