Abstract
This study was undertaken to establish rat embryonic stem (ES) cells from parthenogenetically developing blastocysts. Ten blastocysts were prepared by treatment of ovulated rat oocytes with ionomycin and cycloheximide, and three alkaline phosphatase-positive ES cell lines were established using the N2B27 medium supplemented with mitogen activated protein kinase kinase inhibitor PD0325901, glycogen synthase kinase 3 inhibitor CHIR99021, rat leukemia inhibitory factor, and forskolin. Expression of stem cell marker genes (Oct-4, rNanog, Fgf-4, and Rex-1) was confirmed in all three ES cell lines by reverse transcriptase-polymerase chain reaction (RT-PCR). Combined bisulfite restriction analysis showed that the differentially methylated region locus of five imprinted genes (H19, Meg3IG, Igf2r, Peg5, and Peg10) in these ES cells remained to be demethylated or was hypomethylated, which was similar to that in control ES cells established from normal blastocysts. Characteristics of the parthenogenetic blastocyst-derived ES cells were successfully transmitted to the next generation through a chimeric rat for one of the three ES cell lines. This is the first report on germline-competent (genuine) ES cells derived from parthenogenetically developing rat blastocysts.
Introduction
The reverse genetic approach, such as precise and conditional replacements (knock-in) or loss of gene function (knock-out) at a specific locus, has been considered impossible in rats because protocols to establish stem cell lines conventionally used for mice [1,2] are not applicable to rats. However, functional germline-competent embryonic stem (ES) cell lines [3,4] have been recently established from rats by applying inhibitors of mitogen activated protein kinase kinase (MEK), glycogen synthase kinase 3 (GSK3), and/or the fibroblast growth factor receptor in differentiation-related signaling pathways.
In mice [5,6], monkeys [7,8], and humans [9,10], pluripotent ES cell lines have been established from the inner cell mass of parthenogenetically developing blastocysts. All genes of parthenogenetic blastocyst-derived ES (pES) cells with uniparental disomy are homozygous. Therefore, pES cells can provide a source of histocompatible tissues that avoid immune rejection upon transplantation [11]. Despite the fact that rats have been used as model animals for organ transplantation [12], rat pES cell lines have not been reported to date. The present study was conducted to establish a germline-competent (genuine) ES cell line from parthenogenetic rat blastocysts.
Materials and Methods
Animals
All procedures for animal experimentation were reviewed and approved by the Animal Care and Use Committee of the National Institute for Physiological Sciences (Okazaki, Japan). Parthenogenetic blastocysts for establishment of pES cells were prepared from homozygous CAG/venus-transgenic (original strain, Crlj:WI; Charles River Japan, Inc., Kanagawa, Japan) female rats [13] maintained in our facility. Blastocyst donors and recipients for production of chimeric rats were 8–13-week-old specific pathogen-free Crlj:WI female rats. All rats were housed in an environmentally controlled room with a 12-h dark/12-h light cycle at 23°C±2°C with 55%±5% humidity, and given free access to a laboratory diet (CE-2; CLEA Japan, Inc., Tokyo, Japan) and filtered water.
Parthenogenetic blastocysts
Female rats were superovulated by intraperitoneal injections of 150 IU/kg equine chorionic gonadotropin (Asuka Pharmacies Co., Tokyo, Japan) and 75 IU/kg human chorionic gonadotropin (Asuka Pharmacies) at an interval of 46–50 h. Oocytes were collected from oviductal ampullae at 14–17 h after the hCG injection, and were freed from cumulus cells by incubation for 5–10 min with 0.1% hyaluronidase (w/v; Sigma-Aldrich, St. Louis, MO) dissolved in the mR1ECM medium [14]. The oocytes were chemically activated by incubation with 5 μM ionomycin (Sigma-Aldrich) for 5 min and 5 μg/mL cycloheximide+2 mM 6-dimethylaminopurine (Sigma-Aldrich) for 4 h at 37°C with 5% CO2 as described previously [15,16] with slight modifications. At 6 h after activation, parthenogenetic zygotes with two visible pronuclei were transferred into the oviducts of recipient rats that had been previously mated with a vasectomized male rat. At ∼98 h after the transfer, blastocysts were harvested from the uteri of recipient females with the HER medium [17] supplemented with 18% fetal bovine serum (GIBCO, Auckland, New Zealand).
Establishment of pES cells
Zona-free parthenogenetic blastocysts were plated on mitomycin-treated mouse embryonic fibroblasts (MEFs) in the 2iF medium [N2B27 medium containing 1 μM MEK inhibitor PD0325901 (Stemgent, Cambridge, MA), 3 μM GSK3 inhibitor CHIR99021 (Axon Ligands, Groningen, the Netherlands), 1,000 U/mL ESGRO® (Millipore Co., Billerica, MA), and 10 μM forskolin (Sigma-Aldrich)] as described previously [18]. Outgrowths from parthenogenetic blastocysts were disaggregated and transferred to new culture vessels containing MEFs and the 2iF medium. When ES cell-like colonies had emerged, they were trypsinized in a 0.25% trypsin/0.6 mM EDTA solution (Sigma-Aldrich), and then passaged. The tentative pES cell lines were maintained under MEF/2iF conditions with medium exchanges every other day and passaging every 3 days. Histochemical assays of pES cells were conducted with a Leukocyte Alkaline Phosphatase Kit (Sigma-Aldrich) according to the manufacturer's instructions. At passages 5 and 6, pES cells were cryopreserved until subsequent use. Chimeric rats were generated with each of pES cell lines at passage 7. Using passage 8 cells, karyotype analysis (chromosome number: 50 cells and G-band staining: 20 cells, per pES cell line) was performed by a commercial company (Nihon Gene Research Laboratories, Miyagi, Japan), according to their standard protocols.
Expression of stem cell marker genes
The expression of stem cell marker genes (Oct-4, rNanog, Fgf-4, and Rex-1) and of reference genes (Gata6 and β-actin) was examined by reverse transcriptase-polymerase chain reaction (RT-PCR) analysis. A trophectoderm-specific marker gene, Cdx2, was also examined. The primer sets used are shown in Table 1. Total RNA was extracted from each sample using a RNeasy® mini Kit (Qiagen, Germantown, MD). The cDNA was prepared using SuperScript™ III First-Strand Synthesis System (Invitrogen, Carlsbad, CA) and amplified with TaKaRa LA Taq® (Takara Bio, Shiga, Japan) by 30 cycles at 94°C for 20 s, 55°C for 30 s, and 72°C for 60 s.
Table 1.
Primers Used for PCR Amplification
| Marker gene | Primer sequence | Product size (bp) | |
|---|---|---|---|
|
Oct-4 |
Forward |
5′-GGGATGGCATACTGTGGAC-3′ |
412 |
| |
Reverse |
5′-CTTCCTCCACCCACTTCTC-3′ |
|
|
rNanog |
Forward |
5′-GCCCTGAGAAGAAAGAAGAG-3′ |
356 |
| |
Reverse |
5′-CGTACTGCCCCATACTGGAA-3′ |
|
|
Fgf-4 |
Forward |
5′-CGGGGTGTGGTGAGCATCTTC-3′ |
202 |
| |
Reverse |
5′-CCTTCTTGGTCCGCCCGTTC-3′ |
|
|
Rex-1 |
Forward |
5′-TTCTTGCCAGGTTCTGGAAGC-3′ |
297 |
| |
Reverse |
5′-TTTCCCACACTCTGCACACAC-3′ |
|
|
Cdx2 |
Forward |
5′-CCGAATACCACGCACACCATC-3′ |
394 |
| |
Reverse |
5′-CTTTCCTTGGCTCTGCGGTTC-3′ |
|
|
Gata6 |
Forward |
5′-TCATCACGACGGCTTGGACTG-3′ |
467 |
| |
Reverse |
5′-GCCAGAGCACACCAAGAATCC-3′ |
|
|
ß-actin |
Forward |
5′-CATGGCATTGTGATGGACT-3′ |
427 |
| Reverse | 5′-ACGGATGTCAACGTCACACT-3′ |
PCR, polymerase chain reaction.
Analysis of imprinted genes
The degree of methylation at the differentially methylated region (DMR) locus of five potentially imprinted genes (H19, Meg3IG, Igf2r, Peg5, and Peg10) was analyzed by combined bisulfite restriction analysis (COBRA). These regions of each gene have been assessed for their imprinted status in the rat genome by Shinohara et al. [19]. Genomic DNA from each sample was treated with 3.09 M sodium bisulfite, which deaminates unmethylated cytosines to uracils, but does not affect 5-methylated cytosines, and 0.5 mM hydroquinone. The DMRs were amplified by PCR (43 cycles of 95°C for 30 s, 55°C–59°C for 30 s, and 72°C for 1 min) using specific primers (Table 2), and then digested with restriction enzymes as described by Kanatsu-Shinohara et al. [20]. The intensity of digested bands was quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Table 2.
Sequences of Primers Used For COBRA
| Genes | Forward | Reverse |
|---|---|---|
|
H19 |
GAGGGTTTTAAATTTTATTAGGAGGG |
ACACCCCAAAAATCATTAACATCTA |
|
Meg3IG |
GGGTTTATTGTGTATAAGGATTGTG |
AATTCCATCAAAAATTTCAAAACTC |
|
Igf2r |
ATAGTATAATAGGAATTATATTAAAAATTT |
ACTCCAACTAAAAATTCCAAACTAC |
|
Peg5 |
GGTATATATTTTGTTTTTTGAGAGATTTT |
AAATACCCTCCTACCACCTTAATAC |
| Peg10 | TTTTATGTTTTTAGTGTATTAATGGG | CAAAACTCCATTTTATCTACCACC |
COBRA, combined bisulfite restriction analysis.
Immunofluorescence microscopy
The pES cells were cultured with MEFs on the cover-grass in tissue culture grade four-well plates. After 2 days, the cells were washed twice in the 2iF medium and fixed in freshly prepared 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1 h at room temperature. The cells were incubated in 100% EtOH for 30 min at −20°C, rinsed in PBS and blocking buffer (10% goat serum in PBS), and permeabilized with 0.1% Triton X-100 in the blocking buffer for 1 h at room temperature. Then, the cells were incubated 4 h at 4°C with each specific first antibody. Primary antibodies used include Oct-4 (1:200 for 4 h; Santa Cruz Biotechnology, Santa Cruz, CA), SSEA-1 (1:200 for 4 h; Santa Cruz Biotechnology), Sox2 (1:100 for 3 days; Santa Cruz Biotechnology), and Nanog (1:200 for 4 h; Abcam®, Cambridge, United Kingdom). Alexa Fluor 568 secondary antibodies (1:1,000 for 1 h; Invitrogen) were counterstained with 0.2 μg/mL 4′,6-diamidino-2-phenylindole in PBS, and then the samples were enclosed by Prolong Gold. Fluorescent images were taken under a Nikon microscope using appropriate filters.
Chimeric rat production and G1 transmission
Using a micromanipulation system with a piezo-driving unit (PMM-150FU; Prime Tech, Ibaraki, Japan) and pulse controller (PMAS-CT150; Prime Tech), 10–15 pES cells were microinjected into the blastocoelic cavity of host blastocysts using a blunt-ended injection pipette with an outer diameter of 15 μm as described previously [21]. The reblastulated embryos at 1–2 h after microinjection were transferred into the uteri of pseudopregnant Crlj:WI recipients (5–10 embryos per uterine horn), and allowed to develop into full-term offspring. When the offspring were found to be chimeric rats by venus expression, the chimerism of the rats was determined. Briefly, peripheral blood cells were collected from the retro-orbital venous plexus of chimeric rats at >10 weeks old. Leukocytes isolated by osmotic lysis of erythrocytes were stained with an APC-conjugated mouse anti-rat CD45 antibody (BD Biosciences, San Diego, CA), and then analyzed by flow cytometry using a FACSCanto II (BD Biosciences). To examine the germline competency of each pES cell line to the F1 generation, chimeric female rats were bred with wild-type male rats. Three chimeric rats per pES cell line were used for the procedure, and three litters per chimeric rat were analyzed for venus expression.
Statistical analysis
Three replicates were performed for COBRA. Data on DNA methylation data were analyzed by the Tukey significant difference test after the two-way ANOVA. A value of P<0.05 was chosen as an indication of the statistical significance.
Results and Discussion
A total of 59 oocytes were chemically activated, and all of them parthenogenetically developed into zygotes with two pronuclei. At 4 days after transfer of the zygotes, 10 parthenogenetically developing blastocysts and 30 degenerated zygotes were harvested from recipients. Using the MEF/2iF system, three blastocysts (30%) showed outgrowth (Fig. 1a), resulting in establishment of three tentative pES cell lines (Fig. 1b). Attachment of the pES cell colonies to feeder cells was not very strong, which is the same characteristic observed with normal blastocyst-derived rat ES cells (rESWIv3i-1; XX karyotype) [13]. The morphological appearance of rat pES cell colonies was also similar to that of typical rat ES cell colonies. When normal blastocysts harvested from CAG/venus transgenic rats were used to establish ES cell lines, the efficiency of establishing rat ES cell lines was 60% (9/15; unpublished data). The decreased likelihood of establishing rat pES cell lines versus ES cell lines is consistent with a previous finding by Liu et al. [6], in which mouse pES and ES cell lines were established at efficiencies of 18% and 44%, respectively. The efficiencies of establishing mouse pES cell lines appear to be generally low, ranging from 9% to 27% [4–6,22].
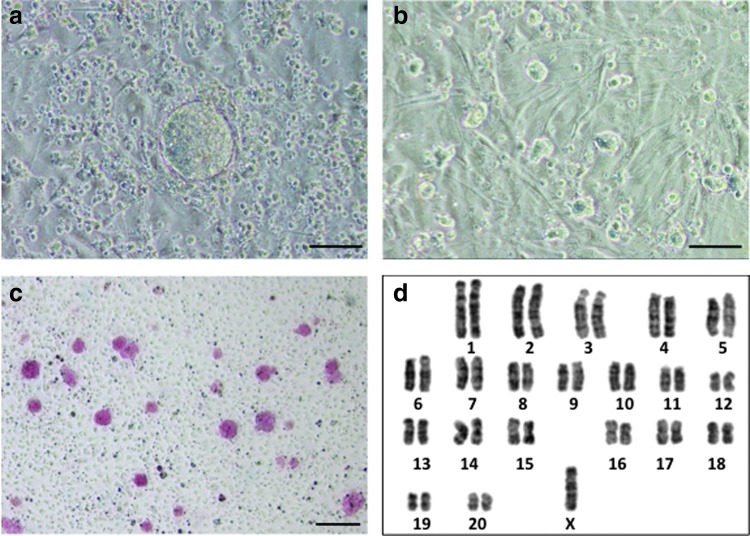
FIG. 1.
Phenotypic characterization of rat pES cells. (a) Outgrowth of a parthenogenetic blastocyst on a MEF feeder layer after 5 days of culture. (b) Colony formation of pES cells on MEFs at passage 6. (c) Alkaline phosphatase-positive colonies of rpESWIv2iF-5 at passage 7. (d) X-chromosome monosomy of rpESWIv2iF-5 cells at passage 8 by G-band staining. Scale bars: (a, b) 100 μm and (c) 200 μm. pES, parthenogenetic blastocyst-derived embryonic stem; MEF, mouse embryonic fibroblast. Color images available online at www.liebertpub.com/scd
Three rat pES cell lines (named rpESWIv2iF-2, rpESWIv2iF-5, and rpESWIv2iF-10) exhibited positive signals for alkaline phosphatase at passage 7 (Fig. 1c). Karyotypic analysis at passage 8 showed that cells had a chromosomal number of either 41 or 42. A normal diploid set of chromosomes (=42) was observed only in approximately half of the analyzed cells (27/50, 26/50, and 23/50 cells in rpESWIv2iF-2, rpESWIv2iF-5, and rpESWIv2iF-10, respectively). G-band staining showed that monosomy of X chromosome was encountered in approximately half of the pES cells (10/20, 9/20, and 10/20 cells in rpESWIv2iF-2, rpESWIv2iF-5, and rpESWIv2iF-10, respectively; Fig. 1d). It is unknown why half of the rat pES cells lacked one X chromosome, despite the normal number of autosomes in every line. Instability of the X chromosome has been characterized in a single deletion event occurring at early passages during the establishment of pES lines, and the resulting pES cells possessed only one X chromosome [23]. The XO genotype in mouse pES cells derived by the nuclear transfer method (NT-pES) was observed at passage 30, and normal XX cell lines have been established after repeated passages of the NT-pES cells for more than 2 months [24].
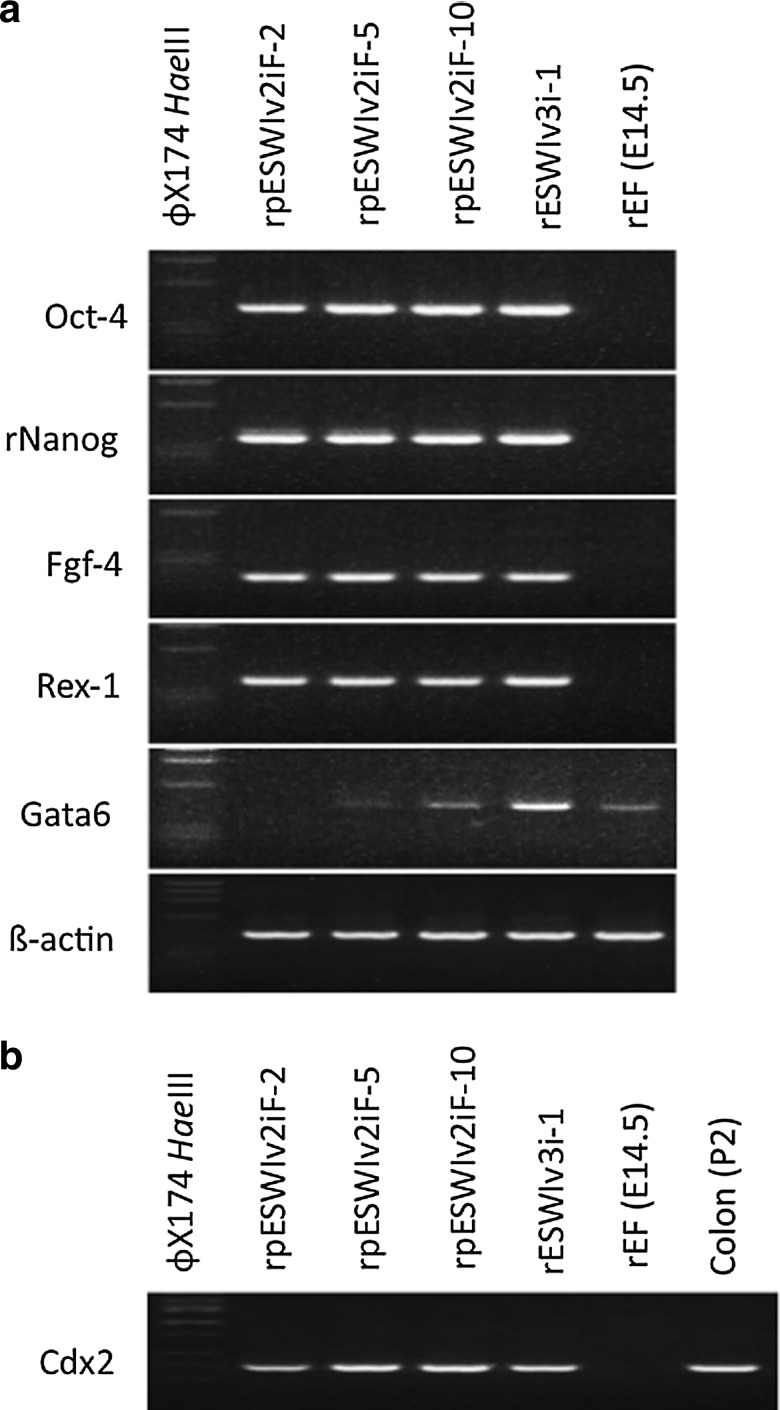
In RT-PCR analysis, stem cell marker genes such as Oct-4, rNanog, Fgf-4, and Rex-1 were expressed in the three rat pES cell lines (Fig. 2a). Weak expression of Gata6 was detected in two pES cell lines (rpESWIv2iF-5 and rpESWIv2iF-10) and one ES cell line (rESWIv3i-1). Buehr et al. [3], a pioneer of rat ES cell establishment, and Hirabayashi et al. [21] also observed a certain expression of Gata6 in rat ES cells. Extraembryonic endoderm stem (XEN) cell-like colonies expressed Gata6 [25,26], and XEN cell-like colonies were observed during passages of rESWIv3i-1 cell line (data not shown). However, such colonies did not appear in any rat pES cell cultures in the present study. In contrast, cells from all the three rat pES cell lines and rESWIv3i-1 line expressed Cdx2 (Fig. 2b). Although Cdx2 gene expression does not occur in mouse ES cells, Hong et al. [27] reported the expression of Cdx2 in genuine rat ES cells. The expression of Gata6 and Cdx2 may be a unique characteristic of the rat stem cells.
FIG. 2.
RT-PCR analysis. (a) Three rat pES cell lines (rpESWIv2iF-2, rpESWIv2iF-5, and rpESWIv2iF-10), a rat ES cell line (rESWIv3i-1), and rat embryonic fibroblasts at E14.5 (rEF; negative control for stem cell markers). The three pES cell lines expressed stem cell marker genes, Oct-4, rNanog, Fgf-4, and Rex-1. (b) Expression of Cdx2, a trophectoderm-specific marker gene. Colon from day-2 rat offspring and rEF were used as positive and negative controls, respectively. RT-PCR, reverse transcriptase-polymerase chain reaction.
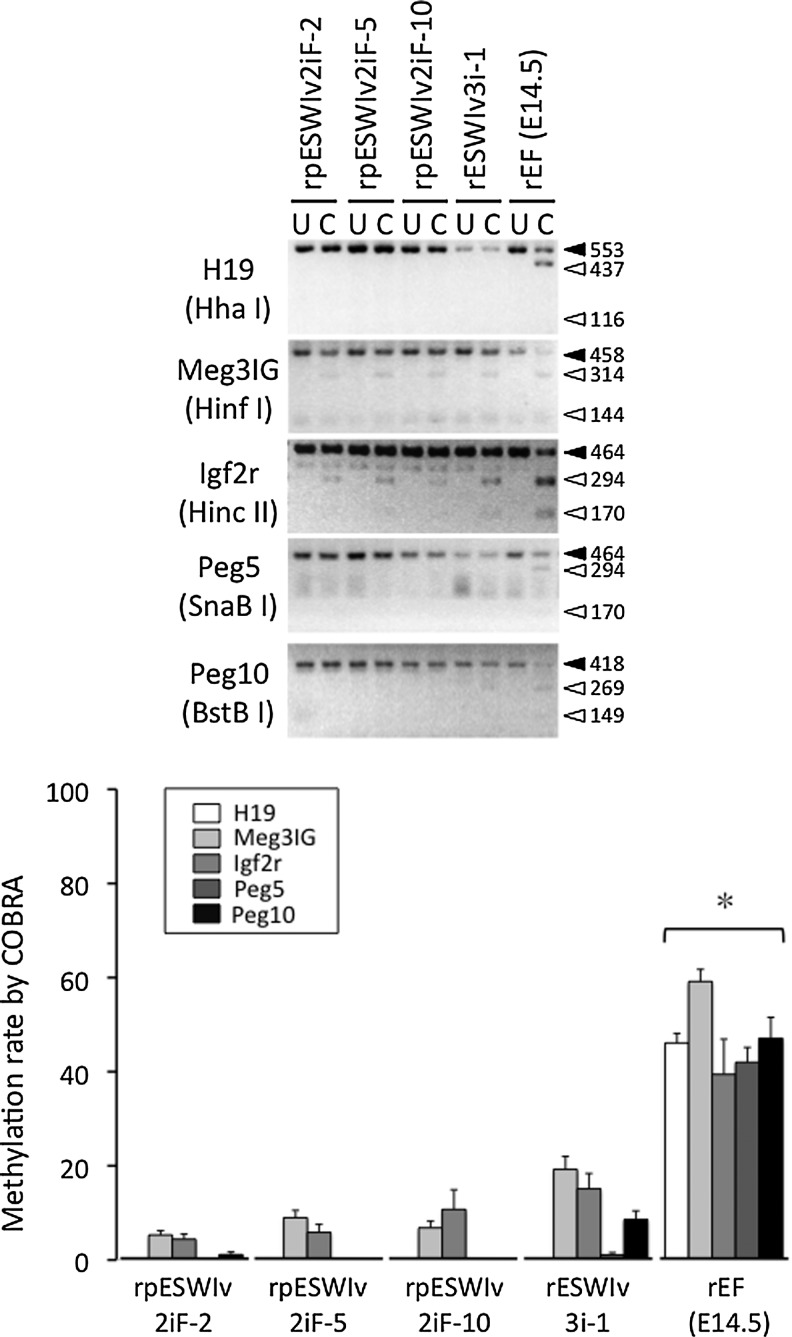
The degree of methylation at the DMR locus of five imprinted genes in rat pES cells is shown in Figure 3. COBRA showed that the DMR locus of five imprinted genes (H19, Meg3IG, Igf2r, Peg5, and Peg10) in rat pES cells remained to be demethylated or was hypomethylated, which was also observed in control ES cells (rESWIv3i-1) established from a normal blastocyst of the same transgenic rat colony. Theoretically, in pES cells, it is expected that paternally imprinted genes (eg, H19 and Meg3IG) remain demethylated, while maternally imprinted genes (eg, Igf2r, Peg5, and Peg10) are highly methylated. Jiang et al. [5] reported methylation of H19 and Gtl2 genes, both of which are paternally imprinted genes, in mouse pES cells. Liu et al. [6] also reported an abnormal methylation pattern of imprinted genes as shown by the highly methylated status of the maternally imprinted Snrpn gene detected in mouse oocytes, which was not maintained in pES cells. In addition, the methylation level in the Snrpn gene of pES cells was comparable with that in control ES cells established from normal blastocysts. In monkey pES cells, maternally imprinted Snurf/Snurpn IC genes are highly methylated, but the paternally imprinted H19 gene is more or less methylated [8]. Horii et al. [28] reported that full methylation marks of maternally imprinted genes (Peg1, Snrpn, and Igf2r) at the 8-cell stage of parthenogenetic mouse embryos are reduced to 60%–70% at the blastocyst stage. In addition, loss of such imprinting occurs in mouse pES cells established from those blastocysts [28]. Further research is necessary to understand the underlying mechanism and significance of DMR imprinting in pES cells.
FIG. 3.
DNA methylation status of imprinted genes (H19, Meg3IG, Igf2r, Peg5, and Peg10) in rat pES cell lines (rpESWIv2iF-2, rpESWIv2iF-5, and rpESWIv2iF-10), a rat ES cell line (rESWIv3i-1), and rat embryonic fibroblasts at E14.5 (rEF). DNA was PCR amplified by specific primers and the products were digested with restriction enzymes as indicated in parentheses. U, uncleaved; C, cleaved. *Significantly different from rpESWIv2iF-2, rpESWIv2iF-5, rpESWIv2iF-10, and rESWIv3i-1 (P<0.05). Mean±SEM.
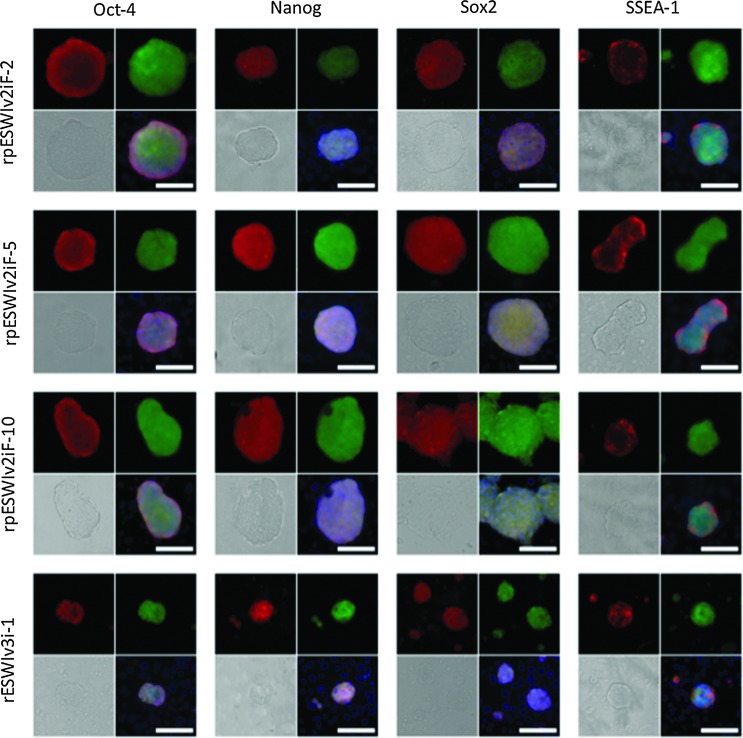
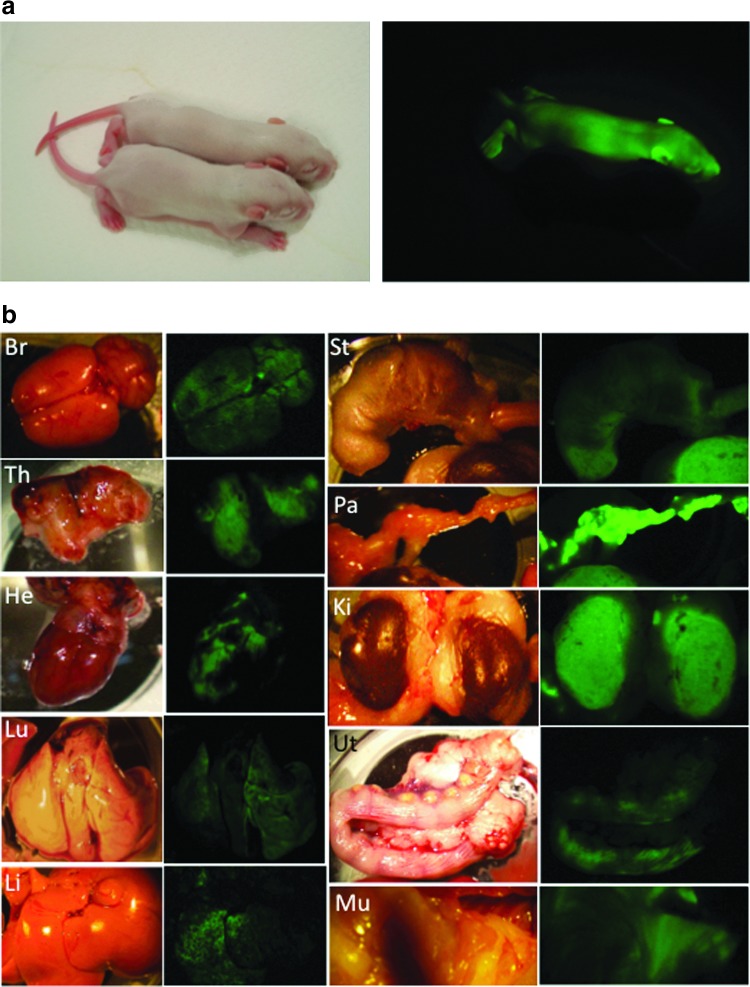
All the three rat pES cells lines expressed surface markers for pluripotency such as Oct-4, Nanog, Sox2, and SSEA-1 (Fig. 4). After microinjection of pES cells into rat blastocysts, a total of 124 blastocysts were transferred to recipients, resulting in the birth of 57 offspring (46%), including 37 venus-positive chimeric offspring (30%) as shown in Table 3. The overall low efficiency of producing chimeric rats with pES cells was comparable to that reported for mouse pES cells (4%–38%) [4–6,22,29]. However, chimeric rats were produced more efficiently (50%, 94/187) when normal blastocyst-derived ES cells were microinjected [13]. Thirty-one chimeric rats were able to develop to adulthood. Three female chimeric rats were selected from each of the three pES cell lines and used to examine their potential for germline transmission to the next generation. Only one chimeric rat (ID #4, rpESWIv2iF-5 cell line) transmitted the venus gene through the germline at a rate of 8% (3/38; three litters) (Fig. 5a). A low proportion of successful germline transmission has also been reported for mouse pES cells (25% vs. 78% for normal ES cells) by Liu et al. [6]. The leukocyte-based chimerism of these nine chimeric rats, which ranged from 25% to 71%, did not reflect the result of germline competency. Allen et al. [22] reported that differentiation of mouse pES cells to endodermal cells, but not to mesodermal or ectodermal cells, is relatively difficult because teratomas produced by transplantation of pES cells contain very little skeletal muscle tissues. However, in the present study, venus expression was detected in all of the main organs (brain, thymus, heart, lung, liver, stomach, intestine, pancreas, kidney, uterus, and skin) as well as the muscle of chimeric rats (ID #1, #7, and #9) during their postmortem autopsy (Fig. 5b).
FIG. 4.
Expression of pluripotency markers in rat pES cells. Oct-4, Nanog, Sox2, and SSEA-1. Upper left: pluripotency marker, upper right: venus, bottom left: bright field, bottom right: merge. Scale bars: 100 μm. Color images available online at www.liebertpub.com/scd
Table 3.
Production of Chimeric Rats with pES Cells and Potential of Germline Transmission
| |
No. (%) of blastocysts |
|
|
No. (%) of F1 pups |
|||
|---|---|---|---|---|---|---|---|
| Rat pES cell line | Injected | Developed to pups | Chimerasa | Female chimeras analyzed: ID | Chimerism (%) | Delivered | Venus+ |
| rpESWIv2iF-2 |
54 |
29 (54) |
17 (31) |
#1 |
71 |
38 |
0 (0) |
| |
|
|
|
#2 |
32 |
33 |
0 (0) |
| |
|
|
|
#3 |
33 |
38 |
0 (0) |
| rpESWIv2iF-5 |
25 |
12 (48) |
9 (36) |
#4 |
69 |
38 |
3 (8) |
| |
|
|
|
#5 |
67 |
36 |
0 (0) |
| |
|
|
|
#6 |
25 |
46 |
0 (0) |
| rpESWIv2iF-10 |
45 |
16 (36) |
11 (24) |
#7 |
25 |
40 |
0 (0) |
| |
|
|
|
#8 |
34 |
34 |
0 (0) |
| #9 | 66 | 30 | 0 (0) | ||||
Percentages for chimeras were calculated from the number of injected blastocysts.
pES, parthenogenetic blastocyst-derived embryonic stem.
FIG. 5.
In vivo differentiation of rat pES cells. (a) F1 generation germline chimera pups ubiquitously expressed the venus gene in a rat ES cell line (rpESWIv2iF-5). (b) Venus expression patterns of main organs in rat pES-derived chimeric rat. Br, brain; Th, thymus; He, heart; Lu, lung; Li, liver; St, stomach; Pa, pancreas; Ki, kidney; Ut, uterus; Mu, muscle. Left: bright field, right: fluorescent image. Color images available online at www.liebertpub.com/scd
In conclusion, a genuine rat pES cell line was successfully established from a parthenogenetic blastocyst. A pES cell line with uniparental disomy has unique advantages for model systems of transplantation therapies because their derivative cells can be transplanted into both homozygous and heterozygous recipients without immune rejection.
Acknowledgments
The authors thank Mrs. Kiyo Itokawa and Mrs. Keiko Yamauchi (National Institute for Physiological Sciences) for their assistance with animal care and preparation. This work was supported, in part, by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS; 22300147 and 25290037 to M.H.). H.H. is a JSPS Research Fellow.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Evans MJ. and Kaufman MH. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156 [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78:7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL. and Smith A. (2008). Capture of anthentic embryonic stem cells from rat blastocysts. Cell 135:1287–1298 [DOI] [PubMed] [Google Scholar]
- 4.Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, et al. (2008). Germline competent embryonic stem cells derived from rat blastocysts. Cell 135:1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, Sun B, Wang W, Zhang Z, Gao F, Shi G, Cui B, Kong X, He Z, et al. (2007). Activation of paternally expressed imprinted genes in newly derived germline-competent mouse parthenogenetic embryonic stem cell lines. Cell Res 17:792–803 [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Hu Z, Pan X, Li M, Togun TA, Tuck D, Pelizzola M, Huang J, Ye X, et al. (2011). Germline competency of parthenogenetic embryonic stem cells from immature oocytes of adult mouse ovary. Hum Mol Genet 20:1339–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cibelli JB,, Grant KA, Chapman KB, Cunniff K, Worst T, Green HL, Walker SJ, Gutin PH, Vilner L, et al. (2002). Parthenogenetic stem cells in nonhuman primates. Science 295:819. [DOI] [PubMed] [Google Scholar]
- 8.Dighe V, Clepper L, Pedersen D, Byrne J, Ferguson B, Gokhale S, Penedo MC, Wolf D. and Mitalipov S. (2008). Heterozygous embryonic stem cell lines derived from nonhuman primate parthenotes. Stem Cells 26:756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mai Q, Yu Y, Li T, Wang L, Chen MJ, Huang SZ, Zhou C. and Zhou Q. (2007). Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res 17:1008–1019 [DOI] [PubMed] [Google Scholar]
- 10.Revazova ES,, Turovets NA, Kochetkova OD, Kindarova LB, Kuzmichev LN, Janus JD. and Pryzhkova MV. (2007). Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells 9:432–449 [DOI] [PubMed] [Google Scholar]
- 11.DeChiara TM,, Robertson EJ. and Efstratiadis A. (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849–859 [DOI] [PubMed] [Google Scholar]
- 12.Charreau B, Tesson L, Soulillou JP, Pourcel C. and Anegon I. (1996). Transgenesis in rats: technical aspects and models. Transgenic Res 5:223–234 [DOI] [PubMed] [Google Scholar]
- 13.Hirabayashi M, Kato M, Kobayashi T, Sanbo M, Yagi T, Hochi S. and Nakauchi H. (2010). Establishment of rat embryonic stem cell lines that can participate in germline chimerae at high efficiency. Mol Reprod Dev 77:94. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi K, Abeydeera LR, Okuda K. and Niwa K. (1995). Effects of osmolarity and amino acids in a chemically defined medium on development of rat one-cell embryos. J Reprod Fertil 103:27–32 [DOI] [PubMed] [Google Scholar]
- 15.Hirabayashi M, Kato M. and Hochi S. (2008). Factors affecting full-term development of rat oocytes microinjected with fresh or cryopreserved round spermatids. Exp Anim 57:401–405 [DOI] [PubMed] [Google Scholar]
- 16.Hirabayashi M, Kato M, Kitada K, Ohonami N, Hirao M. and Hochi S. (2009). Activation regimens for full-term development of rabbit oocytes injected with round spermatids. Mol Reprod Dev 76:573–579 [DOI] [PubMed] [Google Scholar]
- 17.Ogawa S, Satoh K, Hamada M. and Hashimoto H. (1971). In-vitro culture of rabbit ova from the single cell to the blastocyst stage. Nature 233:422–424 [DOI] [PubMed] [Google Scholar]
- 18.Hirabayashi M, Tamura C, Sanbo M, Kato-Itoh M, Kobayashi T, Nakauchi H. and Hochi S. (2013). A retrospective analysis of germline competence in rat embryonic stem cell lines. Transgenic Res 22:411–416 [DOI] [PubMed] [Google Scholar]
- 19.Shinohara T, Kato M, Takehashi M, Lee J, Chuma S, Nakatsuji N, Kanatsu-Shinohara M. and Hirabayashi M. (2006). Rats produced by interspecies spermatogonial transplantation in mice and in vitro microinsemination. Proc Natl Acad Sci USA 103:13624–13628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanatsu-Shinohara M, Kato-Itoh M, Ikawa M, Takehashi M, Sanbo M, Morioka Y, Tanaka T, Morimoto H, Hirabayashi M. and Shinohara T. (2011). Homologous recombination in rat germline stem cells. Biol Reprod 85:208–217 [DOI] [PubMed] [Google Scholar]
- 21.Hirabayashi M. and Hochi S. (2011). Rat embryonic stem cells: establishment and their use for transgeniesis In: Methodological Advances in the Culture, Manipulation and Utilization of Embryonic Stem Cells for Basic and Practical Applications. Atwood CA, ed. InTech, Rijeka, pp. 397–410 [Google Scholar]
- 22.Allen ND,, Barton SC, Hilton K, Norris ML. and Surani MA. (1994). A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development 120:1473–1482 [DOI] [PubMed] [Google Scholar]
- 23.Robertson EJ,, Evans MJ. and Kaufman MH. (1983). X-chromosome instability in pluripotential stem cell lines derived from parthenogenetic embryos. J Embryol Exp Morphol 74:297–309 [PubMed] [Google Scholar]
- 24.Hikichi T, Wakayama S, Mizutani E, Takashima Y, Kishigami S, Van Thuan N, Ohta H, Bui HT, Nishikawa S. and Wakayama T. (2011). Differentiation potential of parthenogenetic embryonic stem cells is improved by nuclear transfer. Stem Cells 25:46–53 [DOI] [PubMed] [Google Scholar]
- 25.Galat V, Binas B, Iannaccone S, Postovit LM, Debeb BG. and Iannaccone P. (2009). Developmental potential of rat extraembryonic stem cells. Stem Cells Dev 18:1309–1318 [DOI] [PubMed] [Google Scholar]
- 26.Demers SP,, Desmarais JA, Vincent P. and Smith LC. (2011). Rat blastocyst-derived stem cells are precursors of embryonic and extraembryonic lineages. Biol Reprod 84:1128–1138 [DOI] [PubMed] [Google Scholar]
- 27.Hong J, He H. and Weiss ML. (2012). Derivation and characterization of embryonic stem cells lines derived from transgenic Fischer 344 and Dark Agouti rats. Stem Cells Dev 21:1571–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horii T, Kimura M, Morita S, Nagao Y. and Hatada I. (2008). Loss of genomic imprinting in mouse parthenogenetic embryonic stem cells. Stem Cells 26:79–88 [DOI] [PubMed] [Google Scholar]
- 29.Chen Z, Liu Z, Huang J, Amano T, Li C, Cao S, Wu C, Liu B, Zhou L, et al. (2009). Birth of parthenote mice directly from parthenogenetic embryonic stem cells. Stem Cells 27:2136–2145 [DOI] [PubMed] [Google Scholar]