Abstract
We investigated whether Notch signaling plays a role in regulating macrophage responses to inflammation. In a wound healing assay, macrophage recruitment was decreased in Notch1+/− mice, and the wounds were characterized by decreased TNF-α expression. As wound healing progressed, Notch1+/− wounds had increased vascularization and collagen deposition compared with wild-type wounds. In mice with myeloid-specific Notch1 deletion, wounds had decreased macrophage recruitment as well as decreased TNF-α expression, indicating the specific role of Notch1 in the inflammatory response in these cells. In vitro, we found that vascular endothelial growth factor receptor-1 (VEGFR-1) was upregulated in macrophages in response to LPS/IFN-γ and that this upregulation depended on Notch signaling. Furthermore, macrophages from Notch1+/− mice had decreased expression of VEGFR-1 compared with macrophages from wild-type mice, whereas VEGFR-1 expression in Notch4−/− macrophages was normal. Inhibition of Notch signaling decreased induction of the inflammatory cytokines IL-6, IL-12, CXCL10, MCP-1, monokine induced by IFN-γ, and TNF-α in macrophages in response to LPS/IFN-γ. Additionally, macrophages from Notch1+/− mice demonstrated decreased induction of IL-6, IL-12, and TNF-α in response to stimulation compared with wild-type mice. Thus, both pharmacological inhibition of Notch and genetic analysis demonstrate that Notch1 regulates VEGFR-1 and cytokine expression in macrophages. We have also established that Notch1 is important for the inflammatory response during wound healing in mice.
Notch receptors are highly conserved transmembrane proteins that are required for normal embryonic development. In addition to a role during development, Notch signaling plays an important role in angiogenesis and vascular homeostasis in both physiological and pathological settings (1).In mammals, there are four Notch proteins (Notch1–4) that act as receptors for five ligands (Delta-like 1, 3, and 4 and Jagged1 and 2). Upon ligand binding, Notch protein is subject to a series of proteolytic cleavages by the A disintegrin and metalloproteinase family of metalloproteases and presenillin/γ-secretase. Cleavage releases the intracellular domain of Notch, which translocates to the nucleus and functions as a transcriptional activator in complex with the transcription factor CSL, Mastermind, and histone acetyltransferaces. In endothelial cells, Notch is downstream of vascular endothelial growth factor (VEGF), and signaling of the Notch receptor through Delta-like 4 regulates the expression of VEGF receptor-1 and −2 (VEGFR-1 and −2) to restrict endothelial cell function and sprouting angiogenesis (2–4). In addition to endothelial cells, VEGFR-1 is expressed on monocytes and macrophages and plays a role in their recruitment and function at areas of angiogenesis and inflammation (5–7). Whether Notch regulates expression of VEGFR-1 in macrophages in a similar manner as in endothelial cells has yet to be elucidated. Moreover, although Notch has been implicated in regulating endothelial cell responses to inflammatory cytokines (8, 9), a role for Notch in the inflammatory response in other cell types has not been fully defined.
There has been increasing interest in the role of Notch signaling in the monocyte/macrophage lineage with respect to myeloid differentiation and inflammation (10, 11). Studies using macrophage cell lines and primary monocytes have demonstrated expression of Notch1–4 as well as the Notch ligands Jagged1, Jagged2, and Deltalike 4 in these cells (10, 12–14). Macrophages have diverse functions in inflammation, tissue remodeling, and angiogenesis. To describe the plasticity of these cells, a simplified paradigm of the polarization of macrophages toward either an M1 “classically activated” or an M2 “alternatively activated” phenotype is useful (15). M1 macrophages are activated in response to IFN-γ, LPS, and TNF-α and produce large amounts of inflammatory cytokines and reactive oxygen intermediates. As such, they exhibit cytotoxic activity that includes clearing infection and mediating resistance against tumors. Notch signaling is upregulated in macrophages in response to LPS and IFN-γ, and it may regulate genes involved in inflammation and Ag presentation (13, 16, 17). In contrast to the M1 phenotype, M2 macrophages are induced in response to IL-4, IL-10, and IL-13, express high levels of scavenger receptors, and promote cell growth via arginine metabolism. M2 macrophages have an immunoregulatory function and promote tissue repair and remodeling. Whether Notch signaling is activated in response to M2 cytokines has not been established.
We hypothesized that Notch signaling may affect macrophage function and expression of inflammatory cytokines in the setting of wound healing, where the inflammatory and trophic functions of macrophages are intimately related. In a wound healing assay, we found delayed recruitment of macrophages and decreased TNF-α expression in wounds in Notch1+/− mice compared with wild-type mice. Interestingly, Notch1+/− wounds also had increased collagen deposition and angiogenesis compared with wild-type wounds. In mice with myeloid-specific Notch1 deletion, wounds were characterized by decreased macrophage recruitment and TNF-α expression, indicating a specific role of Notch1 in these cells during the inflammatory response. In light of this, we isolated bone marrow-derived macrophages (BMM) from mice and characterized Notch signaling in the context of stimulation. We found that VEGFR-1 and the inflammatory cytokines IL-6, IL-12, CXCL10 (IP-10), MCP-1, monokine induced by IFN-γ (MIG), and TNF-α were downstream of Notch in response to stimulation with LPS/ IFN-γ. Furthermore, macrophages from Notch1+/− mice exhibited decreased VEGFR-1 expression and a diminished ability to induce VEGFR-1 and inflammatory cytokines in response to stimulation. Taken together, our data suggest a role for Notch signaling in recruitment and function of macrophages at sites of inflammation via regulation of VEGFR-1 and inflammatory cytokines.
Materials and Methods
Reagents
The γ-secretase inhibitor compound E was obtained from the Korean Research Institute of Chemical Technology and was used in vitro at a concentration of 400 nM, with DMSO as a control. Macrophages were stimulated with 100 ng/ml LPS (Calbiochem, San Diego, CA), 100 U/ml recombinant murine IFN-γ (PeproTech, Rocky Hill, NJ), or 5 ng/ml recombinant murine IL-4 (R&D Systems, Minneapolis, MN).
Mice
Notch1 and Notch4 mutant mice have been described (18, 19). Mice with a conditional allele of Notch1 (Notch1flox/flox) (20) and the myeloid-specific Cre recombinase driver line (LysMCre) (21) were obtained from The Jackson Laboratory (Bar Harbor, ME). Transgenic Notch reporter mice, harboring an enhanced GFP sequence under the control of four tandem copies of the CBF1 binding site consensus sequence (22), were also obtained from The Jackson Laboratory. We maintained all mice in the C57BL/6 background. All procedures were carried out according to approved protocols and guidelines established by the Columbia University Institutional Animal Care and Use Committee.
BMM culture and lentiviral infections
Macrophages were differentiated from hematopoietic stem cells from mouse femurs as described (23) and were cultured in RPMI 1640 supplemented with 10% FBS and 20% conditioned medium from L929 cells (Sigma-Aldrich, St. Louis, MO) on petri dishes. Intracellular domain of Notch1 (N1IC) or GFP constructs were cloned into the lentiviral vector pCCL (provided by Stefano Rivella, Memorial Sloan-Kettering). Lentivirus was produced in transfected 293T cells and used to transduce macrophages as described (24). Conditioned Jag1Fc medium was produced by CHO cells transduced with Jagged1ECDFc or Fc control plasmids. Serum-free medium from confluent plates was collected after 48 h of incubation and used to treat BMM.
Determination of inducible NO synthase activity and arginase expression
BMM culture supernatants from overnight stimulation were aliquoted to a 96-well plate and incubated with Griess reagent as previously described (25). NO production was assessed using a Bio-Rad (Hercules, CA) microplate reader at 550 nm. To determine arginase expression, whole-cell lysates from BMM were harvested using TENT lysis buffer, and lysates were run with SDS-PAGE according to standard protocols. Membranes were probed for arginase II (H-64; Santa Cruz Biotechnology, Santa Cruz, CA) or alpha-tubulin (Sigma-Aldrich).
Reverse transcription and quantitative RT-PCR
BMM were stimulated overnight and RNA was isolated using RNeasy Mini kit (Qiagen, Valencia, CA). Reverse transcription of RNA was performed using SuperScript II (Invitrogen, Carlsbad, CA). Quantitative PCR was performed using primers for mouse ribosomal protein P0, arginase, inducible NO synthase (iNOS), Notch1, Jagged1, VEGFR-1, and Hey1 (primer sequences available upon request) using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and the 7300 Real-Time PCR System (Applied Biosystems). All reactions were performed in triplicate and normalized to expression of P0.
Flow cytometry of peripheral blood and BMM
BMM were harvested from petri dishes after overnight stimulation using cold PBS. Whole blood was harvested from mice via cardiac puncture or retroorbital bleeding. Packed blood was washed with HBSS, and RBCs were lysed using BD FACS lysing solution (BD Biosciences, San Jose, CA) prior to staining. Peripheral blood leukocytes or BMM were incubated for 30 min with Fc Block (anti-mouse CD16/CD32; BD Biosciences), followed by anti-Flt1 Ab (C-17; Santa Cruz Biotechnology) or FITC-conjugated anti-CD11b Ab (BD Biosciences). After washing, cells were incubated with allophycocyanin-conjugated anti-rabbit secondary Ab (Jackson ImmunoResearch Laboratories, West Grove, PA). For detection of GFP in Notch reporter cells, peripheral blood leukocytes were fixed with 2% paraformaldehyde and then permeabilized with 0.12% Triton X-100 in PBS. Cells were stained with FITC-conjugated anti-GFP Ab (BD Biosciences). Flow cytometry was performed using FACSCalibur and Cell-Quest Pro acquisition software (BD Biosciences) or FlowJo v7.5 flow cytometry analysis software.
Analysis of cytokine secretion
BMM culture supernatants were harvested after overnight stimulation. Cytokine concentration in supernatants was assessed using a mouse 20-plex cytokine detection panel (Invitrogen) according to the manufacturer’s protocol. Briefly, samples were incubated with Ab-coated capture beads, washed, and then incubated with detector Ab. Samples were analyzed using the Luminex (Austin, TX) 100 IS system. All reactions were performed in duplicate for each sample.
Wound-healing assay
Mice were anesthetized with isoflurane for the duration of the procedure. Hair was shaved from the dorsum of the mice, and skin was prepared with iodine solution. Full-thickness excisional wounds of 2.25 cm2 were created using a template on the dorsal skin of Notch1 mutant mice or control littermates. Wounds were dressed with benzoin (Henry Schein, Melville, NY) and Tegaderm (3M Health Care, St. Paul, MN), and mice were kept in individual cages for the duration of the assay. To harvest wound tissue, mice were sacrificed on postinjury day 5, 7, or 14. The excisional wounds, together with a peripheral rim of normαl skin and the deep muscle layer, were excised, divided along the longitudinal axis, and fixed in 4% paraformaldehyde for further histological study. Wound healing assays were conducted using 18 Notch1 mutant mice and 8 control littermates.
Immunohistochemistry
Tissue was fixed in 4% paraformaldehyde overnight at 4°C and then embedded in paraffin. Five-micrometer serial sections were deparaffinized with xylene, and heat-mediated Ag retrieval was performed using Dako (Glostrup, Denmark) target retrieval solution. Tissue was stained with H&E according to standard protocols or was stained with anti-F4/80 Ab (Abcam, Cambridge, MA), anti–TNF-α (Abcam), or anti-CD31 (BD Biosciences) and appropriate secondary Abs (Vector Laboratories, Burlingame, CA). Following incubation with an ABC Elite kit (Vector Labroatories), slides were developed with a diaminobenzidine substrate kit (Vector Laboratories) and counterstained with hematoxylin. For immuno-fluorescent costaining, slides were incubated with anti-rat Alexa Fluor 488 or anti-rabbit Alexa Fluor 594 (Invitrogen). Slides were analyzed using a Nikon Eclipse E800 microscope and a Nikon DXM 1200 camera, with ImagePro Plus software. Quantification of immunohistochemical staining was performed ImagePro Plus software. Images of five randomly selected ×20 fields of wound tissue from each mouse were used for quantification of staining, and staining intensity was averaged for each genotype.
Statistical analysis
Data are expressed as mean ± SEM. Statistical analysis was performed by a two-tailed Student t test. A p value <0.05 (indicated with an asterisk) was considered significant. All data are representative of at least three independent experiments.
Results
Decreased macrophage recruitment and decreased TNF-α expression in wounds in Notch1+/− mice
Wound healing is a setting in which macrophages play a wide-ranging and important role. The process of wound healing is divided into three overlapping phases: inflammation, proliferation, and remodeling. The role of macrophages is particularly pronounced during the inflammatory and proliferative phases (26). Previous studies have shown that Notch1 signaling is upregulated in macrophages in response to inflammatory stimuli and may regulate genes involved in inflammation (13). We hypothesized that macrophage function during wound healing may be compromised in mice mutant for Notch1. To test this, dorsal excisional wounds were created in Notch1+/− mice or wild-type littermates, and wound tissue was harvested to assess leukocytic infiltrate, collagen deposition, and neovascularization. Homozygous mutation of Notch1 results in embryonic lethality (18, 19), but Notch1 heterozygous mice survive without gross developmental defects. Additionally, we did not detect changes in the number of myeloid cells in the spleen and peripheral blood of Notch1 heterozygous mice compared with wild-type mice (data not shown). However, we have seen an ∼75% decrease in Notch1 transcript levels in BMM of Notch1+/− mice, suggesting a positive feedback mechanism for Notch1 expression (data not shown). We observed decreased leukocyte recruitment to wounds in Notch1+/− mice at postinjury day 5, and staining for the macrophage marker F4/80 revealed a >30% decrease in macrophage density in Notch1+/− wounds compared with wild-type wounds (Fig. 1). The decrease in macrophage recruitment was not due to changes in VEGF expression, as similar levels of VEGF expression were detected in Notch1+/− and wild-type wounds by immunohistochemistry (data not shown). Because macrophages are an important source of the proinflammatory cytokine TNF-α during the inflammatory phase of wound healing (27), we determined whether TNF-α expression was altered in Notch1+/− wounds. We found that staining for TNF-α was decreased, or that TNF-α–expressing macrophages were decreased in number, in day 5 wounds from Notch1+/− mice compared with wild-type littermates (Fig. 2A–C). We wanted to determine whether the decrease in TNF-α we observed in Notch1+/− wounds was due to decreased expression in macrophages. We found colocalization of TNF-α with F4/80 in both wild-type and Notch1+/− wounds (Fig. 2D, 2E). However, the number of TNF-α–expressing macrophages was decreased in wounds in Notch1+/− mice relative to wild-type littermates (Fig. 2D, 2E). Additionally, we detected decreased TNF-α staining in macrophages in Notch1+/− wounds relative to macrophages in wild-type wounds (Fig. 2F, 2G). Thus, decreased TNF-α expression in Notch1+/− wounds is due to decreased TNF-α expression in Notch1-deficient macrophages.
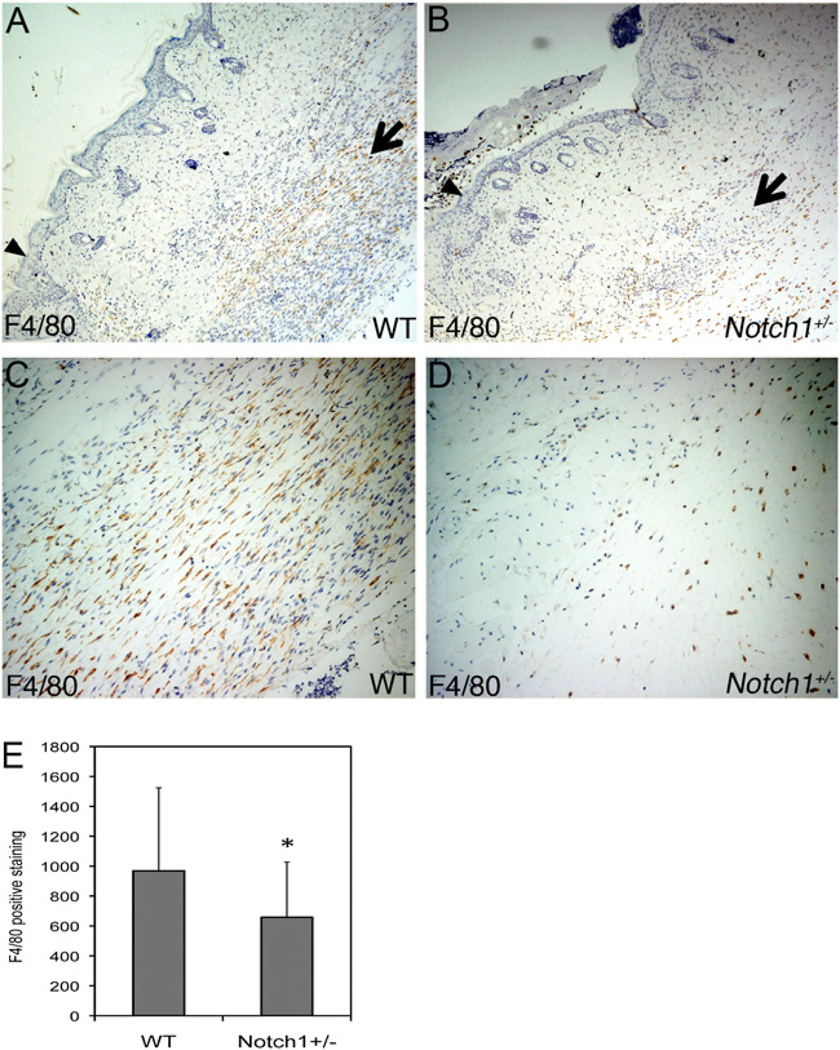
FIGURE 1.
Decreased macrophage recruitment to wounds in Notch1+/− mice. Full-thickness excisional wounds were made on the dorsum of mice. Wounds were harvested for immunohistochemistry at day 5 postinjury from WT (A, C) or Notch1+/− (B, D) mice. Representative images of F4/80 staining of wound tissue at edge of re-epithelialization are shown. Original magnification ×6 (A, B) or ×25 (C, D). Arrowheads indicate wound edge. Arrows indicate areas of macrophage infiltration. E, Quantification of F4/80 staining of day 5 wound tissue. *p < 0.05 relative to control. WT, wild-type.
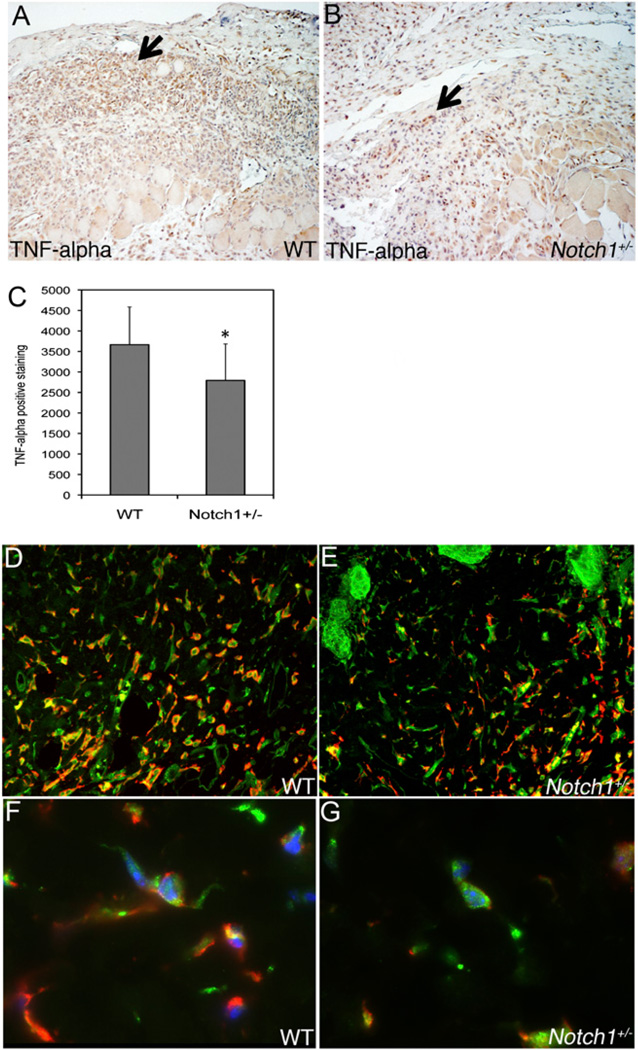
FIGURE 2.
Decreased TNF-α expression in wounds in Notch1+/− mice. TNF-α staining of granulation tissue of day 5 WT (A) or Notch1+/− (B) wounds shown. Original magnification ×20. Arrows indicate areas of TNF-α–expressing cells. C, Quantification of TNF-α staining of day 5 wound tissue. D–G, F4/80 (green) and TNF-α (red) costaining of day 5 wound tissue shown. Original magnification ×25 (D, E) and ×120 (F, G). DAPI staining is shown in blue (F, G). *p < 0.05 relative to control. WT, wild-type.
Increased vascularity and collagen deposition in wounds in Notch1+/− mice
On postinjury day 7, macrophage recruitment to wounds in Notch1+/− mice was still less than that in wild-type littermates, although the magnitude of the difference in macrophage density between mutant and wild-type was not as large as seen at day 5 (Fig. 3A, 3B). By day 7, collagen deposition, as assessed by trichrome staining, was denser in Notch1+/− wounds compared with wild-type wounds (Fig. 3C, 3D). Granulation tissue formation was robust in Notch1+/− wounds, and these tissues had less shearing during wound harvest compared with wild-type wounds (data not shown). At day 14, Notch1+/− wound tissue continued to be more organized than in wild-type littermates, with robust granulation tissue and less shearing during wound harvest compared with wild-type littermates. Notch1+/− wounds also displayed many more large, RBC-filled neovessels in wound tissue compared with wild-type wounds, as assessed by CD31 staining (Fig. 3E–H). The robust vascularity in Notch1+/− wounds was observed as early as day 5 (data not shown). Altered angiogenesis in Notch1+/− wound tissue may be due to decreased Notch signaling in endothelial cells, which has been previously reported to lead to increased vascular proliferation (2, 4, 28, 29). However, decreased inflammation in wounds due to decreased leukocyte infiltration or decreased expression of proinflammatory cytokines such as TNF-α can also affect angiogenesis and collagen deposition (27, 30).
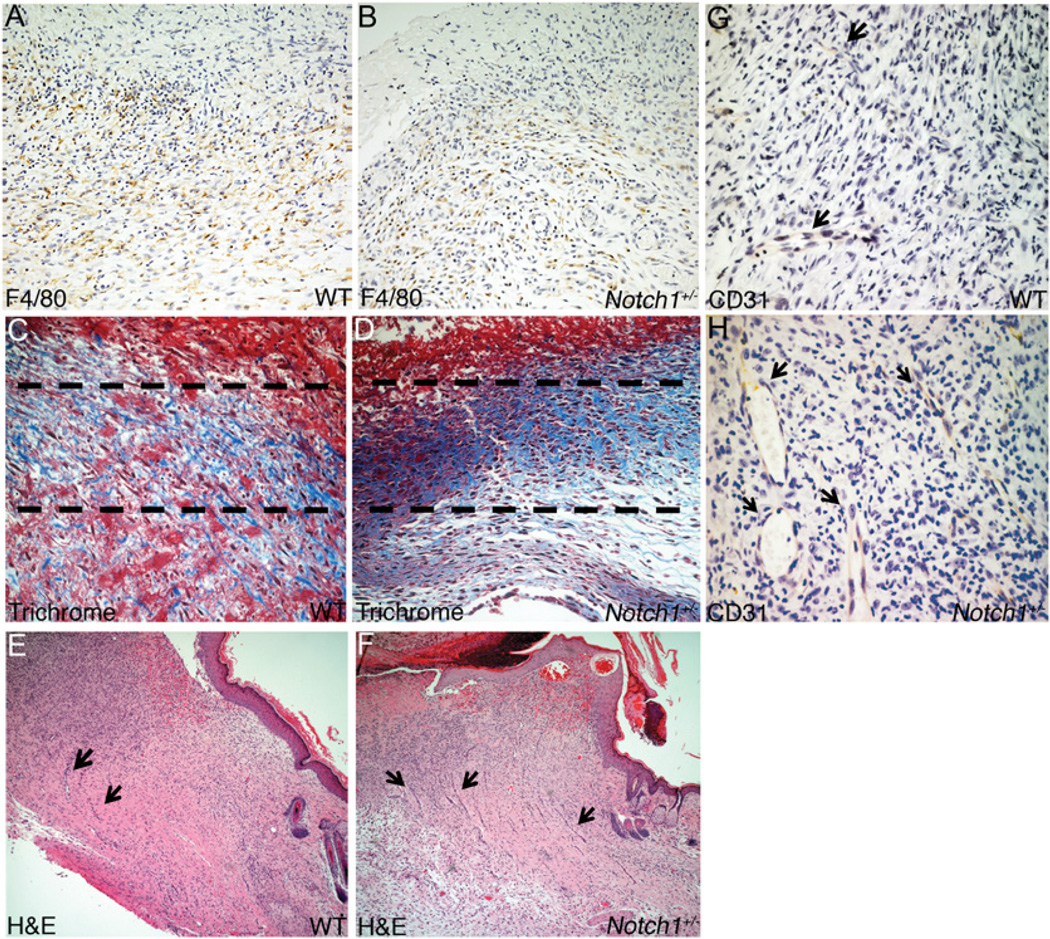
FIGURE 3.
Increased neovascularization and collagen deposition in wounds in Notch1+/− mice. Day 7 wound tissue from WT (A, C) and Notch1+/− (B, D) mice. A and B, Representative images of F4/80 staining of wound tissue at edge of re-epithelialization. Original magnification ×20. C and D, Trichrome staining of day 7 wounds showing collagen deposition (stained in blue) in regions directly below re-epithelialization (indicated by dashed lines). Day 14 wound tissue shown from WT (E, G) and Notch1+/− (F, H) mice. E and F, Visualization of wound tissue by H&E staining. Original magnification ×6. G and H, CD31 staining of wound tissue. Original magnification ×40. RBC-filled neovessels indicated by arrows. WT, wild-type.
Decreased macrophage recruitment and decreased TNF-α expression in wounds of mice with myeloid-specific Notch1 deletion
Because of the possibility that Notch may function in many cell types that effect wound healing, we obtained mice expressing a conditional allele of Notch1 (Notch1flox/flox) (20) and crossed them with mice expressing Cre recombinase under the control of the myeloid-specific lysozyme M promoter (LysMCre) (21) to induce myeloid-specific Notch1 deletion. LysMCre;Notch1flox/flox mice are viable, survive to adulthood, and have no gross developmental defects or deficiency in offspring viability compared with wild-type littermates. We performed a wounding assay with LysMCre;Notch1flox/flox mice and control littermates that express the conditional allele of Notch1 but not the LysMCre transgene. We observed a nearly 40% decrease in macrophage recruitment to wound tissue at postinjury day 5 in LysMCre;Notch1flox/flox mice compared with control (Fig. 4A–C). Furthermore, wounds in LysMCre;Notch1flox/flox mice were characterized by decreased TNF-α expression compared with control (Fig. 4D–F). We costained for F4/80 and TNF-α in wound tissue and found decreased infiltration of TNF-α+ macrophages in wounds in LysMCre; Notch1flox/flox mice compared with control (Fig. 4G, 4H). In contrast to wounds in Notch1+/− mice where angiogenesis and collagen deposition increased (Fig. 3), angiogenesis and collagen deposition was relatively unchanged in wounds in LysMCre;Notch1flox/flox mice compared with control (data not shown), suggesting that Notch functions in nonmyeloid cells to affect these processes. However, these data demonstrate that Notch1 expression in the myeloid lineage in mice contributes to both macrophage recruitment and TNF-α expression during wound healing.
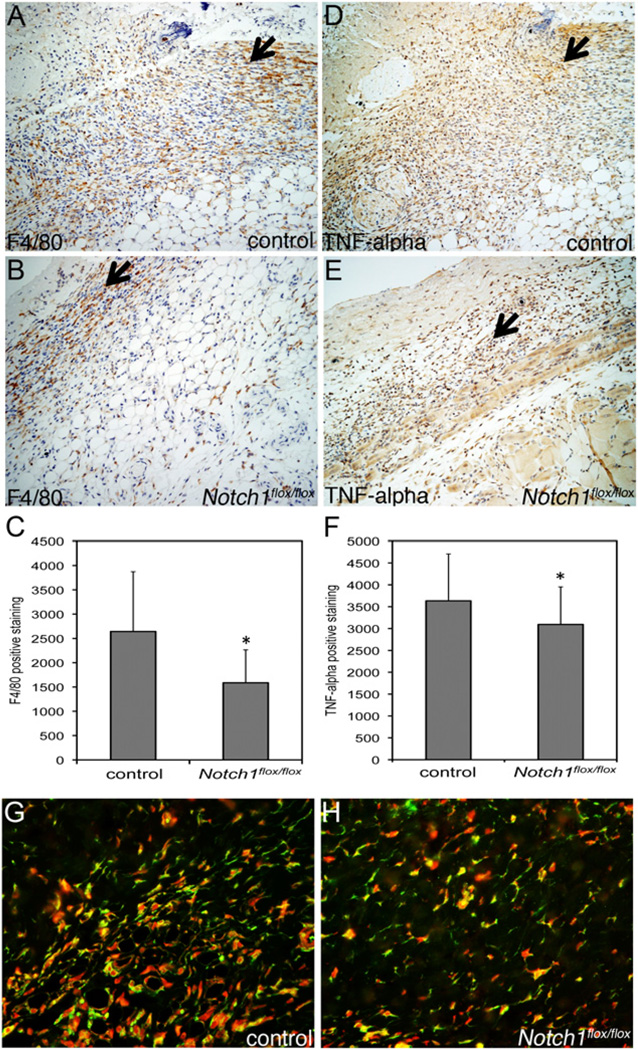
FIGURE 4.
Decreased macrophage recruitment and TNF-α expression in wounds from mice with myeloid-specific loss of Notch1. Day 5 wound tissue was harvested from LysMCre;Notch1flox/flox (B, E, H) or control (A, D, G) mice. Representative images of wound tissue staining for F4/80 (A, B) and TNF-α (D, E) at edge of re-epithelialization. Original magnification ×20. Arrows indicate areas of F4/80 and TNF-α staining. Quantification of F4/80 (C) and TNF-α (F) staining. G and H, Costaining of F4/80 (green) and TNF-α (red) in day 5 wound tissue. Original magnification ×20. *p < 0.05 relative to control.
Notch signaling is induced in BMM response to LPS/IFN-γ or IL-4
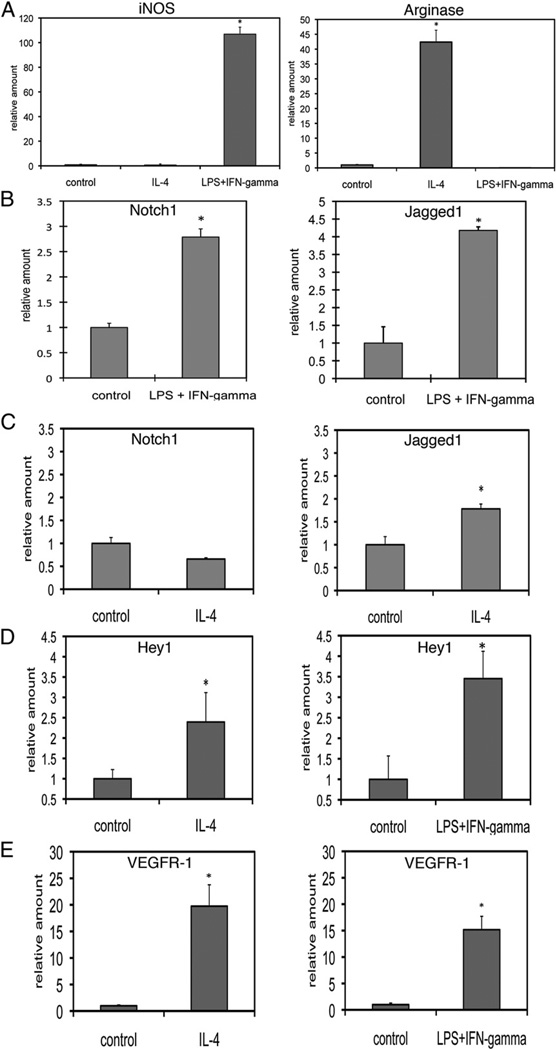
Because we observed changes in macrophage recruitment and function in Notch-deficient mice, we wanted to characterize Notch expression and signaling in isolated macrophages in the context of stimulation. Previously published studies used the immortalized mouse macrophage cell line RAW 264.7 and showed that LPS and IFN-γ increased expression of Notch1 and Jagged1 (13). Because RAW 264.7 cells are immortalized, and Notch signaling has been reported to interact with proteins that regulate the cell cycle (31, 32), we chose to explore the role of Notch signaling in cultured primary mouse BMM. We used LPS/IFN-γ or IL-4 to induce BMM toward differentiation states that approximate the M1 (classically activated) or M2 (alternatively activated) phenotype, respectively. After overnight incubation, RNA was harvested for reverse transcription and quantitative RT-PCR. LPS/IFN-γ–mediated differentiation of BMM toward the M1 phenotype was associated with upregulation of iNOS, whereas differentiation of BMM toward the M2 phenotype by IL-4 was associated with an increase in arginase expression (Fig. 5A). We found that transcript levels of Notch1 and Jagged1 were induced in response to LPS/ IFN-γ in BMM, whereas only Jagged1 was induced in response to IL-4 (Fig. 5B, 5C). Activation of Notch signaling is associated with upregulation of Notch target genes, which include members of the hairy and enhancer of split (HES) and Hey family of transcriptional repressors (1). We detected increased expression of Hey1 in response to both LPS/IFN-γ and IL-4 (Fig. 5D), but expression of HES1 or Hey2 was below the level of detection (data not shown). As Hey1 is a target of Notch/CSL transactivation, these results suggest that stimulation of BMM with LPS/IFN-γ or IL-4 is associated with activation of canonical Notch signaling.
FIGURE 5.
Notch signaling is induced in BMM in response to LPS/IFN-γ and IL-4. BMM were stimulated with 100 ng/ml LPS and 100 U/ml IFN-γ or 5 ng/ml IL-4 overnight .Transcripts were assessed by quantitative RT-PCR. A, iNOS (left) and arginase (right) expression in response to LPS/IFN-γ or IL-4 stimulation. B, Stimulation with LPS/IFN-γ induced expression of Notch1 and Jagged1. C, Stimulation with IL-4 induced expression of Jagged1, but not Notch1. D, Both IL-4 (left) and LPS/IFN-γ (right) induced expression of the Notch target gene Hey1. E, Both IL-4 (left) and LPS/IFN-γ (right) induced expression VEGFR-1. Quantitative RT-PCR results represent reactions performed in triplicate and normalized to expression of P0 (6SD). Data are representative of at least three independent experiments. *p < 0.05 relative to control.
Additionally, transcript levels of VEGFR-1, which has been shown to be downstream of Notch signaling in endothelial cells (2–4) and is the only VEGF receptor expressed in monocytes and macrophages (5, 33), was also upregulated in response LPS/IFN-γ and IL-4 (Fig. 5E). Thus, evidence of Notch signal activation is seen in both classically activated (induced by LPS/IFN-γ) and alternatively activated (induced by IL-4) macrophages.
Notch mediates VEGFR-1 induction in response to LPS/IFN-γ, but not IL-4
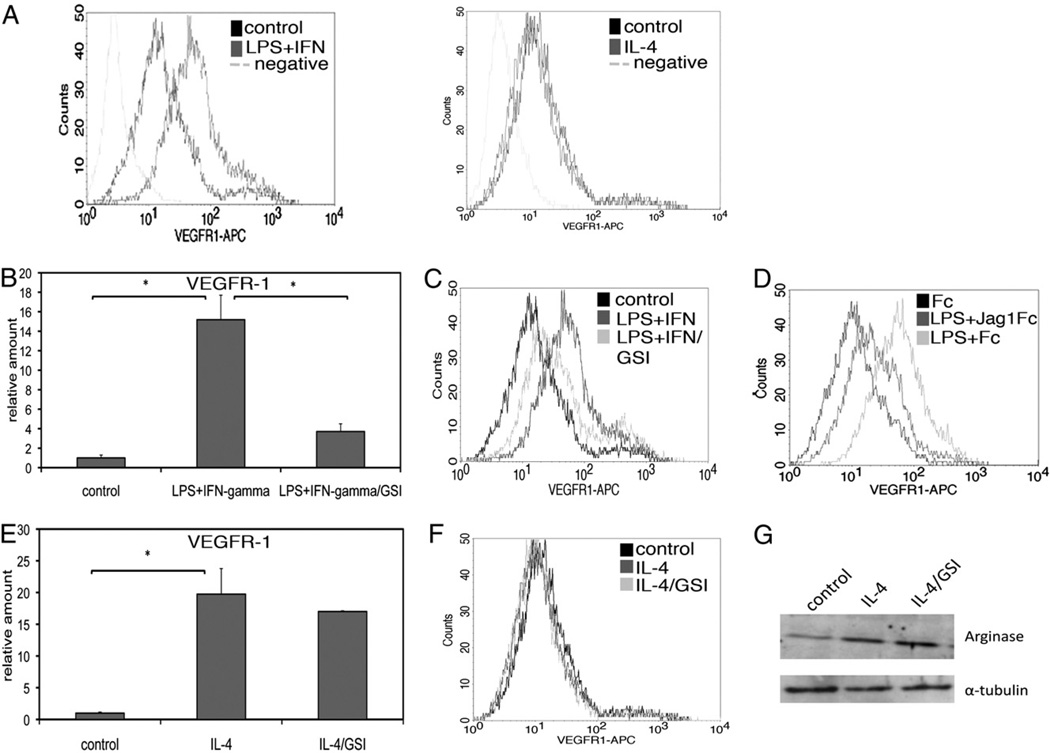
VEGFR-1 is important for recruitment and function of macrophages in areas of angiogenesis and inflammation (5–7). Our data demonstrating decreased macrophage recruitment to wounds in Notch1-deficient mice and the fact that Notch regulates VEGFR-1 expression in endothelial cells (2, 3) provided a rationale for determining whether Notch signaling regulates VEGFR-1 expression in mαcrophages. Despite similar induction of VEGFR-1 transcripts (Fig. 5E), surface expression of VEGFR-1 was increased in BMM treated with LPS/IFN-γ, but not those treated with IL-4, as shown by flow cytometry (Fig. 6A). We investigated whether VEGFR-1 is downstream of Notch signaling using compound E, a γ-secretase inhibitor (GSI) that prevents cleavage and nuclear translocation of Notch receptors, thus inhibiting Notch signaling. Induction of transcripts and surface expression of VEGFR-1 by LPS/IFN-γ was decreased when BMM were coincubated with 400 nM GSI (Fig. 6B, 6C). Because of the possibility of GSI affecting substrates other than Notch, we also treated BMM with conditioned medium containing the extracellular domain of Jagged1 (Jag1Fc), which prevents endogenous ligand binding and inhibits Notch signaling, as verified by a luciferase reporter assay using a CSL-dependent construct (data not shown). Incubation of BMM with Jag1Fc inhibited the induction of VEGFR-1 in response to LPS (Fig. 6D), indicating that Notch receptor-ligand interactions are necessary for VEGFR-1 expression in response to stimulation in these cells. This is also supported by the observation that cells plated at high density, where cell–cell contact is maximized, have enhanced induction of VEGFR-1, compared with cells seeded at lower densities (data not shown). In contrast, transcript levels of VEGFR-1 in response to IL-4 were unaffected by GSI (Fig. 6E), and surface expression of VEGFR-1 was only slightly decreased (Fig. 6F). Protein expression of arginase was induced in response to IL-4 but unaffected by Notch inhibition with GSI (Fig. 6G). These results suggest that protein expression of VEGFR-1 is not associated with the M2 phenotype. Although Notch signaling is induced by IL-4 treatment (Fig. 5D), the induction of VEGFR-1 transcripts by IL-4 is not dependent on Notch signal activation. However, Notch signaling is used to regulate VEGFR-1 downstream of LPS/IFN-γ.
FIGURE 6.
Notch mediates VEGFR-1 induction in response to LPS/IFN-γ, but not IL-4. A, Flow cytometry of BMM after overnight stimulation showed increased surface expression of VEGFR-1 in response to LPS/IFN-γ (left), but not IL-4 (right). Coincubation of BMM with 400 nM GSI inhibited induction of transcript (B) and protein (C) levels of VEGFR-1 after overnight stimulation with LPS/IFN-γ. D, Incubation with Jag1Fc-conditioned medium inhibited induction of VEGFR-1 in response to LPS. E, Induction of transcript levels of VEGFR-1 in response to IL-4 was unaffected by GSI. F, Surface expression of VEGFR-1 was unaffected by IL-4, but coincubation with GSI led to a slight decrease in VEGFR-1 expression. G, Immunoblotting for arginase expression in whole-cell protein extracts from IL-4–stimulated BMM, with or without GSI (upper panel). The same membrane was probed for α-tubulin as a loading control (lower panel). Quantitative RT-PCR results represent reactions performed in triplicate and normalized to expression of P0 (±SD). Data are representative of at least three independent experiments. *p < 0.05 relative to control.
Notch signaling regulates baseline levels of VEGFR-1 in BMM
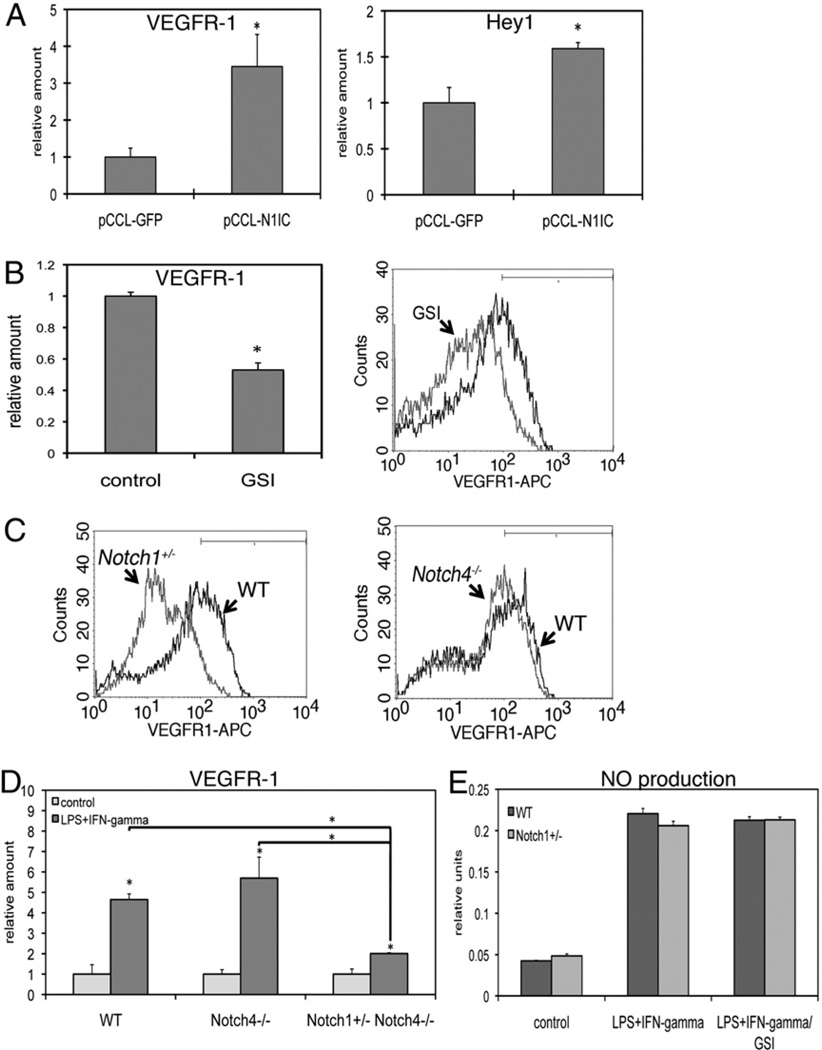
To determine if Notch signal activation could induce expression of VEGFR-1, we used a lentiviral vector to express the N1IC in BMM. This N1IC construct results in cleavage-independent, constitutive activation of Notch signaling. Quantitative RT-PCR demonstrated that expression of N1IC in BMM induced expression of VEGFR-1 as well as the Notch target gene Hey1, further indicating that VEGFR-1 is downstream of Notch1 signaling (Fig. 7A). We next investigated whether Notch signaling maintains levels of VEGFR-1 in BMM in the absence of stimulation. We treated unstimulated BMM with GSI and found decreased VEGFR-1 expression at both the transcript and protein level compared with control (Fig. 7B). Thus, maintenance of VEGFR-1 expression in resting macrophages is dependent on γ-secretase activity, suggesting a dependency on Notch signaling.
FIGURE 7.
Notch1, not Notch4, regulates baseline levels of VEGFR-1 in BMM. A, A lentiviral construct encoding the constitutively active N1IC was transduced into BMM and transcripts were assessed by quantitative RT-PCR 48 h postinfection. N1IC trans-duction led to increased VEGFR-1 expression (left), as well as increased expression of the Notch target gene Hey1 (right). B, Incubation of resting BMM with GSI overnight led to decreased transcript (left) and protein (right) levels of VEGFR-1. C, BMM from Notch mutant mice were analyzed for VEGFR-1 expression by flow cytometry. Notch1+/− BMM had decreased surface expression of VEGFR-1 (left), whereas VEGFR-1 expression in Notch4−/− BMM was largely unchanged (right) compared with WT BMM. D, Quantitative RT-PCR demonstrated that BMM from Notch4−/− mice induce VEGFR-1 in response to LPS/IFN-γ to a similar degree as do WT BMM, whereas loss of Notch1 leads to diminished induction of VEGFR-1. E, Production of NO in culture supernatant from WT or Notch1+/− BMM was assessed by Griess reagent. Cells were stimulated overnight with LPS/IFN-γ. Some cells were coincubated with GSI. Quantitative RT-PCR results represent reactions performed in triplicate and normalized to expression of P0 (±SD). Data are representative of at least three independent experiments. *p < 0.05 relative to control. WT, wild-type.
Notch1, not Notch4, maintains levels of VEGFR-1 in BMM
We next wanted to determine whether genetic loss of Notch affects VEGFR-1 expression in macrophages. To do this, we cultured BMM from Notch mutant mice and assessed expression of VEGFR-1. BMM from hematopoietic stem cells isolated from Notch mutant mice were able to be cultured (data not shown). We detected decreased expression of VEGFR-1 in unstimulated BMM from Notch1+/− mice compared with wild-type littermates by flow cytometry (Fig. 7C, left). However, BMM from Notch4−/− mice had only slightly decreased expression of VEGFR-1 compared with wild-type littermates (Fig. 7C, right), demonstrating that Notch4 is dispensable for maintaining VEGFR-1 expression in these cells. Furthermore, while induction of VEGFR-1 in response to LPS/IFN-γ in Notch4−/− BMM was comparable to wild-type BMM, induction of VEGFR-1 in Notch1+/−Notch4−/− BMM was significantly attenuated (Fig. 7D). However, NOS2 activity, as demonstrated by production of NO, was similar between wild-type and Notch1+/− BMM and was unaffected by coincubation with GSI (Fig. 7E). Thus, both pharmacological (Fig. 6) and genetic (Fig. 7) evidence demonstrated that Notch1 regulates VEGFR-1 expression in macrophages and is necessary for induction of VEGFR-1 in response to LPS/IFN-γ.
Decreased Notch signaling and VEGFR-1 expression in circulating monocytes from Notch1+/− mice
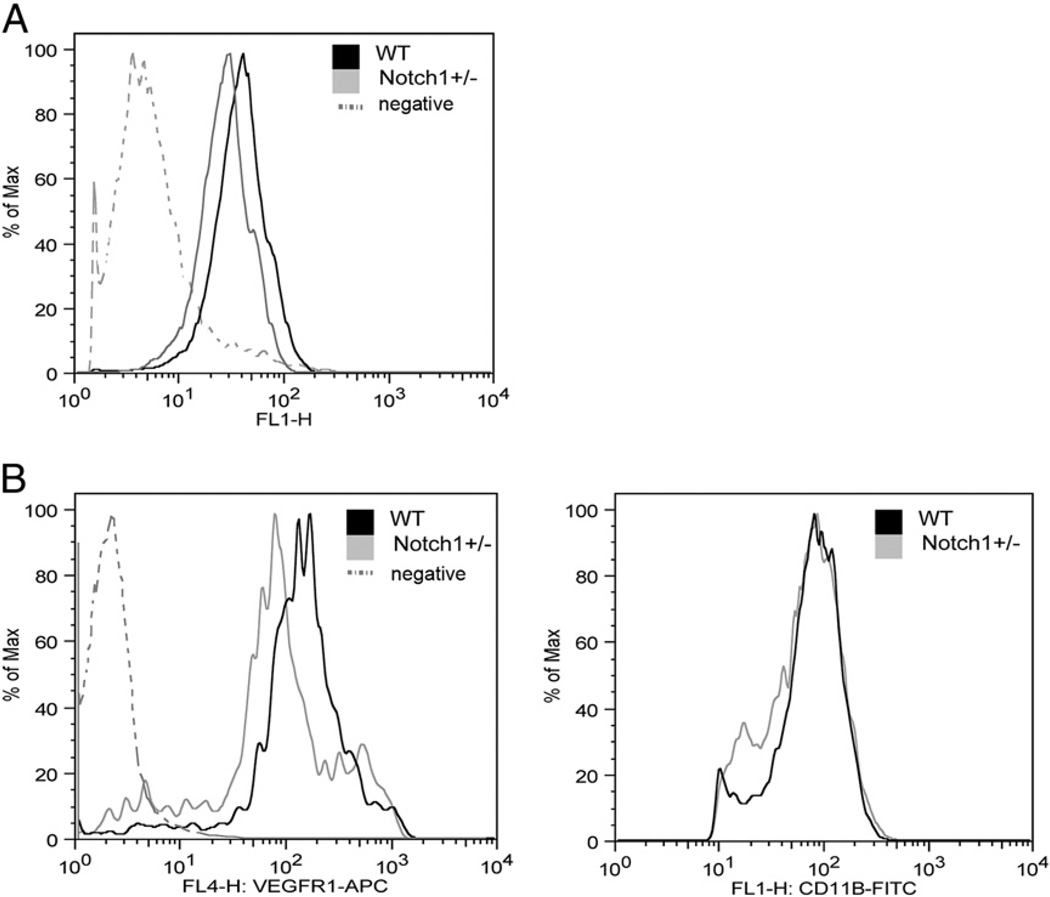
We next established that Notch signaling is active in peripheral blood leukocytes by performing flow cytometry on cells from transgenic Notch reporter mice that express GFP under the control of a CSL-responsive promoter (22). We crossed these transgenic mice into the Notch1 mutant background and found that GFP expression was decreased in peripheral blood leukocytes from Notch1+/− transgenic mice compared with leukocytes from transgenic mice with both wild-type Notch1 alleles (Fig. 8A). Based on our results in BMM, we hypothesized that circulating monocytes in the peripheral blood of Notch1+/− mice would have decreased VEGFR-1 expression. To assess this, peripheral blood leukocytes from Notch1+/− mice or wild-type littermates were stained for VEGFR-1 and assessed by flow cytometry. Using gating for the monocytic population by side and forward scatter properties, we found that circulating monocytes from Notch1+/− mice express less VEGFR-1 than do monocytes from wild-type littermates (Fig. 8B, left). As a control, we found that expression of the myeloid marker CD11b was unchanged (Fig. 8B, right). The number of circulating monocytes in Notch1+/− mice was not affected (data not shown). Thus, peripheral blood leukocytes from in Notch1+/− mice display reduced Notch reporter activity and decreased VEGFR-1 compared with wild-type cells.
FIGURE 8.
Decreased Notch signaling and VEGFR-1 expression in circulating monocytes from Notch1+/− mice. A, Notch signaling was assessed in peripheral blood leukocytes from Notch reporter mice harboring a CSL-responsive GFP transgene using flow cytometry to detect GFP expression. B, Circulating monocytes from Notch1+/− mice expressed decreased VEGFR-1 compared with WT littermates (left), whereas expression of CD11b was unchanged (right). WT, wild-type.
Notch signaling regulates inflammatory cytokines in BMM
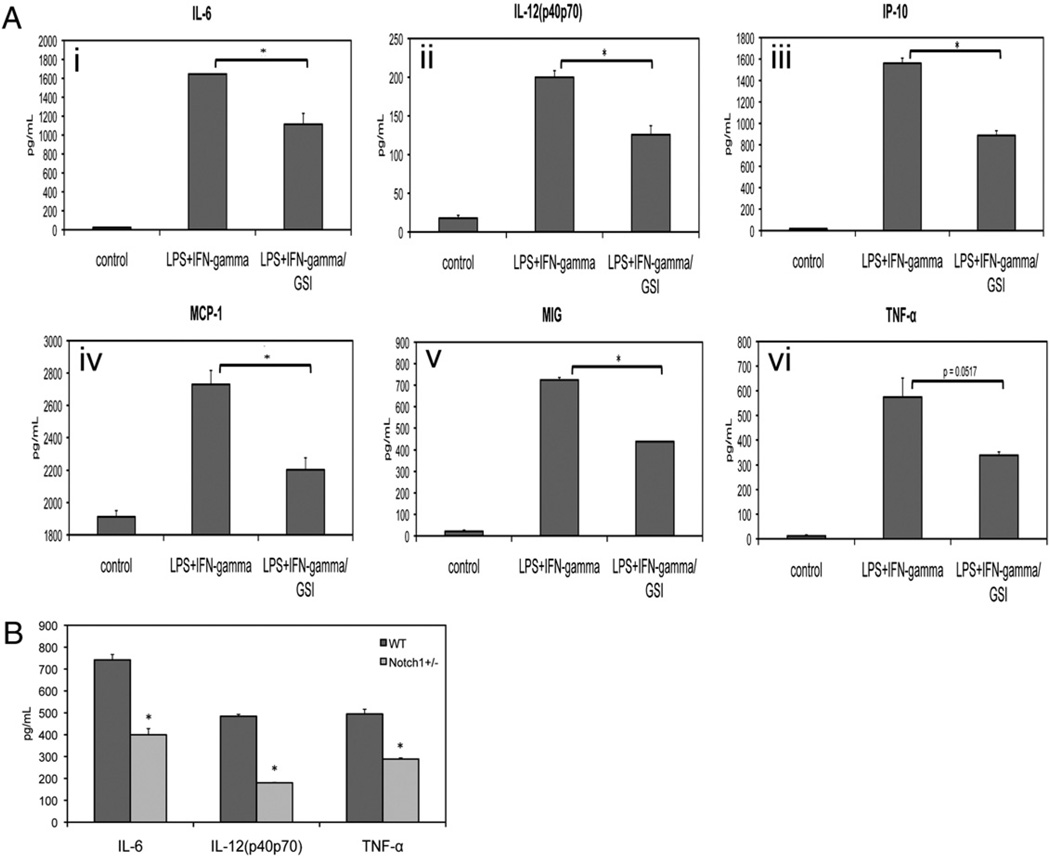
Our wounding assays showed decreased expression of TNF-α in wounds in Notch1 mutant mice compared with wild-type littermates. We used BMM to explore the effect of Notch inhibition on a broad panel of inflammatory cytokines. We stimulated BMM with LPS/IFN-γ, then collected culture supernatants and assessed cytokine levels using a bead-based ELISA assay. Increased levels of IL-6, IL-12, CXCL10 (IP-10), MCP-1, MIG, and TNF-α were detected in the supernatant of stimulated BMM. The induction of these cytokines upon stimulation was diminished when Notch signaling was inhibited with GSI (Fig. 9A), establishing a role for Notch in the regulation of IL-6, IL-12, IP-10, MCP-1, MIG, and TNF-α. Levels of IL-1-b, IL-10, CXCL1, and MIP-1a were also increased in response to LPS/IFN-γ stimulation, but secretion of these cytokines was not affected by Notch inhibition with GSI (data not shown). None of these inflammatory cytokines was upregulated in response to IL-4 (data not shown). We next investigated whether induction of inflammatory cytokines would be decreased in BMM from Notch1+/− mice. We found that levels of LPS/IFN-γ–induced IL-6, IL-12, and TNF-α were lower in culture supernatants from Notch1+/− BMM compared with BMM from wild-type littermates (Fig. 9B). These results confirm and extend previous data demonstrating a role for Notch signaling in regulating the inflammatory response (16, 34) and provide genetic evidence that signaling by the Notch1 receptor is necessary for optimal inflammatory cytokine response in macrophages.
FIGURE 9.
Notch signaling regulates inflammatory cytokines in BMM. A, BMM were stimulated with LPS/IFN-γ overnight and culture supernatants were analyzed for cytokine secretion using bead-based ELISA. Coincubation of stimulated BMM with GSI led to decreased secretion of (i) IL-6, (ii) IL-12, (iii) IP-10, (iv) MCP-1, (v) MIG, and (vi) TNF-α. B, BMM from Notch1+/−mice had decreased induction of IL-6, IL-12, and TNF-α in response to LPS/IFN-γ. Data are representative of at least three independent experiments. Analysis of cytokine secretion was performed in duplicate for each sample. *p < 0.05 relative to control.
Discussion
We explored the functional consequence of loss of Notch1 expression or activity in macrophages in the context of inflammation. In an excisional wound healing assay, we found that mice mutant for Notch1 have an altered inflammatory response during wound healing compared with wild-type littermates. Normal wound healing goes through three overlapping phases: inflammation, proliferation, and remodeling (26). We focused on time points during the inflammatory and proliferative phases, where macrophage function is most pronounced. Macrophage recruitment to wounds was delayed at postinjury day 5 in Notch1+/− mice, and we observed decreased expression of TNF-α in macrophages in Notch1+/− wounds. Interestingly, several aspects of wound healing improved over time when compared with wild-type mice. We found that wounds in Notch1+/− mice had increased collagen deposition and angiogenesis, and they exhibited less shearing during wound harvest. Thus, we provide the first evidence, to our knowledge, of decreased inflammatory response and increased collagen deposition and angiogenesis in wounds in Notch1+/− mice.
The effects of loss of Notch1 on wound healing are likely to be multifactorial in this model, as Notch may affect the function of multiple cell types that would be expected to play a role in wound healing. This is validated by the finding that mice with myeloid-specific deletion of Notch1 display decreased macrophage recruitment and decreased TNF-α expression in wounds, but unchanged vascular density or collagen content. It is likely that the changes in angiogenesis observed in Notch1+/− wounds are due to loss of Notch1 in endothelial cells, where Notch function in endothelial cell sprouting and growth has been well documented (2, 4, 28, 29). These findings are consistent with a model whereby Notch signaling results in reduced growth of endothelium as part of a feedback loop downstream of VEGF signaling (reviewed by Thurston and Kitajewski in Ref. 35). In contrast, a previously published report showed delayed wound healing and decreased angiogenesis in mice expressing a Notch1 antisense sequence, or mice treated topically with the g-secretase inhibitor DAPT (36). Unlike evaluation of reduced Notch1 activity due to heterozygosity or myeloid-specific knockout reported in this study, treatment of mice with antisense sequences or γ-secretase inhibitors may have phenotypes associated with targets other than Notch. This discrepancy could be also due to differences in experimental methods or to less efficient decrease in Notch1 expression in the skin of Notch1 antisense mice (∼50% of wild-type levels) (36). Importantly, our finding that wounds in Notch1+/− mice have increased collagen content is the first genetic evidence, to our knowledge, of a role for Notch in extracellular matrix homeostasis during inflammation. Thus, Notch may function in additional cell types, such as fibroblasts, that are associated with wound healing. Taken together, our genetic models have established a specific role for Notch1 in macrophage recruitment and function during wound healing, but further study is needed to elaborate on how Notch1 in other cell types may contribute to wound resolution.
We also assessed Notch activity in macrophages in response to classical (M1) or alternative (M2) activation. Classically activated macrophages induced by LPS/IFN-γ upregulated surface expression of VEGFR-1 in a Notch-dependent manner. Although macrophages in culture express both Notch1 and Notch4 (10), we found that Notch1 is important for the regulation of VEGFR-1 expression in response to stimulation and in maintaining baseline levels of VEGFR-1, whereas Notch4 is dispensable for these functions. Notch signaling was not required for production of NO in response to LPS/IFN-γ. In contrast to classically activated macrophages, we found that alternatively activated macrophages induced by IL-4 have evidence of Notch signal activation but did not upregulate surface expression of VEGFR-1, suggesting that this is not a target of Notch activation in this setting. Additionally, upregulation of arginase in response to IL-4 was not compromised by Notch inhibition. Our results support the hypothesis that increased expression of VEGFR-1 is indicative of classically activated macrophages, but not alternatively activated macrophages. However, Notch activity was not found to be solely indicative of the M1 or M2 phenotype, but it appears to be activated in both settings based on evaluation of the Notch target gene Hey1.
We have provided evidence for the importance of Notch signaling for cytokines expressed in classically activated macrophages. Our data show that Notch inhibition results in diminished induction of IL-6, IL-12, IP-10, MCP-1, MIG, and TNF-α in response to LPS/IFN-γ. Previous reports have indicated cooperation between TLR and Notch pathways and have suggested an additional regulatory mechanism by which IFN-γ mediates a feedback loop that inhibits further activation of Notch targets (16). Nonetheless, stimulation with both LPS and IFN-γ induced Notch activation in our assays, as has been demonstrated by others (13, 34), and inhibition of Notch signaling with GSI was able to partially block the expression of target genes in response to stimulation. The cytokine response to Notch activation in macrophages is clearly complicated, with regulation of several distinct cytokines. It will be important to determine which, if any, of these genes are directly regulated by Notch, the HES/Hey genes, or by interactions with other pathways.
Our data provide genetic evidence that Notch1 plays a role in regulating the inflammatory response in macrophages, and that macrophages from Notch1 mutant mice are deficient in their ability to respond to inflammatory cues. We hypothesize that the decrease in macrophage recruitment to wounds in Notch1 mutant mice is due, at least in part, to decreased VEGFR-1 and inflammatory cytokine expression. Notch1, and not Notch4, is more important for these functions, as expression of VEGFR-1 and macrophage recruitment to wounds were largely unaltered in Notch4−/− mice (Fig. 7 and data not shown). Studies in mice have shown that VEGFR-1 signaling is critical for macrophage recruitment to areas of inflammation and angiogenesis and is also necessary for secretion of cytokines and macrophage function at these sites (6, 37). Our studies offer the possibility that therapeutic inhibition of Notch signaling may result in decreased macrophage recruitment and altered macrophage function in a variety of settings.
Although several studies have documented that decreased macrophage recruitment to wounds impairs tissue repair (38–40), other reports have raised questions as to whether macrophage-mediated inflammation at the wound site is necessary or conducive for wound healing (30, 41, 42). Because of the release of inflammatory cytokines, accumulation of macrophages at the wound site may be associated with tissue destruction that prevents the proliferation and remodeling that is necessary for wound resolution. Accordingly, the activation state of infiltrating macrophages plays an important role in whether they promote or prevent wound healing, and inflammation, although necessary to prevent infection, may do more harm than good in terms of wound resolution (26). Using Notch1 heterozygous mice as well as mice with myeloid-specific loss of Notch1, we have shown that Notch1 is important for recruitment of TNF-α– expressing inflammatory macrophages to wounds. Our data suggest that Notch1 plays an important role in classically activated M1 macrophages, a finding that has implications in a variety of settings, including in tumor-associated macrophages, where Notch signaling has been implicated for antitumor function (43). Given the widespread expression of Notch ligands and receptors, it is likely that interactions between macrophages and other cells in their micro-environment, including with each other, may induce activation of Notch signaling and expression of a variety of downstream targets that affect macrophage function.
Acknowledgments
We thank Carrie Shawber for assistance in editing this manuscript and the Diabetes and Endocrinology Research Center Phenotyping Core at Columbia University for Luminex analysis.
This work was supported in part by National Institutes of Health Grants R01HL62454 and R01CA136673 (to J.K.) and F31HL090032 (to H.H.O.).
Abbreviations used in this paper
- BMM
bone marrow-derived macrophage
- GSI
γ-secretase inhibitor
- HES
hairy and enhancer of split
- iNOS
inducible NO synthase
- MIG
monokine induced by IFN-γ
- N1IC
intracellular domain of Notch1
- VEGF
vascular endothelial growth factor
- VEGFR-1
vascular endothelial growth factor receptor-1
- VEGFR-2
vascular endothelial growth factor receptor-2
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler. Thromb. Vasc. Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 2.Funahashi Y, Shawber CJ, Vorontchikhina M, Sharma A, Outtz HH, Kitajewski J. Notch regulates the angiogenic response via induction of VEGFR-1. J. Angiogenes Res. 2010;2:3. doi: 10.1186/2040-2384-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington LS, Sainson RC, Williams CK, Taylor JM, Shi W, Li JL, Harris AL. Regulation of multiple angiogenic pathways by Dll4 and Notch in human umbilical vein endothelial cells. Microvasc. Res. 2008;75:144–154. doi: 10.1016/j.mvr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Suchting S, Freitas C, le Noble F, Benedito R, Brèant C, Duarte A, Eichmann A. The Notch ligand Delta-like 4 negatively regulates endo-thelial tip cell formation and vessel branching. Proc. Natl. Acad. Sci. USA. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barleon B, Sozzani S, Zhou D, Weich HA, Mantovani A, Marmé D. Migration of human monocytes in response to vascular endothelial growth factor (VEGF) is mediated via the VEGF receptor flt-1. Blood. 1996;87:3336–3343. [PubMed] [Google Scholar]
- 6.Luttun A, Tjwa M, Moons L, Wu Y, Angelillo-Scherrer A, Liao F, Nagy JA, Hooper A, Priller J, De Klerck B, et al. Revascularization of ischemic tissues by PlGF treatment, and inhibition of tumor angiogenesis, arthritis and atherosclerosis by anti-Flt1. Nat. Med. 2002;8:831–840. doi: 10.1038/nm731. [DOI] [PubMed] [Google Scholar]
- 7.Murakami M, Zheng Y, Hirashima M, Suda T, Morita Y, Ooehara J, Ema H, Fong GH, Shibuya M. VEGFR1 tyrosine kinase signaling promotes lymphangiogenesis as well as angiogenesis indirectly via macrophage recruitment. Arterioscler. Thromb. Vasc. Biol. 2008;28:658–664. doi: 10.1161/ATVBAHA.107.150433. [DOI] [PubMed] [Google Scholar]
- 8.Johnston DA, Dong B, Hughes CC. TNF induction of jagged-1 in endothelial cells is NFkB-dependent. Gene. 2009;435:36–44. doi: 10.1016/j.gene.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams CK, Segarra M, Sierra Mde. L, Sainson RC, Tosato G, Harris AL. Regulation of CXCR4 by the Notch ligand Delta-like 4 in endothelial cells. Cancer Res. 2008;68:1889–1895. doi: 10.1158/0008-5472.CAN-07-2181. [DOI] [PubMed] [Google Scholar]
- 10.Fung E, Tang SM, Canner JP, Morishige K, Arboleda-Velasquez JF, Cardoso AA, Carlesso N, Aster JC, Aikawa M. Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation. 2007;115:2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, Li LW, Yan Q, Petryniak B, Man Y, Su C, Shim J, Chervin S, Lowe JB. Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood. 2008;112:308–319. doi: 10.1182/blood-2007-11-115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.JÖnsson JI, Xiang Z, Pettersson M, Lardelli M, Nilsson G. Distinct and regulated expression of Notch receptors in hematopoietic lineages and during myeloid differentiation. Eur. J. Immunol. 2001;31:3240–3247. doi: 10.1002/1521-4141(200111)31:11<3240::aid-immu3240>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 13.Monsalve E, Pe´rez MA, Rubio A, Ruiz-Hidalgo MJ, Baladro´n V, Garcı´a-Ramírez JJ, GÓmez JC, Laborda J, Díaz-Guerra MJ. Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J. Immunol. 2006;176:5362–5373. doi: 10.4049/jimmunol.176.9.5362. [DOI] [PubMed] [Google Scholar]
- 14.Singh N, Phillips RA, Iscove NN, Egan SE. Expression of Notch receptors, Notch ligands, and fringe genes in hematopoiesis. Exp. Hematol. 2000;28:527–534. doi: 10.1016/s0301-472x(00)00146-6. [DOI] [PubMed] [Google Scholar]
- 15.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, Tateya T, Kang YJ, Han J, Gessler M, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-γ pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monsalve E, Ruiz-García A, Baladrόn V, Ruiz-Hidalgo MJ, Sánchez-Solana B, Rivero S, García-Ramírez JJ, Rubio A, Laborda J, Díaz-Guerra MJ. Notch1 upregulates LPS-induced macrophage activation by increasing NF-κB activity. Eur. J. Immunol. 2009;39:2556–2570. doi: 10.1002/eji.200838722. [DOI] [PubMed] [Google Scholar]
- 18.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 19.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 22.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 23.Zhao W, Cha EN, Lee C, Park CY, Schindler C. Stat2-dependent regulation of MHC class II expression. J. Immunol. 2007;179:463–471. doi: 10.4049/jimmunol.179.1.463. [DOI] [PubMed] [Google Scholar]
- 24.Tung JJ, Hobert O, Berryman M, Kitajewski J. Chloride intracellular channel 4 is involved in endothelial proliferation and morphogenesis in vitro. Angiogenesis. 2009;12:209–220. doi: 10.1007/s10456-009-9139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Classen A, Lloberas J, Celada A. Macrophage activation: classical versus alternative. Methods Mol. Biol. 2009;531:29–43. doi: 10.1007/978-1-59745-396-7_3. [DOI] [PubMed] [Google Scholar]
- 26.Martin P, Leibovich SJ. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 2005;15:599–607. doi: 10.1016/j.tcb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J. Invest. Dermatol. 2007;127:514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 28.Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 29.Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 30.Mori R, Kondo T, Ohshima T, Ishida Y, Mukaida N. Accelerated wound healing in tumor necrosis factor receptor p55-deficient mice with reduced leukocyte infiltration. FASEB J. 2002;16:963–974. doi: 10.1096/fj.01-0776com. [DOI] [PubMed] [Google Scholar]
- 31.Noseda M, Niessen K, McLean G, Chang L, Karsan A. Notch-dependent cell cycle arrest is associated with downregulation of minichromosome maintenance proteins. Circ. Res. 2005;97:102–104. doi: 10.1161/01.RES.0000174380.06673.81. [DOI] [PubMed] [Google Scholar]
- 32.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawano A, Iwai S, Sakurai Y, Ito M, Shitara K, Nakahata T, Shibuya M. Flt-1, vascular endothelial growth factor receptor 1, is a novel cell surface marker for the lineage of monocyte-macrophages in humans. Blood. 2001;97:785–791. doi: 10.1182/blood.v97.3.785. [DOI] [PubMed] [Google Scholar]
- 34.Palaga T, Buranaruk C, Rengpipat S, Fauq AH, Golde TE, Kaufmann SH, Osborne BA. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur. J. Immunol. 2008;38:174–183. doi: 10.1002/eji.200636999. [DOI] [PubMed] [Google Scholar]
- 35.Thurston G, Kitajewski J. VEGF and Delta-Notch: interacting signalling pathways in tumour angiogenesis. Br. J. Cancer. 2008;99:1204–1209. doi: 10.1038/sj.bjc.6604484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chigurupati S, Arumugam TV, Son TG, Lathia JD, Jameel S, Mughal MR, Tang SC, Jo DG, Camandola S, Giunta M, et al. Involvement of Notch signaling in wound healing. PLoS ONE. 2007;2:e1167. doi: 10.1371/journal.pone.0001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami M, Iwai S, Hiratsuka S, Yamauchi M, Nakamura K, Iwakura Y, Shibuya M. Signaling of vascular endothelial growth factor receptor-1 tyrosine kinase promotes rheumatoid arthritis through activation of monocytes/ macrophages. Blood. 2006;108:1849–1856. doi: 10.1182/blood-2006-04-016030. [DOI] [PubMed] [Google Scholar]
- 38.Goren I, Allmann N, Yogev N, Schürmann C, Linke A, Holdener M, Waisman A, Pfeilschifter J, Frank S. A transgenic mouse model of inducible macrophage depletion: effects of diphtheria toxin-driven lysozyme M-specific cell lineage ablation on wound inflammatory, angiogenic, and contractive processes. Am. J. Pathol. 2009;175:132–147. doi: 10.2353/ajpath.2009.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirza R, DiPietro LA, Koh TJ. Selective and specific macrophage ablation is detrimental to wound healing in mice. Am. J. Pathol. 2009;175:2454–2462. doi: 10.2353/ajpath.2009.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagaoka T, Kaburagi Y, Hamaguchi Y, Hasegawa M, Takehara K, Steeber DA, Tedder TF, Sato S. Delayed wound healing in the absence of intercellular adhesion molecule-1 or L-selectin expression. Am. J. Pathol. 2000;157:237–247. doi: 10.1016/S0002-9440(10)64534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin P, D’Souza D, Martin J, Grose R, Cooper L, Maki R, McKercher SR. Wound healing in the PU.1 null mouse: tissue repair is not dependent on inflammatory cells. Curr. Biol. 2003;13:1122–1128. doi: 10.1016/s0960-9822(03)00396-8. [DOI] [PubMed] [Google Scholar]
- 42.Ashcroft GS, Yang X, Glick AB, Weinstein M, Letterio JL, Mizel DE, Anzano M, Greenwell-Wild T, Wahl SM, Deng C, Roberts AB. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat. Cell Biol. 1999;1:260–266. doi: 10.1038/12971. [DOI] [PubMed] [Google Scholar]
- 43.Wang YC, He F, Feng F, Liu XW, Dong GY, Qin HY, Hu XB, Zheng MH, Liang L, Feng L, et al. Notch signaling determines the M1 versus M2 polarization of macrophages in antitumor immune responses. Cancer Res. 2010;70:4840–4849. doi: 10.1158/0008-5472.CAN-10-0269. [DOI] [PubMed] [Google Scholar]