Abstract
The National Institute on Alcohol Abuse and Alcoholism (NIAAA) was founded 40 years ago to help elucidate the biological underpinnings of alcohol dependence, including the potential contribution of genetic factors. Twin, adoption, and family studies conclusively demonstrated that genetic factors account for 50 to 60 percent of the variance in risk for developing alcoholism. Case–control studies and linkage analyses have helped identify DNA variants that contribute to increased risk, and the NIAAA-sponsored Collaborative Studies on Genetics of Alcoholism (COGA) has the expressed goal of identifying contributing genes using state-of-the-art genetic technologies. These efforts have ascertained several genes that may contribute to an increased risk of alcoholism, including certain variants encoding alcohol-metabolizing enzymes and neurotransmitter receptors. Genome-wide association studies allowing the analysis of millions of genetic markers located throughout the genome will enable discovery of further candidate genes. In addition to these human studies, genetic animal models of alcohol’s effects and alcohol use have greatly advanced our understanding of the genetic basis of alcoholism, resulting in the identification of quantitative trait loci and allowing for targeted manipulation of candidate genes. Novel research approaches—for example, into epigenetic mechanisms of gene regulation—also are under way and undoubtedly will further clarify the genetic basis of alcoholism.
Keywords: Alcohol dependence, alcoholism, genetics and heredity, genetic theory of alcohol and other drug (AOD) use, genetic causes of AOD use, abuse and dependence (genetic AOD), genetic risk and protective factors, hereditary versus environmental factors, genetic mapping, Collaborative Studies on Genetics of Alcoholism, human studies, animal studies
Evidence from archeological artifacts indicates that fermented beverages existed as early as 10,000 B.C. The excessive consumption of alcohol, however, results in dangers to the health and well being of the drinker and those around him or her. Today, the World Health Organization estimates that alcohol causes 1.8 million deaths (3.2 percent of all deaths) worldwide and 58.3 million (4 percent of total) disability-adjusted life-years (DALYs)1 lost to disease (http://www.who.int/substance_abuse/facts/alcohol/en/index.html). In the United States, alcohol dependence (i.e., alcoholism) is a major health problem, affecting 4 to 5 percent of the population at any given time, with a lifetime prevalence of 12.5 percent (Hasin et al. 2007).
The National Institute on Alcohol Abuse and Alcoholism (NIAAA) was founded 40 years ago to further understanding of the biological underpinnings of alcohol dependence. Early genetic studies were focused on delineating whether environmental factors, genetic factors, or both contributed to the risk for alcohol dependence. Once it was apparent that genetics did indeed play a role in alcohol dependence, NIAAA began to fund studies seeking to identify relevant genes. Since then, studies in humans and animals have used complementary approaches to understand the genetics of alcohol use and dependence. This overview summarizes the evidence supporting a role for genetic factors in alcoholism and describes how new genetic findings could affect our understanding of the causes and factors contributing to this debilitating disease and could potentially guide the development of improved treatments.
Evidence of a Genetic Contribution to Alcohol Dependence
Several study designs, including twin, family, and adoption studies, are used to determine whether relatively common diseases, such as alcohol dependence, are caused at least in part by genetic factors and to estimate the magnitude of the overall genetic contribution. Twin studies compare the similarity in disease status (i.e., concordance2) between identical (i.e., monozygotic) and fraternal (i.e., dizygotic) twins. If risk for a disease (e.g., alcohol dependence) is determined at least in part by genetic factors, monozygotic twins, who have identical genetic material (i.e., genomes), would be expected to have a higher concordance rate for alcohol dependence than dizygotic twins, who on average share only half their genome. Studies by several groups have indeed shown higher concordance rates for alcohol dependence among monozygotic than among dizygotic twins (Agrawal and Lynskey 2008). Family studies, which evaluate the members of a family (both alcoholic and nonalcoholic members) for the presence of the disease, also have provided convincing evidence that the risk for alcohol dependence is determined partly by genetic influences (Gelernter and Kranzler 2009). Overall, family, adoption,3 and twin studies provide convergent evidence that hereditary factors play a role in alcohol dependence, with variations in genes estimated to account for 50 to 60 percent of the total variance in risk. These estimates suggest that although genetic factors are important, nongenetic factors also contribute significantly to the risk for alcohol dependence.
Strategies for Identifying Genes Contributing to Alcohol Dependence
Researchers have developed several strategies to identify genes that contribute to differences in the risk for alcohol dependence, including case–control studies and linkage analyses. These strategies depend on the premise that for a particular position in the DNA of these genes, more than one possible form exists. Each of these forms is termed an allele. The study methods used to identify genes that affect the risk for alcohol dependence assume that the presence of certain alleles increases the risk of alcoholism. These variants that affect risk can be located either directly within a gene or near a gene.
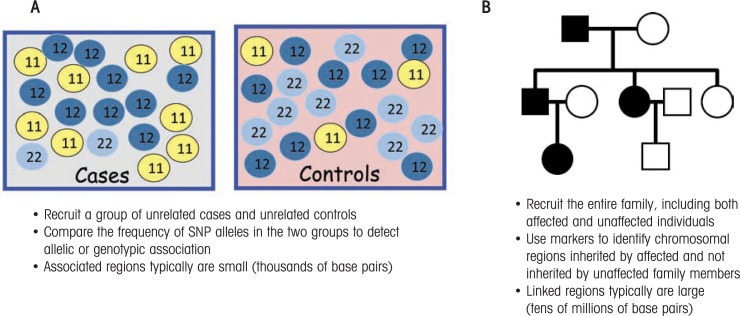
Case–control studies compare allele frequencies in a sample of alcoholic and control subjects. Because DNA is inherited from both parents, every person carries two copies of the DNA at a given position in the genome—one allele that was inherited from the father and one allele that was inherited from the mother. The genotype describes the variation at a particular position within the genome and is defined by the allele inherited from the father and the allele inherited from the mother. If a given allele contributed to the risk for alcohol dependence, one would expect the allele and/or genotype frequencies to differ between the case and the control subjects (see figure 1A).
Figure 1.
Approaches to identifying genes contributing to the risk of alcoholism. A) Case–control association study design. Each circle represents a person who is either an alcoholic (case subject) or not an alcoholic (control subject). The study assesses the role of a single-nucleotide polymorphism (SNP)* that exists in two different variants (i.e., alleles)—allele 1 and allele 2. Because each person inherits two copies of the SNP from their parents, the numbers in the circles represent the three possible genotypes (11, 12, and 22). Many more case than control subjects carry at least one copy of allele 1 (i.e., have the 11 and 12 genotypes), suggesting that people with allele 1 may be more likely to develop alcoholism. B) Linkage study design. A three-generation family tree (pedigree) is shown. Squares represent male subjects and circles represent female subjects. Shaded symbols represent alcoholic individuals and unshaded symbols represent nonalcoholic individuals. In this pedigree, there are alcoholic individuals in each generation, and both men and women are affected.
NOTE: *An SNP is a DNA sequence variation occurring when a single nucleotide in a DNA marker (or other genetic sequence) differs between members of a species or between the chromosome pairs in an individual.
Initially, case–control studies often were performed using small numbers of alcoholic and control subjects and examined the role of a single gene, frequently testing only for a single variation. This approach has limited power, and many results could not be replicated. The most robust result from these early studies was the demonstration that the genes encoding two alcohol-metabolizing enzymes—alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH)—played an important role in determining alcoholism risk (this will be discussed in more detail in the next section).
With the advances of molecular genetics technologies, it then became possible to scan the genome using a type of genetic variation called microsatellites. In this approach, called linkage analysis, the pattern of transmission of a disease (e.g., alcoholism) in families with multiple affected members is compared with the pattern of transmission of certain microsatellites (see figure 1B). The underlying hypothesis is that alcoholics within a family share many risk alleles; therefore, genes containing alleles that increase the risk for alcoholism reside within chromosomal regions that are inherited by most or all alcoholic family members. Unfortunately, however, the chromosomal regions that were identified using this approach often contained hundreds or even thousands of genes, making it very challenging to determine which specific gene(s) contribute to the risk for alcoholism.
The Collaborative Studies on Genetics of Alcoholism Study
Another major advancement in the search for genes contributing to the risk for alcoholism was the initiation in 1989 of the NIAAA-funded Collaborative Studies on Genetics of Alcoholism (COGA), a family study with the expressed goal of identifying contributing genes using newly available genetic technologies (Begleiter et al. 1995; Bierut et al. 2002; Edenberg 2002). The study was groundbreaking in several ways, including its size, emphasis on families, and extensive characterization of subjects. In the process, COGA researchers developed a novel assessment instrument, the Semi-Structured Assessment of the Genetics of Alcoholism (SSAGA), which since has been translated into nine languages and is used by over 237 investigators worldwide in studies of alcohol use and dependence.
Families were obtained by recruiting alcohol-dependent probands (i.e., index cases) who were in treatment and who gave permission to contact their family members. This approach generated a dataset of 1,857 families consisting of 16,062 individuals as of March 2010. Moreover, the researchers identified a genetically informative subset comprising 262 families with at least three first-degree relatives who met lifetime criteria for both Diagnostic and Statistical Manual of Mental Disorders, Third Edition, Revised (DSM–III–R) (American Psychiatric Association 1987) alcohol dependence and Feighner definite alcoholism;4 this subset became the focus of genetic analyses. The extensive characterization of subjects also allowed analysis of the role of hereditary characteristics (i.e., endophenotypes) that often are associated with alcoholism but are not direct symptoms of alcoholism, such as certain electrophysiological traits, drug dependence, other related psychiatric conditions, and personality measures (Edenberg 2002).
Genetic analyses in this subsample of the COGA dataset have implicated several different chromosomal regions as possibly containing one or more genes contributing to alcohol dependence; to related clinical characteristics (i.e., phenotypes) such as smoking, depression, suicidal behavior, conduct disorder, and the largest number of drinks within a 24-hour period; and to neurobiological endophenotypes such as event-related potentials and brain oscillations in electrophysiological activity (Edenberg 2002; Edenberg and Foroud 2006). Despite much progress, however, identification of the specific genes contributing to these phenotypes remains a challenging task because they lie within broad linkage regions that often encompassed 10 to 30 million base pairs.
In addition to COGA, NIAAA has supported several other large family studies designed to identify genes contributing to the risk for alcohol dependence. These include a large study in Ireland that is recruiting siblings (Kendler et al. 1996; Prescott et al. 2005), a family study of both alcohol dependence and alcohol-related endophenotypes (including electro-physiological measures, similar to COGA) (Hill 1998), and a study of Mission Indian families (Ehlers et al. 2004). Twin studies also have remained a focus of several NIAAA-funded research projects (Jacob et al. 2001; Madden et al. 2000). Moreover, a study of offspring of alcoholic fathers has expanded into a longitudinal, multigenerational genetic study that is focused on better understanding the factors contributing to the initiation of alcohol use as well as the long-term risk for alcohol dependence (Schuckit 1991). Finally, studies also have examined African-American alcohol-dependent families ascertained on the basis of cocaine or opioid dependence (Gelernter and Kranzler 2009). Together, these approaches, although by no means completed, already have resulted in the identification of some genes that impact the risk for alcohol dependence. Some of these genes and the proteins they encode are discussed in the next section.
Genes Contributing to Alcohol Dependence
Genes Encoding Alcohol-Metabolizing Enzymes
Classic studies, which have been replicated in many populations, have demonstrated that certain coding variations in two genes affecting alcohol metabolism have a strong protective effect—that is, they both substantially lower the risk for alcoholism. These variants affect a gene called ADH1B, which encodes a variant of ADH, and a gene called ALDH2, which encodes a variant of ALDH (Edenberg 2000, 2007; Hurley et al. 2002) (figure 2). The protective variant in the ALDH2 gene, known as ALDH2*2, involves a point mutation that results in the exchange of the amino acid glutamate at position 487 of the ALDH protein for the amino acid lysine. This mutation acts in a nearly dominant manner to render the enzyme almost inactive: even people who inherit only one copy of ALDH2*2 and one “normal” copy of the gene (i.e., people who are heterozygous for this mutation) produce an ALDH enzyme with extremely low enzyme activity (Crabb et al. 1989). As a result, these individuals exhibit highly elevated levels of acetaldehyde, which produces aversive reactions, including flushing, elevated heart rate (i.e., tachycardia), and nausea after consuming even a small amount of alcohol (Eng et al. 2007). Similarly, coding variations in the ADH1B gene (called ADH1B*2 and ADH1B*3) that encode highly active enzymes which increase the rate at which acetaldehyde is produced also are strongly protective and reduce the risk for alcohol dependence (Edenberg 2007; Thomasson et al. 1991).
Figure 2.

The main steps of alcohol metabolism. Alcohol first is metabolized to acetaldehyde by the enzyme alcohol dehydrogenase (ADH), which is encoded by several genes, each of which may exist in several variants (i.e., alleles). Certain alleles encode ADH molecules that result in the metabolism of alcohol (denoted by the red arrow above ADH). As a result, buildup of acetaldehyde occurs (denoted by the upward-pointing arrow), leading to such aversive effects as nausea, flushing, and accelerated heart beat (i.e., tachycardia). The acetaldehyde then is metabolized to acetate by the enzyme aldehyde dehydrogenase (ALDH), which also is encoded by several genes existing in different alleles. Certain alleles in the ALDH2 gene, which encodes a key ALDH enzyme, can result in very low activity of the enzyme (denoted by the black arrow with a red line through it), again causing acetaldehyde accumulation and the resulting aversive effects.
These gene variations have been selected for in different populations. For example, the ALDH2*2 variant is common only among people from east Asia, the ADH1B*2 variant is common among people from east Asia and the Middle East, and the ADH1B*3 variant is common in people from Africa (Edenberg 2007; Eng et al. 2007; Li et al. 2007, 2009). All of these variations have strikingly strong effects on risk; thus, in Asian populations, ALDH2*2 and ADH1B*2 each can lower risk by two- to sevenfold. No other known gene variations have such a strong effect on risk for alcoholism.
The influence of ADH variations on risk was further investigated through linkage studies performed in non-Asian families. These analyses detected linkage of alcoholism to a broad region on chromosome 4q that included the ADH gene cluster (Long et al. 1998; Prescott et al. 2006; Reich 1996; Reich et al. 1998; Williams et al. 1999). Given the strong prior evidence for the role of the ADH genes in alcoholism susceptibility, the COGA investigators initially focused on the 262 families from the study with a very strong history of alcoholism. In these families, they determined the genotype for 110 DNA markers known as single-nucleotide polymorphisms (SNPs), which were distributed throughout the ADH gene cluster. These analyses detected significant evidence of association of alcoholism with 12 SNPs located in and around the ADH4 gene (Edenberg et al. 2006) and modest evidence of association with noncoding SNPs5 in ADH1A and ADH1B. Moreover, the analyses provided evidence that the ADH1B*3 allele was protective among African-American families (Edenberg et al. 2006). The association of several noncoding ADH polymorphisms with alcohol dependence has been replicated in other studies (Edenberg 2007; Macgregor et al. 2009).
Genes Encoding γ-Aminobutyric Acid Receptors
The brain-signaling molecule (i.e., neurotransmitter) γ-aminobutyric acid (GABA), by interacting with a molecule called the GABA-A receptor, mediates several effects of alcohol, including alcohol’s sedative and anxiety-reducing (i.e., anxiolytic) effects, motor incoordination, tolerance, and dependence (Kumar et al. 2009). Several genes that encode subunits of the GABA-A receptor are associated with an increased risk for alcoholism. For example, significant evidence suggests that a gene called GABRA2, which with other GABA-A receptor genes is located in a cluster on chromosome 4, is associated with alcoholism (Edenberg et al. 2004). This finding has been replicated in many (but not all) case–control studies in Europeans, Australians, and Plains Indians (Edenberg and Foroud 2006; Gelernter and Kranzler 2009). In several samples, the association with GABRA2 was greatest among those alcohol-dependent people who also were dependent on nicotine (Philibert et al. 2009) or illicit drugs (Agrawal et al. 2006; Philibert et al. 2009); the latter subgroup is characterized by greater severity of alcohol problems in general (Dick et al. 2007). In addition, another gene within the chromosome 4 GABA-A cluster, GABRG1, also may influence the risk for alcoholism (Covault et al. 2008; Enoch et al. 2009).
Finally, GABA-A genes on other chromosomes, including GABRG3 on chromosome 15 (Dick et al. 2004) and GABRA1 on chromosome 5 (Dick et al. 2006), also have been associated with alcoholism. However, these associations have not yet been replicated in other samples and therefore must be considered tentative.
Genes Encoding Acetylcholine Receptors
Another neurotransmitter system involved in the actions of alcohol is acetylcholine, which can interact with different types of receptors, including muscarinic and nicotinic receptors. As with the GABA-A receptor, the subunits for each of these receptors are encoded by different genes that have several different alleles (i.e. code for different forms of the receptor subunit), and certain alleles have been associated with an increased risk for alcoholism. For example, the gene that encodes the muscarinic acetylcholine receptor subtype 2, called CHRM2, appears to be an important risk factor for alcohol dependence. The receptor encoded by this gene is a G-protein–coupled receptor6 involved in many functions. In the COGA study, SNPs in CHRM2 were associated with alcohol dependence, a finding that was replicated in an independent study (Edenberg and Foroud 2006).
Extensive research also has examined the neuronal nicotinic acetylcholine receptors (nAChRs), which are affected by both nicotine and alcohol. DNA variation in the genes that encode the subunits of these receptors may play a role in the susceptibility to alcohol dependence and nicotine addiction. Similar to the GABA-A receptors, the genes encoding these receptors are found in clusters on several chromosomes. Studies have reported an association of SNPs in CHRNA5–CHRNA3 (Wang et al. 2009) and CHRNA6–CHRNB3 (Hoft et al. 2009) gene clusters with alcohol dependence or alcohol consumption.
Genome-wide Association Studies
In the past few years, it has become possible to genotype up to a million SNPs throughout the genome in a single experiment—an approach called genome-wide association studies (GWASs). This technique, which is based on the assumption that common genetic variation contributes to disease risk, allows a comprehensive test of association across the genome, rather than testing only one gene at a time. It has been used for many different diseases, with varying success. In particular, the relatively low statistical power of GWASs is a significant hurdle. Thus, the analyses require very large samples because most variations only have small effects; moreover, the multiple testing involved in a GWAS reduces the statistical power to detect associations.
Several studies recently have reported GWAS results from case–control studies comparing alcohol-dependent case subjects to nondependent control subjects. The first published study, conducted in Germany, compared 487 men in inpatient treatment for alcohol dependence to 1,358 control subjects (Treutlein et al. 2009). The study identified several SNPs in a region on chromosome 2 that previously had been linked to alcohol dependence, as well as SNPs in a gene called CDH13 that is located on chromosome 16 and the ADH gene ADH1C on chromosome 4.
Recently, COGA reported results of a GWAS that included 847 alcohol-dependent case and 552 control subjects (Edenberg et al. 2010). The combined evidence from this case–control study, a follow-up in families, and gene expression data provided strongest support for the association with alcohol dependence of a cluster of genes on chromosome 11.7 However, the associations detected in the COGA GWAS did not reach the threshold for statistical significance for this type of analysis, and therefore additional studies must be conducted to further define the associated genes. Several SNPs nominated as candidates in the earlier German GWAS also were replicated in the COGA sample, including SNPs in or near the genes CPE, DNASE2B, SLC10A2, ARL6IP5, ID4, GATA4, SYNE1, and ADCY3.
Another recent report (Bierut et al. 2010) described a GWAS using an overlapping set of COGA subjects as well as additional subjects recruited as part of other addiction research projects. This sample included both African-American and European-American subjects, and the primary analysis sought to identify association with alcohol dependence using a case–control design. Although none of the detected associations met genome-wide criteria for statistical significance, there was some evidence to support the previously reported association in GABRA2 as well as in a gene called ERAP1, which encodes the enzyme endoplasmic reticulum aminopeptidase 1 (Bierut et al. 2010). Finally, a GWAS in a sample of twins and their families recruited in Australia is currently being analyzed.
Genetic Animal Models of Alcohol’s Effects and Alcohol Use
Since the earliest days of alcohol research, the use of animal models has featured strongly in attempts to understand genetic contributions to the mechanisms through which alcohol exerts its biological effects and to individual differences in risk for alcohol dependence. The main advantage of animal models for these genetic analyses is that they allow researchers to more tightly control environmental influences, thereby making it easier to identify genetic risk factors.
In 1959, inbred mouse strains first were shown to differ in their tendency to drink alcohol (McClearn and Rodgers 1959), and studies with inbred strains continue to this day. Each inbred strain possesses a random collection of genes (i.e., genotype), but all the animals within a strain are genetically identical. This reduction in the genetic variation among the animals studied could increase the power to identify genes contributing to alcohol-related traits.
Another commonly used type of animal model involves selectively bred lines. Starting in the late 1940s, researchers in Chile bred rats that preferred to drink alcohol-containing solutions as well as rats that avoided alcohol (Mardones and Segovia-Riquelme 1983). Such selective breeding has been repeated numerous times with rats and mice, resulting in pairs of animal lines that differ with respect to a particular alcohol-related trait. A list of currently available rodent selected lines is shown in table 1. Studies with the high- and low-drinking selected lines in particular have been a major focus of NIAAA-sponsored research efforts (for a review, see Crabbe et al. 2010; other reviews were published in a special issue of Addiction Biology, Vol. 11[3–4], 2006). Animals have been selected for many alcohol-related traits, including preference for alcohol, tolerance or sensitivity to alcohol’s effects, and withdrawal severity. New selection projects also are emerging; for example, researchers are breeding mice that exhibit binge-like drinking (Crabbe et al. 2009).
Table.
Selectively Bred Rat and Mouse Lines With Differential Responses to Alcohol*
| Lines | Abbreviation | Selected Trait | |
|---|---|---|---|
| Rats | University of Chile Alcohol Drinker and nondrinker | UChB/UChA | High/low drinking, 10 percent ethanol vs. water |
| ALKO Alcohol and Nonalcohol | AA/ANA | High/low drinking, 10 percent ethanol vs. water | |
| Alcohol Preferring and Nonpreferring | P/NP | High/low drinking, 10 percent ethanol vs. water | |
| Sardinian Alcohol Preferring and Nonpreferring | sP/sNP | High/low drinking,10 percent ethanol vs. water | |
| Marchigian Sardinian Alcohol Preferring (derived from 13th generation sP rats) | msP | High drinking, 10 percent ethanol vs. water | |
| High/Low Alcohol Drinking | HAD-1/LAD-1 HAD-2/LAD-2 |
High/low drinking,10 percent ethanol vs. water | |
| High/Low Addiction Research Foundation | HARF/LARF | High/low drinking, 12 percent ethanol during a 20-minute period of limited access | |
| High Alcohol Sensitive and Low Alcohol Sensitive | HAS-1/LAS-1 HAS-2/LAS-2 |
Long/short duration of loss-of-righting reflex after high-dose ethanol injection | |
| Mice | High/Low Alcohol Preference | HAP-1/LAP-1 HAP-2/LAP-2 HAP-3/LAP-3 |
High/low drinking,10 percent ethanol vs. water |
| High Drinking in the Dark | HDID-1 HDID-2 |
High blood alcohol levels after drinking 20 percent ethanol in a single-bottle, limited access exposure | |
| Long Sleep and Short Sleep | LS/SS | Long/short duration of loss-of-righting reflex after high-dose ethanol injection | |
| FAST and SLOW | FAST-1/SLOW-1 FAST-2/SLOW-2 |
Sensitivity/resistance to low-dose ethanol stimulation of activity | |
| Withdrawal Seizure Prone and Withdrawal Seizure Resistant | WSP-1/WSR-1 WSP-2/WSR-2 |
Severe/mild handling-induced convulsions after exposure to ethanol vapor inhalation for 72 hours |
SOURCE:
In press.
Studies with these selected lines have contributed a great deal to understanding the neurobiological bases for alcohol’s myriad effects. For example, researchers consistently have observed low levels of the neurotransmitter serotonin in certain brain areas (i.e., the limbic system) and other indications of dysregulation of the serotonin system in animal lines bred for high alcohol drinking (Crabbe 2008). Other studies with selected lines have shown dysregulation of the GABA and glutamate systems in animals bred to exhibit severe withdrawal. (Finn et al. 2004).
Contributions of Genetic Animal Model Research
Enhanced Understanding of Alcohol’s Pharmacology and Neurobiology
Animal research has been invaluable for discovering how alcohol exerts its biological effects. For example, numerous studies have shown an important role for GABA neurotransmission in mediating alcohol’s acute and chronic effects (Finn et al. 2004; Lobo and Harris 2008; Kumar et al. 2009). Additional animal studies have demonstrated that alcohol’s pharmacology involves nearly all major neurotransmitter targets, including the glutamate/NMDA,8 serotonin, dopamine, norepinephrine, and cannabinoid receptor systems (Kelai et al. 2006; Smith et al. 2008; Vengeliene et al. 2005). By acting on all these signaling systems, alcohol ultimately exerts its effects through modulation of intracellular signaling cascades (Newton and Messing 2006). Without animal models, researchers could not have gained an understanding of these highly complex mechanisms underlying alcohol’s diverse effects, and genetic animal models in particular have aided in understanding individual differences in sensitivity to these effects.
Gene Identification and Quantitative Trait Locus Mapping
Animal models also have been exploited for many years in attempts to identify specific gene variations associated with increased sensitivity to alcohol’s effects. These gene-mapping studies, which commenced in the early 1990s, have used methods similar to those described above for human studies (e.g., linkage analyses). They primarily have sought to identify quantitative trait loci (QTLs)—DNA regions that are associated with characteristics (i.e., quantitative traits) which vary in the degree to which they are present (e.g., sensitivity to alcohol or height). Such traits typically are determined by multiple genes and each QTL may contain one or more of these genes. Compared with humans, studies with rats and mice have the distinct advantage that researchers can use individuals with defined genotypes and control patterns of mating, making it much easier to localize the chromosome region of interest (i.e., the “locus” of the QTL). The most recent systematic review (Crabbe et al. 1999) of the alcohol-related QTL data for the various alcohol-related traits being mapped, which now is out of date, listed the likely locations of several genes affecting alcohol withdrawal severity, preference for drinking, and sensitivity to alcohol’s effects. Researchers at the Oregon Health & Science University now maintain a much more recent update of mouse alcohol QTL locations for these and other alcohol-related traits, which can be accessed via the Portland Alcohol Research Center Web site (http://www.ohsu.edu/parc/).
The greatest success story for alcohol-related QTL mapping in rodents has been the discovery of a quantitative trait gene (QTG)9 that affects acute withdrawal severity from both alcohol and pentobarbital in mice. Originally, investigators mapped several QTLs contributing to this trait to locations on various mouse chromosomes (Buck et al. 1997). Subsequent studies with a variety of specifically created genetic animal models gradually narrowed down the size of the DNA region (i.e., reduced the confidence interval) around one of these QTLs until only a few genes remained within the confidence interval. Functional studies then demonstrated that the most likely gene contributing to the trait was Mpdz, which encodes a protein containing multiple structural components known as PDZ-domains (Shirley et al. 2004). Studies of this gene’s pattern of expression in the brain and of the functions of the MPDZ protein continue, as do studies to identify the receptor molecules with which MPDZ interacts (e.g., the serotonin 2C receptor) (Chen et al. 2008a; Reilly et al. 2008).
Additional mapping studies aim to narrow other QTLs for alcohol responses, both in animals (Bennett et al. 2007, 2008; Hitzemann et al. 2009) and in humans. A recent comparison of data from mouse and human QTL mapping identified a promising region of human chromosome 1 that was linked to alcohol dependence and which overlapped with an area of mouse chromosome 1 that has been linked to an alcohol withdrawal QTL (Ehlers et al. 2010).10 However, as described by Ehlers and colleagues (2010) a detailed comparison of rodent and human maps to see whether the QTLs from rodent studies identify the same chromosomal regions as the linkage studies in humans is very difficult. Nevertheless, some promising results of cross-species consistency exist, which likely will increase in number as the details of both rodent and human genetic maps improve.
Classical QTL analysis has associated individual differences in gene sequence (or in other genetic markers, such as microsatellites) with differences in the phenotype being mapped. A recent development in rodent QTL mapping has been development of expression QTL (eQTL) mapping. eQTLs are DNA regions that differ not in their gene sequence, but in the level to which the gene becomes active (i.e., is expressed) in individuals differing with respect to certain alcohol-related traits. This information can be gathered from microarray experiments that measure the levels of individual mRNAs. These additional eQTLs greatly expand the pool of potentially informative genes.
The eQTL approach has been used to compare gene expression in brain tissue from several rodent lines and strains genetically predisposed to drink alcohol with control tissue from low-drinking animals. The chromosomal location of differentially expressed genes then was compared with QTL data based on genetic sequence variations (i.e., polymorphisms). This combination of information suggested several candidate genes that may influence alcohol drinking (Mulligan et al. 2006; Weng et al. 2009).
An additional refinement to the gene-finding efforts has been the study of networks of proteins or the genes that encode them. The reasoning is that even if different studies (or studies in different species) do not identify the same specific gene as being involved in a trait, they might identify a network of genes that underlies the genetic “signal” across studies and datasets and which encodes proteins that have similar functions or are involved in similar pathways (e.g., McBride et al. 2009).
Candidate Gene Studies and Gene Targeting
Another important development enhancing the possibilities of genetic animal models of alcoholism was the development of transgenic animals in the late 1980s. These are animals that have been genetically modified so that the expression of a single candidate gene has been selectively inactivated or augmented compared with the parent strain. This approach allows researchers to study the influence of individual genes on risk for alcoholism (or many other diseases or behaviors). By now, more than 100 candidate genes have been studied for their contribution to alcohol’s effects, usually by comparing mice in which a single gene has been inactivated (i.e., knockout mice) with control mice in which the gene still is functional. As reviewed by Crabbe and colleagues (2006), most of the genes thus studied were found to influence some aspect of alcohol sensitivity. For example, of 84 different transgenic animals tested for effects on alcohol self-administration, one-quarter exhibited increased drinking, one-third exhibited decreased drinking, and 40 percent did not differ from control animals (Crabbe et al. 2006). This finding clearly demonstrates the multiplicity of genetic influences on alcohol responses. As gene-targeting technologies allow more specific experimental regulation of genes than simple deactivation or over-expression, these approaches will continue to provide important data. For example, researchers now can manipulate genes so that they are expressed only in certain cell types or during particular developmental periods.
Candidate gene studies also have been valuable in looking for consistency across species in the impact of certain genes or gene variants. Invertebrate models (e.g., the fruit fly Drosophila or the worm Caenorhabditis elegans) offer much more powerful tools to manipulate the genome than do rodents (Lovinger and Crabbe 2005). However, to be able to use such models, researchers first need to demonstrate that corresponding genes exist in these organisms and that they actually have similar functions. One example of such convergence of evidence is the finding that a small signaling molecule called neuropeptide Y (NPY) and its receptors play a role in alcohol intoxication in mice, rats, and Drosophila (Chen et al. 2008b; Gilpin et al. 2004; Thiele et al. 2002). A meta-analysis of human association data, in contrast, found no clear evidence that polymorphisms in the gene encoding a precursor of NPY are associated with alcohol dependence (Zhu et al. 2003). However, some genes encoding NPY receptors may play a role in alcohol dependence and withdrawal (Wetherill et al. 2008). Finally, certain signaling proteins (e.g., epidermal growth factor receptor [EGFR], protein kinase C, protein kinase A, and cyclic AMP [cAMP]) have been implicated in alcohol’s effects across multiple species, including humans, rats, mice, Drosophila, and zebrafish (Corl et al. 2009; Newton and Messing 2006, 2007; Peng et al. 2009).
Studies of Gene–Environment Interaction
Studies clearly have shown that both genetic and environmental factors contribute to the risk for alcohol dependence, and it is likely that the interplay between these factors is critical for determining the risk for alcohol abuse and dependence. Advances in genetic technologies already have allowed researchers to explore the genome in ever greater detail, and with the advent of whole-genome sequencing, complete delineation of genetic variation soon will be available. In contrast, our understanding of the critical environmental factors influencing alcohol use disorders remains inadequate and is an area of active research. One of the challenges is how to define the environment, which may include family, peer, and societal influences; other exposures; personality or psychiatric factors (which also have genetic components); and many more, most of which change over time. Furthermore, the influence of these factors on the risk of alcohol use disorders varies within the lifespan (Sher et al. 2010; van der Zwaluw and Engels 2009).
Animal models offer significant advantages for studies attempting to tease apart genetic and environmental influences on an individual’s risk for alcoholism. Given their methodological power, it is surprising how little research into this area has been done using genetic animal models. One trait that has been found to be genetically determined is alcohol preference of inbred mouse strains. Thus, specific mouse strains have displayed their tendencies to drink more or less alcohol by choice repeatedly across 50 years of studies. In fact, alcohol preference in these animals is even more replicable across studies (and therefore, across environments) than brain weight (Wahlsten et al. 2006), suggesting that it is strongly influenced by genetic effects. Not all alcohol traits are so stable, however, and the combined effects of genetic and environmental manipulations could be exploited more fully using genetic animal models.
A recent review has discussed several important features of gene–environment interaction research (Sher et al. 2010). For example, the social environment plays such a crucial role in shaping drinking behaviors in humans, but it is difficult to identify corresponding rat and mouse behaviors and environmental factors. One example of a study analyzing gene– environment interactions in animals (Hansson et al. 2006) compares the influence of environmental stress in a rat line selectively bred for high alcohol preference (i.e., the Marchigian-Sardinian preferring rats) with their nonselected control group. The investigators found that the genetically “enriched” rats were more sensitive than the control animals to the effects of environmental stress on reinstatement of previously extinguished alcohol drinking (i.e., the alcohol-preferring rats resumed alcohol drinking more easily after being exposed to a stressor). Moreover, the differences resulted, at least in part, from variations between high-drinking and low-drinking animals in a gene encoding a receptor for corticotropin-releasing hormone (CRH) (which is involved in the body’s stress response) and in the expression of that gene (Hansson et al. 2006). Thus, this study demonstrated an interaction between a specific genotype and an environmental factor (i.e., stress).
Analysis of human gene–environment interactions also are complicated by the fact that these interactions are important from adolescent exposure to alcohol and then throughout life. Accordingly, from a developmental perspective, the critical environmental influences are likely to change over time (e.g., the relative influence of family versus peer factors). Studies that follow genetically specified animals prospectively while extracting biological information at different times along the way are a promising area for future research that has not been sufficiently exploited thus far.
Future Directions
Research into the genetics of alcoholism, both in humans and in animal models, has made great strides over the past four decades, and even more approaches are beginning to be evaluated. For example, there is growing interest in studying epigenetic factors—that is, factors which alter certain phenotypes by modifying regulation of gene expression, without, however, changing the gene’s DNA sequence. One such factor that can impact gene expression is methylation of the DNA. Other epigenetic changes alter the packaging of DNA into chromatin. For example, two enzyme families called histone acetyltransferases and deacetylases can be used to alter chromatin structure experimentally, and studies found that when such changes accompany chronic drug administration, they can modify cocaine-related behaviors in rats (Renthal and Nestler 2009). Although similar research on alcohol-related traits still is in its infancy, some studies have found that alcoholic patients exhibited greater levels of DNA methylation of two different genes than nonalcoholics and, consequently, greater reduction in the expression of those genes (Bleich et al. 2006; Bonsch et al. 2005).
MicroRNAs—short RNA molecules naturally encoded by the genome that can bind to certain mRNA molecules, thereby repressing the further processing of these mRNAs—also might be involved in regulating alcohol’s effects (Miranda et al. 2010). These microRNAs also offer a new experimental method for silencing the expression of specifically targeted genes. The expression of microRNAs is sensitive to epigenetic modulation, and turning microRNAs on or off has become feasible in rodent models. Modification of microRNAs may offer a new pathway for identifying critical genes that can then serve as target for new therapeutic drugs for alcoholism treatment.
In summary, the genetics field has undergone a technological revolution, particularly in the past decade, allowing researchers to process large numbers of samples for their genetic studies and to efficiently interrogate the entire genome. Using these strategies, researchers have been able to identify a number of genes in which variations appear to contribute to the susceptibility to alcohol dependence. It is important to note, however, that the individual role of each of these genes, and the SNPs within them, is quite modest. This means that a given allele or SNP that has been found to be associated with alcohol dependence may increase the risk of alcoholism only incrementally. As a result, it would be a gross overinterpretation of the results obtained in human association studies to date to suggest that we currently have a means to identify people at greatest risk for alcohol dependence. With the exception of the strong protective effects of certain ADH and ALDH variants, each gene variant identified to date has a much smaller individual effect on alcoholism risk than, for example, a family history of alcoholism.
Another challenge is to relate the complex human behavioral phenotypes to specific variations in the sequence and expression of specific genes and, perhaps more importantly, to the function of the proteins encoded by these genes. The answers may come from networks of genes that encode proteins of similar function, rather than from specific genes individually. Examining such networks represents another level of complexity that poses a huge quantitative challenge, computationally and statistically. However, researchers also are making substantial progress on this bioinformatics front, and the continuing development of greatly enhanced bioinformatics capacity is increasing the power of studies in both rodent models and humans.
The identification of any genes that appear important in alcoholism susceptibility provides an opportunity to better understand the biological pathways involved in alcohol’s actions. It also may yield important insights that will allow the development of better pharmacological treatments to help those who wish to reduce their alcohol consumption. All such potential new therapies will of course be tested first in animal models (Egli 2005), and the coordination of animal model and human research therefore will continue to be an important theme for alcohol research for many years to come.
Acknowledgments
Preparation of this manuscript was supported by National Institutes of Health grants U10AA008401, P60AA007611, P60AA10760, U01AA13519, R37AA06460, U01AA016660, and R21AA017941 and the Department of Veterans Affairs.
Footnotes
DALYs are a measure of burden of disease. One DALY is equal to 1 healthy year of life lost.
For a definition of this and other technical terms, see the glossary, pp. 161–164.
Adoption studies compare the disease status of adoptees with that of their birth parents (with each of whom they share on average half their genome) and of their adoptive parents (with whom they typically have no genetic relationship and do not share their genome).
These criteria, which were the accepted diagnostic criteria at the time of COGA’s initiation, were based on the definitions established in the DSM–III–R (American Psychiatric Association 1987) and by Feighner and colleagues (1972).
Noncoding SNPs are DNA sequence variations that are located in regions of the ADH gene that do not encode the actual ADH protein.
G-protein–coupled receptors interact with a signaling molecule (e.g., acetylcholine) outside the cell, resulting in the activation of signaling pathways within the cell and thereby inducing a cellular response. Specifically, binding of the receptor to the signaling molecule alters the structure of the receptor so that it can activate an associated G-protein, which in turn can act on other proteins in the cell.
The genes located in this cluster are SLC22A18, PHLDA2, NAP1L4, snora54, CARS, and OSBPL5.
The N-methy-d-aspartate receptor is one of the receptor types for the neurotransmitter glutamate.
In contrast to a QTL, which only identifies a DNA region that is likely to contain a gene contributing to a quantitative trait (but also may contain other, unrelated DNA sequences), a QTG represents the actual gene.
Although humans and mice have different numbers of chromosomes and substantial variation in their genome, there are some parallels between the two genomes. Thus, about 80 percent of genes that are located closely together on a human chromosome also tend to be located in a cluster on a mouse chromosome.
Financial Disclosure
The authors declare that they have no competing financial interests.
References
- Agrawal A, Lynskey MT. Are there genetic influences on addiction: Evidence from family, adoption and twin studies. Addiction. 2008;103(7):1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Edenberg HJ, Foroud T, et al. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behavior Genetics. 2006;36:640–650. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. Third Edition, Revised. Washington, DC: American Psychiatric Association; 1987. [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, et al. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health & Research World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P, Beeson M, et al. Genetic dissection of quantitative trait locus for ethanol sensitivity in long- and short-sleep mice. Genes, Brain, and Behavior. 2008;7:659–668. doi: 10.1111/j.1601-183X.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- Bennett B, Downing C, Carosone-Link P, et al. Quantitative trait locus mapping for acute functional tolerance to ethanol in the L x S recombinant inbred panel. Alcoholism: Clinical and Experimental Research. 2007;31:200–208. doi: 10.1111/j.1530-0277.2006.00296.x. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, et al. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences of the USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Saccone NL, Rice JP, et al. Defining alcohol-related phenotypes in humans. The Collaborative Study on the Genetics of Alcoholism. Alcohol Research & Health. 2002;26:208–213. [PMC free article] [PubMed] [Google Scholar]
- Bleich S, Lenz B, Ziegenbein M, et al. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcoholism: Clinical and Experimental Research. 2006;30:587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16:167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. Journal of Neuroscience. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Kozell LB, Hitzemann R, Buck KJ. Involvement of the limbic basal ganglia in ethanol withdrawal convulsivity in mice is influenced by a chromosome 4 locus. Journal of Neuroscience. 2008a;28:9840–9849. doi: 10.1523/JNEUROSCI.1713-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Y, Shen P. A protein kinase C activity localized to neuropeptide Y-like neurons mediates ethanol intoxication in Drosophila melanogaster. Neuroscience. 2008b;156:42–47. doi: 10.1016/j.neuroscience.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, et al. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, et al. Markers in the 5’-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb DW, Edenberg HJ, Bosron WF, et al. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. Journal of Clinical Investigation. 1989;83:314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Rhodes JS, et al. A line of mice selected for high blood ethanol concentrations shows drinking in the dark to intoxication. Biological Psychiatry. 2009;65:662–670. doi: 10.1016/j.biopsych.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Belknap JK. The complexity of alcohol drinking: Studies in rodent genetic models. Behavior Genetics. 2010 doi: 10.1007/s10519-010-9371-z. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Buck KJ, et al. Identifying genes for alcohol and drug sensitivity: Recent progress and future directions. Trends in Neuroscience. 1999;22:173–179. doi: 10.1016/s0166-2236(99)01393-4. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, et al. Alcohol-related genes: Contributions from studies with genetically engineered mice. Addiction Biology. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Wang JC, et al. Alcohol dependence with comorbid drug dependence: Genetic and phenotypic associations suggest a more severe form of the disorder with stronger genetic contribution to risk. Addiction. 2007;102:1131–1139. doi: 10.1111/j.1360-0443.2007.01871.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, et al. Association of GABRG3 with alcohol dependence. Alcoholism: Clinical and Experimental Research. 2004;28:4–9. doi: 10.1097/01.ALC.0000108645.54345.98. [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, et al. Association between GABRA1 and drinking behaviors in the collaborative study on the genetics of alcoholism sample. Alcoholism: Clinical and Experimental Research. 2006;30:1101–1110. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. Regulation of the mammalian alcohol dehydrogenase genes. Progress in Nucleic Acid Research and Molecular Biology. 2000;64:295–341. doi: 10.1016/s0079-6603(00)64008-4. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The collaborative study on the genetics of alcoholism: An update. Alcohol Research & Health. 2002;26:214–218. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Research & Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. American Journal of Human Genetics. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: Identifying specific genes through family studies. Addiction Biology. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcoholism: Clinical and Experimental Research. 2010 doi: 10.1111/j.1530-0277.2010.01156.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, et al. Association of alcohol dehydrogenase genes with alcohol dependence: A comprehensive analysis. Human Molecular Genetics. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Egli M. Can experimental paradigms and animal models be used to discover clinically effective medications for alcoholism? Addiction Biology. 2005;10:309–319. doi: 10.1080/13556210500314550. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, et al. Genomic screen for loci associated with alcohol dependence in Mission Indians. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 2004;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Walter NA, Dick DM, et al. A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models. Addiction Biology. 2010;15:185–199. doi: 10.1111/j.1369-1600.2009.00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng MY, Luczak SE, Wall TL. ALDH2, ADH1B, and ADH1C genotypes in Asians: A literature review. Alcohol Research & Health. 2007;30:22–27. [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, et al. GABRG1 and GABRA2 as independent predictors for alcoholism in two populations. Neuropsychopharmacology. 2009;34:1245–1254. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feighner JP, Robbins E, Guze SB, et al. Diagnostic criteria for use in psychiatric research. Archives of General Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- Finn DA, Ford MM, Wiren KM, et al. The role of pregnane neurosteroids in ethanol withdrawal: Behavioral genetic approaches. Pharmacology & Therapeutics. 2004;101:91–112. doi: 10.1016/j.pharmthera.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR. Genetics of alcohol dependence. Human Genetics. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin NW, Stewart RB, Elder RL, et al. Sedative and motor-impairing effects of neuropeptide Y and ethanol in selectively bred P and NP rats. Pharmacology, Biochemistry, and Behavior. 2004;78:65–73. doi: 10.1016/j.pbb.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Cippitelli A, Sommer WH, et al. Variation at the rat Crhr1 locus and sensitivity to relapse into alcohol seeking induced by environmental stress. Proceedings of the National Academy of Sciences of the USA. 2006;103:15236–15241. doi: 10.1073/pnas.0604419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Archives of General Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Hill SY. Alternative strategies for uncovering genes contributing to alcoholism risk: Unpredictable findings in a genetic wonderland. Alcohol. 1998;16:53–59. doi: 10.1016/s0741-8329(97)00177-8. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Edmunds S, Wu W, et al. Detection of reciprocal quantitative trait loci for acute ethanol withdrawal and ethanol consumption in heterogeneous stock mice. Psychopharmacology (Berl) 2009;203:713–722. doi: 10.1007/s00213-008-1418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft NR, Corley RP, McQueen MB, et al. SNPs in CHRNA6 and CHRNB3 are associated with alcohol consumption in a nationally representative sample. Genes, Brain, and Behavior. 2009. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Hurley TD, Edenberg HJ, Li TK. The pharmacogenomics of alcoholism. In: Licinio J, Wong M-L, editors. Pharmacogenomics: The Search for Individualized Therapies. Weinheim: Wiley-VCH; 2002. pp. 417–441. [Google Scholar]
- Jacob T, Sher KJ, Bucholz KK, et al. An integrative approach for studying the etiology of alcoholism and other addictions. Twin Research. 2001;4:103–118. doi: 10.1375/1369052012218. [DOI] [PubMed] [Google Scholar]
- Kelai S, Hanoun N, Aufrere G, et al. Cannabinoid-serotonin interactions in alcohol-preferring vs. alcohol-avoiding mice. Journal of Neurochemistry. 2006;99:308–320. doi: 10.1111/j.1471-4159.2006.04054.x. [DOI] [PubMed] [Google Scholar]
- Kendler KS, O’Neill FA, Burke J, et al. Irish study on high-density schizophrenia families: Field methods and power to detect linkage. American Journal of Medical Genetics. 1996;67:179–190. doi: 10.1002/(SICI)1096-8628(19960409)67:2<179::AID-AJMG8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Li H, Borinskaya S, Yoshimura K, et al. Refined geographic distribution of the Oriental ALDH2*504Lys (nee 487Lys) variant. Annals of Human Genetics. 2009;73:335–345. doi: 10.1111/j.1469-1809.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Mukherjee N, Soundararajan U, et al. Geographically separate increases in the frequency of the derived ADH1B*47His allele in eastern and western Asia. American Journal of Human Genetics. 2007;81:842–846. doi: 10.1086/521201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Porcu P, Werner DF, et al. The role of GABA(A) in the acute and chronic effects of ethanol: A decade of progress. Psychopharmacology (Berl) 2009;205:529–564. doi: 10.1007/s00213-009-1562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo IA, Harris RA. GABA(A) receptors and alcohol. Pharmacology, Biochemistry, and Behavior. 2008;90:90–94. doi: 10.1016/j.pbb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an auto-some-wide scan in an American Indian population. American Journal of Medical Genetics. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, Crabbe JC. Laboratory models of alcoholism: Treatment target identification and insight into mechanisms. Nature Neuroscience. 2005;8:1471–1480. doi: 10.1038/nn1581. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, et al. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: An integrated analysis. Human Molecular Genetics. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden PA, Bucholz KK, Martin NG, et al. Smoking and the genetic contribution to alcohol-dependence risk. Alcohol Research & Health. 2000;24:209–214. [PMC free article] [PubMed] [Google Scholar]
- Mardones J, Segovia-Riquelme N. Thirty-two years of selection of rats by ethanol preference: UChA and UChB strains. Neurobehavioral Toxicology and Teratology. 1983;5:171–178. [PubMed] [Google Scholar]
- McBride WJ, Schultz JA, Kimpel MW, et al. Differential effects of ethanol in the nucleus accumbens shell of alcohol-preferring (P), alcohol-non-preferring (NP) and Wistar rats: A proteomics study. Pharmacology, Biochemistry, and Behavior. 2009;92:304–313. doi: 10.1016/j.pbb.2008.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proceedings of the National Academy of Sciences of the USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton PM, Messing RO. Intracellular signaling pathways that regulate behavioral responses to ethanol. Pharmacology & Therapeutics. 2006;109:227–237. doi: 10.1016/j.pharmthera.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Newton PM, Messing RO. Increased sensitivity to the aversive effects of ethanol in PKCepsilon null mice revealed by place conditioning. Behavioral Neuroscience. 2007;121:439–442. doi: 10.1037/0735-7044.121.2.439. [DOI] [PubMed] [Google Scholar]
- Peng J, Wagle M, Mueller T, et al. Ethanol-modulated camouflage response screen in zebrafish uncovers a novel role for cAMP and extra-cellular signal-regulated kinase signaling in behavioral sensitivity to ethanol. Journal of Neuroscience. 2009;29:8408–8418. doi: 10.1523/JNEUROSCI.0714-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Gunter TD, Beach SR, et al. Role of GABRA2 on risk for alcohol, nicotine, and cannabis dependence in the Iowa Adoption Studies. Psychiatric Genetics. 2009;19:91–98. doi: 10.1097/YPG.0b013e3283208026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: Evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Molecular Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Myers JM, et al. The Irish Affected Sib Pair Study of Alcohol Dependence: Study methodology and validation of diagnosis by interview and family history. Alcoholism: Clinical and Experimental Research. 2005;29:417–429. doi: 10.1097/01.alc.0000156085.50418.07. [DOI] [PubMed] [Google Scholar]
- Reich T. A genomic survey of alcohol dependence and related phenotypes: Results from the Collaborative Study on the Genetics of Alcoholism (COGA) Alcoholism: Clinical and Experimental Research. 1996;20(8 Suppl):133A–137A. doi: 10.1111/j.1530-0277.1996.tb01763.x. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, et al. Genome-wide search for genes affecting the risk for alcohol dependence. American Journal of Medical Genetics. 1998;81:207–215. [PubMed] [Google Scholar]
- Reilly MT, Milner LC, Shirley RL, et al. 5-HT2C and GABAB receptors influence handling-induced convulsion severity in chromosome 4 congenic and DBA/2J background strain mice. Brain Research. 2008;1198:124–131. doi: 10.1016/j.brainres.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Histone acetylation in drug addiction. Seminars in Cell & Developmental Biology. 2009;20:387–394. doi: 10.1016/j.semcdb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. A longitudinal study of children of alcoholics. Recent Developments in Alcoholism. 1991;9:5–19. [PubMed] [Google Scholar]
- Sher KJ, Dick DM, Crabbe JC, et al. Consilient research approaches in studying gene x environment interactions in alcohol research. Addiction Biology. 2010;15:200–216. doi: 10.1111/j.1369-1600.2009.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley RL, Walter NA, Reilly MT, et al. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nature Neuroscience. 2004;7:699–700. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- Smith JE, Co C, McIntosh S, Cunningham CC. Chronic binge-like moderate ethanol drinking in rats results in widespread decreases in brain serotonin, dopamine, and norepinephrine turnover rates reversed by ethanol intake. Journal of Neurochemistry. 2008 doi: 10.1111/j.1471-4159.2008.05296.x. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Koh MT, Pedrazzini T. Voluntary alcohol consumption is controlled via the neuropeptide Y Y1 receptor. Journal of Neuroscience. 2002;22:RC208. doi: 10.1523/JNEUROSCI.22-03-j0006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. American Journal of Human Genetics. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, et al. Genome-wide association study of alcohol dependence. Archives of General Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaluw CS, Engels RC. Gene-environment interactions and alcohol use and dependence: Current status and future challenges. Addiction. 2009;104:907–914. doi: 10.1111/j.1360-0443.2009.02563.x. [DOI] [PubMed] [Google Scholar]
- Vengeliene V, Bachteler D, Danysz W, Spanagel R. The role of the NMDA receptor in alcohol relapse: A pharmacological mapping study using the alcohol deprivation effect. Neuropharmacology. 2005;48:822–829. doi: 10.1016/j.neuropharm.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Bachmanov A, Finn DA, Crabbe JC. Stability of inbred mouse strain differences in behavior and brain size between laboratories and across decades. Proceedings of the National Academy of Sciences of the USA. 2006;103:16364–16369. doi: 10.1073/pnas.0605342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Molecular Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Symons MN, Singh SM. Ethanol-responsive genes (Crtam, Zbtb16, and Mobp) located in the alcohol-QTL region of chromosome 9 are associated with alcohol preference in mice. Alcoholism: Clinical and Experimental Research. 2009;33:1409–1416. doi: 10.1111/j.1530-0277.2009.00971.x. [DOI] [PubMed] [Google Scholar]
- Wetherill L, Schuckit MA, Hesselbrock V, et al. Neuropeptide Y receptor genes are associated with alcohol dependence, alcohol withdrawal phenotypes, and cocaine dependence. Alcoholism: Clinical and Experimental Research. 2008;32:2031–2040. doi: 10.1111/j.1530-0277.2008.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, et al. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II: alcoholism and event-related potentials. American Journal of Human Genetics. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Pollak L, Mottagui-Tabar S, et al. NPY Leu7Pro and alcohol dependence in Finnish and Swedish populations. Alcoholism: Clinical and Experimental Research. 2003;27:19–24. doi: 10.1097/01.ALC.0000050642.62233.44. [DOI] [PubMed] [Google Scholar]