Abstract
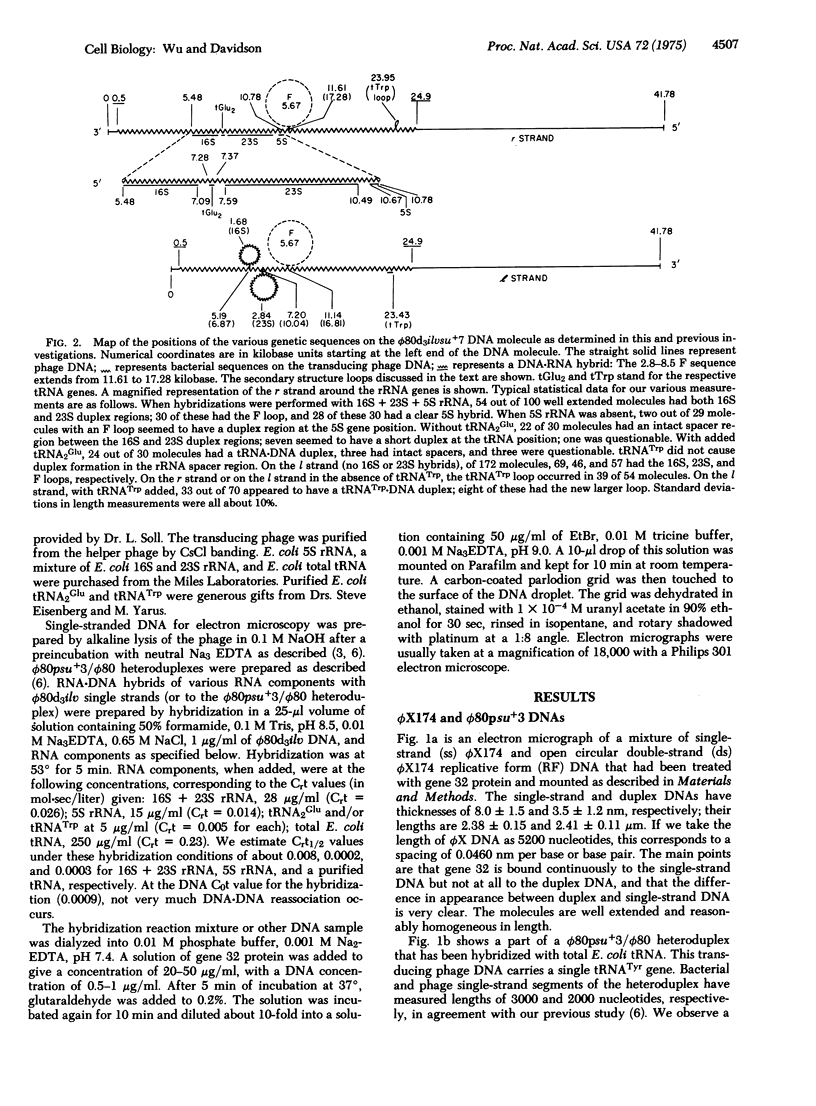
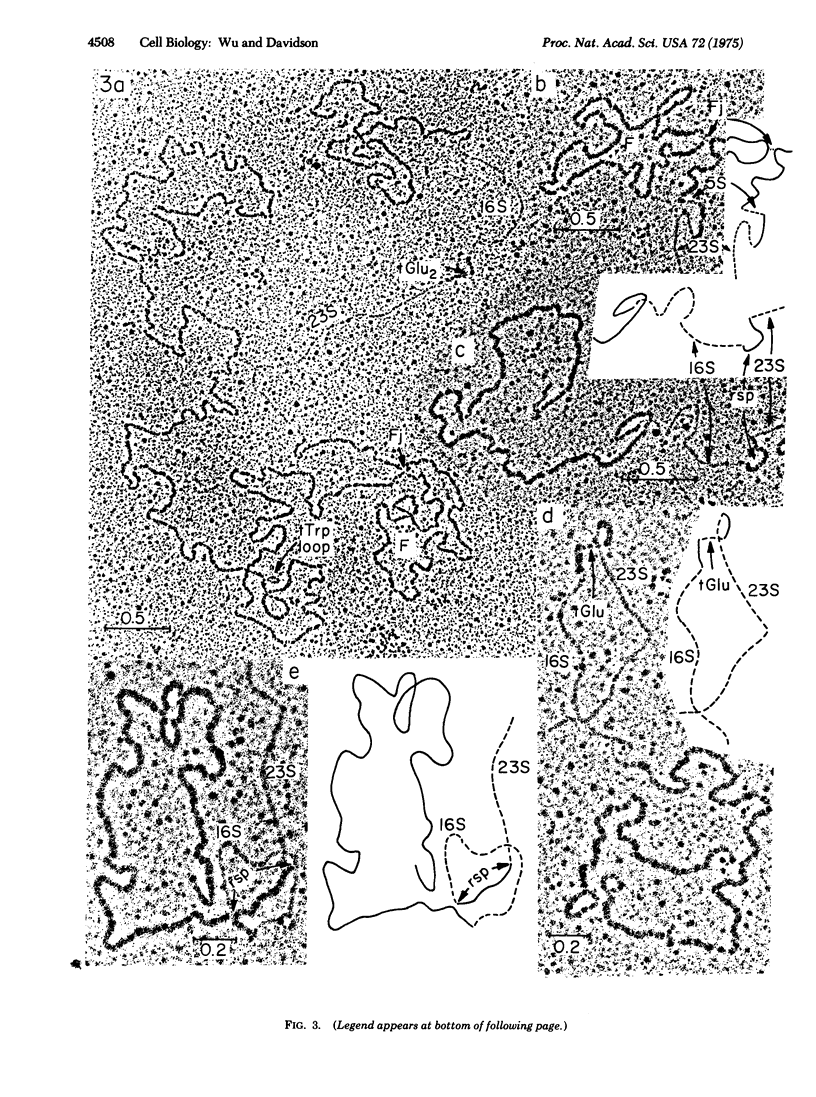
A method for visualizing RNA-DNA duplex regions along a single strand of DNA in the electron microscope is described. A preparation of RNA molecules is hybridized to a long DNA strand containing the coding sequences (genes) for some of the RNAs. T4 gene 32 protein, which binds selectively and cooperatively only to the single-strand regions, is added, followed by glutaraldehyde. The resulting nucleic acid-gene 32 complex is adsorbed to the surface of an electron microscope grid in the presence of ethidium bromide. The single-strand regions are relatively thick (8.5 nm) compared to the duplex (RNA-DNA hybrid) regions (3.5 nm), so that the two kinds of regions are readily recognized by electron microscopy. In favorable cases, tRNA-DNA hybrids of length about 80 nucleotide pairs can be recognized (although with difficulty). The positions of a number of interesting genetic sequences on the DNA of the transducing phage phi80d3ilvsu+7 have been mapped. The r strand contains 16S, 23S, and 5S rRNA coding sequences in that order. The spacer between 16S and 23S genes has a length of 500 nucleotides and contains the coding sequence for a tRNA2Glu gene in agreement with previous biochemical observations. The spacer between the 23S and 5S genes has a length of 180 nucleotides. The su+7 tRNATrp coding sequence has been mapped on the l strand at a position just to the left of the ilv genes. Secondary structure loops due to short inverted repeat sequences flanking the 16S, 23S, tRNATrp, and F sequences in the DNA have been observed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brack C., Bickle T. A., Yuan R. The relation of single-stranded regions in bacteriophage PM2 supercoiled DNA to the early melting sequences. J Mol Biol. 1975 Aug 25;96(4):693–702. doi: 10.1016/0022-2836(75)90146-1. [DOI] [PubMed] [Google Scholar]
- Delius H., Mantell N. J., Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972 Jun 28;67(3):341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- Doolittle W. F., Pace N. R. Transcriptional organization of the ribosomal RNA cistrons in Escherichia coli. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1786–1790. doi: 10.1073/pnas.68.8.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman R. W. Physical mapping of T7 messenger RNA. J Mol Biol. 1971 Oct 28;61(2):369–376. doi: 10.1016/0022-2836(71)90386-x. [DOI] [PubMed] [Google Scholar]
- Koller T., Sogo J. M., Bujard H. An electron microscopic method for studying nucleic acid-protein complexes. Visualization of RNA polymerase bound to the DNA of bacteriophages T7 and T3. Biopolymers. 1974 May;13(5):995–1009. doi: 10.1002/bip.1974.360130514. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Otsubo E., Deonier R. C., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. V. ilv+ Deletion mutants of F14. J Mol Biol. 1974 Nov 15;89(4):585–597. doi: 10.1016/0022-2836(74)90037-0. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Silengo L., Schlessinger D. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3361–3365. doi: 10.1073/pnas.70.12.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo E., Soll L., Deonier R. C., Lee H. J., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. VIII. The structure of bacteriophage phi 80d-3ilv+su+7, including the mapping of the ribosomal RNA genes. J Mol Biol. 1974 Nov 15;89(4):631–646. doi: 10.1016/0022-2836(74)90040-0. [DOI] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Davidson N. A technique for mapping transfer RNA genes by electron microscopy of hybrids of ferritin-labeled transfer RNA and DNA: the phi-80hpsu+3-system. J Mol Biol. 1973 Jun 25;78(1):1–21. doi: 10.1016/0022-2836(73)90424-5. [DOI] [PubMed] [Google Scholar]
- Wu M., Davidson N., Carbon J. Physical mapping of the transfer RNA genes on lambda-h80dglytsu+36. J Mol Biol. 1973 Jun 25;78(1):23–34. doi: 10.1016/0022-2836(73)90425-7. [DOI] [PubMed] [Google Scholar]
- Wu M., Davidson N. Secondary structure in transfer RNA genes. J Mol Biol. 1973 Jun 25;78(1):35–41. doi: 10.1016/0022-2836(73)90426-9. [DOI] [PubMed] [Google Scholar]
- Yaniv M., Folk W. R., Berg P., Soll L. A single mutational modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J Mol Biol. 1974 Jun 25;86(2):245–260. doi: 10.1016/0022-2836(74)90016-3. [DOI] [PubMed] [Google Scholar]






