Abstract
A human anti-HIV monoclonal antibody (mAb), 2909, selected on the basis of its potent neutralizing activity against HIV-1SF162, recognizes a complex epitope V2/V3 present on intact virions but not on soluble gp120. To confirm the quaternary nature of the epitope, 2909 binding was tested against the pseudovirus SF162 wild type (WT) expressing trimers and/or an SF162 mutant expressing monomeric envelope proteins. The construction of the SF162 mutant was made by an alanine substitution of nine hydrophobic residues in the N-terminal heptad repeat region of gp41 molecules that failed to form trimers on the virus surface. Monoclonal Ab 2909 bound only to SF162 WT virions and transfected cells as determined by mmunoprecipitation and flow cytometry, respectively, but showed no reactivity to the SF162 mutant expressing monomeric gp120. The data provide further evidence for the existence of a unique quaternary epitope V2/V3 on the surface of unliganded virus.
Keywords: HIV-1, neutralizing antibody, V2/V3 regions
1. Introduction
Human monoclonal antibody (mAb) 2909 is an example of a mAb selected on the basis of its neutralizing activity, having been selected using a single cycle infectivity assay against an SF162 pseudovirus [6]. A unique quality of mAb 2909 is its capacity to bind to intact virions but not to soluble viral proteins, suggesting that the epitope to which this mAb is directed is formed by a quaternary structure on the virus surface. Prior analyses showed that mAb 2909 recognizes a complex epitope comprised of elements in both the V2 and V3 loops of gp120 [6] and the specificity of this mAb is limited by variation in V2 rather than V3 [10]. In addition, mAb 2909 exhibits remarkably potent activity not achieved by previously described HIV neutralizing human mAbs including 447–52D (447), IgG1b12, 2G12 and 2F5 [6].
Asimilar type of the epitope like 2909 may be present more frequently; however, there is only one published study which describes antibodies (Abs) that appeared to be similar to mAb 2909. These are polyclonal Abs induced in rhesus macaques by simian-human immunodeficiency virus 89.6 (SHIV-89.6P) that recognize a discontinuous epitope formed by both V2 and V3 segments present on the virus envelope (Env) [4].
The fact that mAb 2909 targets both V2 and V3 and has such potent neutralizing activity [6] prompted further studies to elucidate the nature of its epitope. To do this, an SF162 pseudovirus construct was made which bears a mutant form of gp41 which fails to form trimers. The gp120 expressed by this mutant appears as a monomer on the surface of the pseudovirus. The data presented belowdocument the ability ofmAb2909 to bind to the wild type (WT) SF162 pseudovirus but not to the mutant (mut) form, revealing that the V2/V3 epitope of 2909 is formed only by the Env trimer.
2. Production of pseudoviruses with monomeric SF162 Env
The construction of the SF162 mut was based on previous studies of gp41 which included the substitution of nine hydrophobic residues in the N-terminal heptad repeat region that forms a leucine zipper-like structure [2]. An alanine substitution inhibited the formation of multiple hydrophobic contacts between gp41 molecules, resulting in the expression of monomers rather than trimers of gp41 on the virus surface [18]. These substitutions in the N-terminal heptad repeat of the gp41 protein of the SF162 Env sequence were engineered after aligning its sequence with that of HIV-1HXB2 (Fig. 1).
Fig. 1.
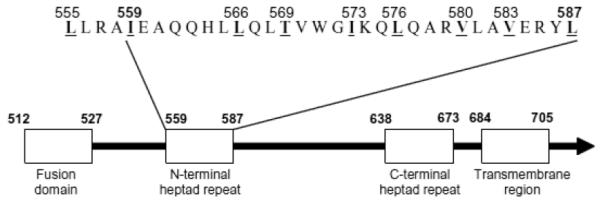
Schematic representation of the gp41 ectodomain of HIV-1HXB2. Four functional regions of gp41 are depicted as boxes. The sequence of the N-terminal heptad repeat of SF162 with four additional amino acids at the N-terminal side is also shown. Amino acid residues are numbered according to the HXB2 Env protein. The hydrophobic residues, which were replaced by alanine residues, are shown numbered, underlined, and in bold type.
The full-length SF162 env gene, digested from pSMSF162 [12], was ligated into pIRES-puro (Stratagene) using EcoRI and BamHI sites to construct a pIRESSF162 wild type (WT) vector. An 1187bp fragment of the SF162 env gene (positions 1048–2252, based on the HXB2 numbering engine) was PCR-amplified with the oligonucleotide primers 5'-CAAGCACAA TTTGGGAATAAAA-3' and 5'-CCATGCACTAATG GACTGGA-3' using Ex Taq DNA polymerase (Takara Bio Inc) and cloned into the pCR2.1-TOPO vector (Invitrogen). The resulting pCR2.1-SF162 vector was used as a template for the following site-directed mutagenesis: Nine amino acids in the amino-terminal heptad repeat region of gp41 (Fig. 1) were substituted with alanine residues using the QuikChange site-directed mutagenesis kit (Stratagene) with three sets of mutagenesis oligonucleotide primers. The resulting pCR2.1-SF162 mutant vector was digested with PpuMI, and the 1060bp fragment was substituted into the corresponding part of the pIRES-SF162 WT vector to construct a pIRES-SF162 mut vector. The pNL4-3.Luc.RE- and pSV-rev vectors were obtained through the NIH, AIDS Research and Reference Reagent Program (ARRRP), contributed by Dr. Nathaniel Landau [3,9], and by Dr. Marie-Louise Hammarskjöld and Dr. David Rekosh [19,21], respectively.
To produce Env pseudotyped viruses, pNL4-3.Luc. R-E-, pSV-rev and pIRES-SF162 env expression vectors were cotransfected into 293T cells (American Type Culture Collection) using the ProFection calcium phosphate transfection reagent (Promega).
3. SF162 mut pseudotyped virus carries only monomeric Env
To test if the SF162 WT and SF162 mut Env proteins were correctly assembled onto pseudotyped viruses, the viruses were analyzed by standard SDSpolyacrylamide gel electrophoresis (PAGE) followed by Western blotting. For standard SDS-PAGE, 15 μl of the virus stocks, equivalent to 200 ng of p24, were mixed with 5 μl of 4X Laemmli loading dye, boiled for 5 minutes, subjected to SDS-PAGE on 12.5% polyacrylamide gels and then transferred to a nitrocellulose membrane. The membranewas incubated with anti-C5 gp120 mAb 1331A [1] followed by goat anti-human IgG conjugated with alkaline phosphatase,and the virus proteins were detected using an enhanced chemiluminescence detection system (Pierce).
Both SF162 WT and SF162 mut viruses carried gp160 as well as gp120 (Fig. 2A) indicating that these Env proteins were appropriately processed, transported and assembled. There is a quantitative difference in the amount of Env proteins – the mutant appears to have less Env protein than the WT – but gp120 and gp160 bands are present in both (Fig. 2A).
Fig. 2.
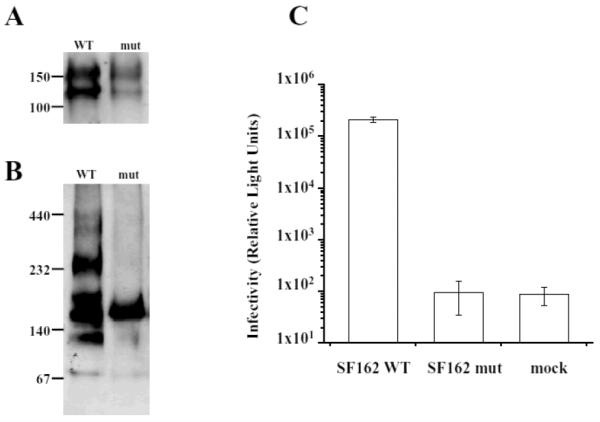
Analysis of pelleted viruses pseudotyped with SF162 WT and SF162 mut Env. (A) SF162 WT and SF162 mut pseudoviruses were analyzed on a standard SDS-PAGE followed by Western blotting using anti-C5 mAb 1331A. Both SF162 WT and SF162 mut had gp120 as well as gp160 proteins. (B) The pseudotyped viruses were subjected to non-reduced, semi-denatured PAGE followed by Western blotting using human anti-gp41 mAb 246-D. Virus pseudotyped with SF162 WT displayed multiple bands in high molecular weight regions of the gel indicative of multimer formation while the SF162 mut showed a single band of 160 kDa, indicative of the unique presence of the monomeric form of the Env protein. (C) The infectivity of pseudoviruses SF162 WT, SF162 mut, and mock virus without Env protein. Infectivity was assessed in a single-round infectivity assay using U87.CD4.CCR5 cells as targets. Infectivity is shown as Relative Light Units (luciferase activity) in the lysed target cells. Each column represents the mean of three experiments +/− S.D.
To analyze whether the SF162 WT and SF162 mut pseudoviruses display the multimeric forms of gp41 and gp160, a non-reduced, semi-denatured PAGE followed byWestern blotting was used (Fig. 2B). The 4X non-reducing loading dye (60 mM Tris-HCl pH 6.8, 0.2 mM EDTA, 10% Sucrose, 2% SDS, and 0.02% Bromphenol Blue) was substituted for the 4X Laemmli loading dye and the samples were not boiled. The samples were then loaded onto 4–13% gradient native polyacrylamide gels [14,20]. Following electrophoresis, the samples were blotted onto a nitrocellulose membrane and incubated with mAb 246-D, which recognizes a linear epitope of gp41 in the cluster I region [22], and treated as described above.
Under non-reduced, semi-denatured conditions, the virus pseudotyped with SF162 WT displayed bands in the high molecular weight regions of the gel, representing multimers, and a band at 160 kDa representing a monomer (Fig. 2B). Previous studies using mAbs against this region have shown that they preferentially recognize oligomeric forms of gp41 that are resolved as trimers or tetramers under semi-denaturing conditions used in Western blots [17,25]. These forms migrate with similar mobilities as gp120 and gp160, and may account for some of the extra bands detected in this region of the gel shown in Fig. 2B. The bands migrating above the 232 kDa marker may represent higher gp41 aggregates or oligomeric forms of gp160. In contrast, the SF162 mut virus showed only a single band representing the 160 kDa monomer (Fig. 2B). Thus, as anticipated, the virus pseudotyped with the SF162 mut Env protein carries only the monomeric form of the Env protein while SF162 WT carries both monomers and multimers.
4. Infectivity of the virus pseudotyped with SF162 mut expressing Env monomers
The infectivity of the pseudotyped SF162 mut virus was tested in a single-round infectivity assay and compared with the infectivity of SF162 WT pseudovirus (Fig. 2C). U87.CD4.CCR5 cells (NIH ARRRP) in 96-well plates were infected with Env pseudotyped viruses at 500 ng of p24 per well. The cells were lysed after 48 hrs, and luciferase activity was measured as previously described [6].
High relative light unit (RLU) levels were obtained from the lysed target U87.CD4.CCR5 cells exposed to pseudotypes with the SF162 WT Env (> 1 × 105 RLU), while cells inoculated with the SF162 mut gave RLU levels equivalent to the mock pseudovirus (~1× 102 RLU) (Fig. 2C). These results indicate that expression of monomeric Env renders the pseudovirions non-infectious.
The lack of infectivity of the mutant pseudovirions demonstrates that this mutation in gp41 blocks virus entry and/or fusion. This could be explained if the monomer does not undergo the same conformational change upon binding of CD4 as the trimer and/or if gp120/CD4 monomers do not form a fully functional coreceptor binding site.
5. Immunoprecipitation analysis of mAb 2909 binding to pseudovirions
For immunoprecipitation, SF162 WT or mutant SF162 intact pseudovirionswere incubated with human mAbs and precipitated with protein G Sepharose (Bio-Rad). After washing, the precipitate was resuspended in Laemmli buffer, boiled for five minutes and analyzed by SDS-PAGE. The samples were transferred onto a nitrocellulose membrane and incubated with sheep antigp120 C5 antibody (Cliniqa Corporation) followed by goat anti-sheep IgG antibody conjugated with alkaline phosphatase as previously described [6].
As shown in Fig. 3A, gp120 could be detected in SF162WTpseudoviruses immunoprecipitated by mAb 2909 but not in 2909 precipitates of the SF162 mut or the mock virus. Human anti-V3 mAb 447, which binds to an epitope present on both virions and soluble gp120 and on both monomers and trimers [7], was used as a positive control; this mAb immunoprecipitated both pseudotyped SF162 WT and SF162 mut. Human anti-parvovirus B19 mAb 1418 [5], which was used as a negative control, did not precipitate either of the viruses. These data demonstrate that mAb 2909 only recognizes the complex V2/V3 epitope present on the trimer of pseudotyped SF162WT; this epitope is absent when the proteins are expressed on the surface of the pseudovirus SF162 mut as monomers.
Fig. 3.
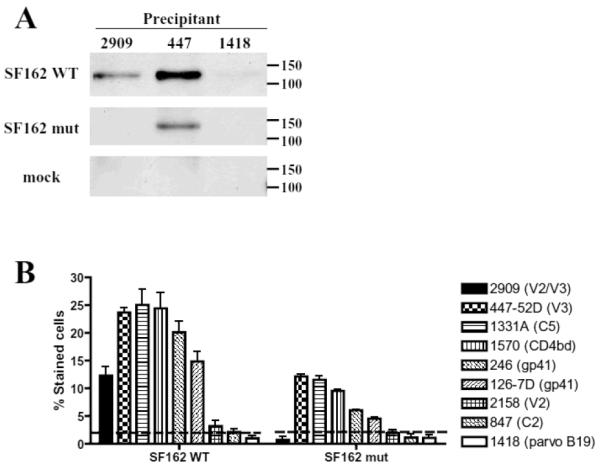
Binding of human mAbs to virions and transfected cells. (A) Monoclonal Ab 2909 immunoprecipitated SF162 WT pseudoviruses but not those expressing SF162 mut or mock virus which does not express Env proteins. Monoclonal Ab 447 (anti-V3) precipitated both viruses and the control anti-parvovirus mAb 1418 did not precipitate any of the pseudoviruses. (B) The binding of 2909 and control mAbs to 293T cells expressing membrane-asociated Env proteins of SF162 WT and SF162 mut on the cell surface was measured by flow cytometry. The control mAbs were specific for V3 (447), C5 (1331A), CD4bd (1570), gp41/cluster I and II (246 and 126-7D, respectively), V2 (2158), C2 (847), and parvovirus B19 (1418; negative control).
Interesting, SF162WT Env proteins were precipitated by both mAbs, however, mAb 447 precipitated more protein than 2909 (Fig. 3A). It indicates that 2909 and 447 epitopes are not equally expressed on the gp120 and possibly the V2/V3 complex epitope for 2909 is more rarely formed due to its requirement for quaternary structure than the 447 epitope that is present on any V3 regions. This observation is confirmed by another binding assay (Fig. 3B).
6. Binding of 2909 to virus transfected cells
The second binding assay measured the reactivity of mAb 2909 and control mAbs to 293T cells expressing membrane-associatedEnv proteins of either SF162WT or SF162 mut pseudoviruses. 293T cells were transfected with the pIRES-SF162 env expression vector and pSV-rev vector and after 48 hrs incubated with 2 μg/ml of human mAbs followed by staining with anti-human IgG conjugated with phycoerythrin (Caltag Laboratories). The cells were then fixed with 2% paraformaldehyde and analyzed by flow cytometry.
Monoclonal Ab 2909 reacted with cells expressing SF162WTEnv but not with those expressing the SF162 mut monomeric Env (Fig. 3B). In contrast, the majority of the control mAbs, anti-V3 mAb 447 [8], anti-C5 mAb 1331A [7], anti-CD4bd mAb1570 [11], antigp41 246 and 126-7D [22], displayed binding to both cells transfected with SF162 mut Env and with SF162 WT (Fig. 3B). The reactivity of control mAbs with the SF162 mut compared to SF162 WT transfected cells was lower displaying less efficient expression of monomeric Env on transfected cells. This may explain the binding of anti-V2 mAb 2158 [16], which bound very weakly to SF162 WT but not at all to SF162 mut transfected cells. Neither anti-C2 mAb 847 [15], nor the negative control mAb 1418 was reactive with either pseudovirus.
It is interesting that there is some discrepancy between binding (Fig. 3A, 3B) and neutralizing activity [6] of 2909 compared to anti-V3 mAb 447. Binding activity to intact pseudovirions SF162 WT (Fig. 3A) and to infectious virions SF162WT [6] as well as reactivity to SF162 WT transfected cells was much lower for 2909 in comparison to 447-52D, while in neutralization assay against SF162 mAb 2909 displayed much higher activity than 447 [6]. It suggests that the complex epitope V2/V3 may not be formed on each trimer on either infectious virions or pseudovirions of SF162 WT resulting in less 2909 molecules binding than 447 that may reacts with the V3 region on all trimers on the virus. But once the V2/V3 epitope is formed and bound by 2909 it may blocks the virus binding to target cells so efficiently that it needs 750 fold lower dose than 447 for 50% neutralization of SF162 [6].
7. Conclusion
The absence of 2909 binding to the gp120 monomers that are present on SF162 mut irions and transfected cells indicates that the 2909 epitope can only be found in the trimer. The trimer specificity of mAb 2909 suggests that the elements of the V2 and V3 regions which constitute the epitope may originate either (a) from one gp120 subunit (cis-epitope) or (b) from two adjacent gp120 molecules (trans-epitope). The models of the virus Env proteins on unliganded HIV-1 and SIV virions, which are based on cryo-electron tomographic studies, offer only limited information regarding the positions of the V1V2 loop and how it may interact with V3 [13,23,24]. One study suggests, however, that the V1/V2 and V3 regions on each monomer are near the apex of the trimer based on an actual fit of the gp120 coordinates to the density map [13] and may eventually make the 2909-like epitope available to antibodies on the unliganded virus.
Production and characterization of additional mAbs targeting the unliganded form of gp120 are needed before conclusions can be drawn about whether such Abs are always limited in their breadth or can target conserved elements on the envelope spike.
Acknowledgements
This study was supported in part by NIH grants HL59725 and AI36085,by the Immunology Core of the NYU Center for AIDS Research (NIH grant AI27742) and by research funds from Department of Veterans Affairs. We thank Dr. Jennifer Fuller for help in the preparation of this manuscript.
References
- [1].Bandres JC, Wang QF, O'Leary J, Baleaux F, Amara A, Hoxie JA, Zolla-Pazner S, Gorny MK. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. doi: 10.1128/jvi.72.3.2500-2504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- [3].Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- [4].Etemad-Moghadam B, Karlsson GB, Halloran M, Sun Y, Schenten D, Fernandes M, Letvin NL, Sodroski J. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J Virol. 1998;72:8437–8445. doi: 10.1128/jvi.72.10.8437-8445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gigler A, Dorsch S, Hemauer A, Williams C, Kim S, Young NS, Zolla-Pazner S, Wolf H, Gorny MK, Modrow S. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. JVirol. 1999;73:1974–1979. doi: 10.1128/jvi.73.3.1974-1979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gorny MK, VanCott TC, Williams C, Revesz K, Zolla-Pazner S. Effects of oligomerization on the epitopes of the Human Immunodeficiency Virus Type 1 envelope glycoproteins. Virology. 2000;267:220–228. doi: 10.1006/viro.1999.0095. [DOI] [PubMed] [Google Scholar]
- [8].Gorny MK, Xu J-Y, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- [9].He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Honnen WJ, Krachmarov C, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Type-Specific Epitopes Targeted by Monoclonal Antibodies with Exceptionally Potent Neutralizing Activities for Selected Strains of Human Immunodeficiency Virus Type 1 Map to a Common Region of the V2 Domain of gp120 and Differ Only at Single Positions from the Clade B Consensus Sequence. J Virol. 2007;81:1424–1432. doi: 10.1128/JVI.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jeffs SA, Gorny MK, Williams C, Revesz K, Volsky B, Burda S, Wang XH, Bandres J, Zolla-Pazner S, Holmes H. Characterization of human monoclonal antibodies selected with a hypervariable loop-deleted recombinant HIV-1(IIIB) gp120. Immunol Lett. 2001;79:209–213. doi: 10.1016/s0165-2478(01)00289-9. [DOI] [PubMed] [Google Scholar]
- [12].Koito A, Harrowe G, Levy JA, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nijtmans LG, Henderson NS, Holt IJ. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- [15].Nyambi PN, Mbah HA, Burda S, Williams C, Gorny MK, Nadas A, Zolla-Pazner S. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J Virol. 2000;74:7096–7107. doi: 10.1128/jvi.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pinter A, Honnen WJ, Tilley SA, Bona C, Zaghouani H, Gorny MK, Zolla-Pazner S. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Poumbourios P, Wilson KA, Center RJ, El Ahmar W, Kemp BE. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic alpha-helical/leucine zipper-like sequence. J Virol. 1997;71:2041–2049. doi: 10.1128/jvi.71.3.2041-2049.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rekosh D, Nygren A, Flodby P, Hammarskjöld ML, Wigzell H. Coexpression of human immunodeficiency virus envelope proteins and tat from a single simian virus 40 late replacement vector. Proc Natl Acad Sci U S A. 1988;85:334–338. doi: 10.1073/pnas.85.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- [21].Smith AJ, Cho MI, Hammarskjold ML, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gagpol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xu J-Y, Gorny MK, Palker T, Karwowska S, Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol. 1991;65:4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-Electron Tomographic Structure of an Immunodeficiency Virus Envelope Complex In Situ. PLoS Pathog. 2006;2:790–797. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhu P, Liu J, Bess J, Jr., Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and threedimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- [25].Zolla-Pazner S, Gorny MK, Pinter A, Honnen W. Reinterpretation of human immunodeficiency virus Western blot patterns. N Engl J Med. 1989;320:1280–1281. doi: 10.1056/NEJM198905113201914. [DOI] [PubMed] [Google Scholar]


