Abstract
The cytoplasmic receptor NOD2 (nucleotide-binding oligomerization domain 2) senses peptidoglycan fragments and triggers host defense pathways that lead to inflammatory immune responses. Dysregulation of NOD2 signaling is associated with inflammatory diseases, such as Crohn’s disease and Blau syndrome. We used a genome-wide, small interfering RNA (siRNA) screen to identify regulators of the NOD2 signaling pathway. Several genes associated with Crohn’s disease risk were identified in the screen, supporting a role for NOD2 and nuclear factor κB (NF-κB) pathways in the pathogenesis of Crohn’s disease. A comparison of hits from this screen with other “omics” data sets revealed interconnected networks of genes implicated in NF-κB signaling. Secondary assays, including the measurement of interleukin-8 secretion, served to validate many of the regulators. Knockdown of putative regulators in HEK293 cells followed by stimulation with tumor necrosis factor α revealed that most of the genes identified were general regulators of NF-κB signaling. Overall, the genes identified here provide a resource to facilitate the elucidation of the molecular mechanisms that regulate NOD2- and NF-κB–mediated inflammation.
INTRODUCTION
To combat microbial infections, mammals mount a sophisticated immune response. Initial recognition of microorganisms by the innate immune system is mediated by various hostencoded pattern recognition receptors (PRRs). In many cases, PRRs stimulate both antimicrobial and pro-inflammatory responses. The nucleotide-binding oligomerization domain (NOD)-like receptor family of intracellular PRRs consists of multidomain scaffolding proteins, many of which are dysregulated in inflammatory diseases (1). NOD2 is found in various cell types, including monocytes and intestinal epithelial cells, and it is capable of sensing bacterial peptidoglycan fragments containing muramyl dipeptide (MDP) (2, 3). Upon sensing MDP, NOD2 undergoes a conformational change that leads to the recruitment of receptor-interacting protein serine-threonine kinase 2 (RIPK2), a step that is essential for the activation of downstream signaling (4). Subsequently, Lys63-linked poly-ubiquitination of RIPK2 and nuclear factor κB (NF-κB) essential modulator (NEMO), a regulatory subunit of the inhibitor of κB (IκB) kinase (IKK) complex, leads to the recruitment of transforming growth factor–β (TGF-β)-activating kinase 1 (TAK1), a kinase that is required for the activation of mitogen-activated protein kinases (MAPKs) and NF-κB signaling (5–7). Together, these pathways trigger a potent antimicrobial and inflammatory host response.
The role of NOD2 in maintaining inflammatory homeostasis is underscored by the association of loss-of-function NOD2 mutations with increased susceptibility to Crohn’s disease, an inflammatory disease of the gastrointestinal tract (8, 9). Conversely, gain-of-function mutations in NOD2 are associated with Blau syndrome and early onset sarcoidosis, two relatively rare disorders characterized by inflammation of the eyes, skin, and joints (10, 11). Genetic variation in the gene encoding NOD2 is, of all of the loci identified, the strongest known genetic risk factor in the development of Crohn’s disease. Because more than 70 loci have been identified that predispose to Crohn’s disease (12), it is possible that some of the genes associated with Crohn’s disease are involved in the regulation of the NOD2 signaling pathway. Thus, increased understanding of the NOD2 signaling pathway may provide insight into the pathogenesis of Crohn’s disease.
Genomewide, small interfering RNA (siRNA) screens are effective tools for identifying previously uncharacterized modulators of biological processes. Using a human cell line expressing human NOD2 and an NF-κB–responsive luciferase reporter, we performed a genome-wide siRNA screen aimed at identifying regulators of the NOD2 signaling pathway. Our screen revealed many regulators of the NOD2 and NF-κB signaling pathways, including previously uncharacterized networks of positive and negative regulators. Additionally, we linked several genes associated with Crohn’s disease risk to the NOD2 signaling pathway. Together, our results provide a framework for understanding how gene networks contribute to the regulation of NOD2-mediated inflammatory signaling and provide insight into the genetic mechanisms of Crohn’s disease pathogenesis.
RESULTS
Genome-wide siRNA screen identifies regulators of NOD2 signaling
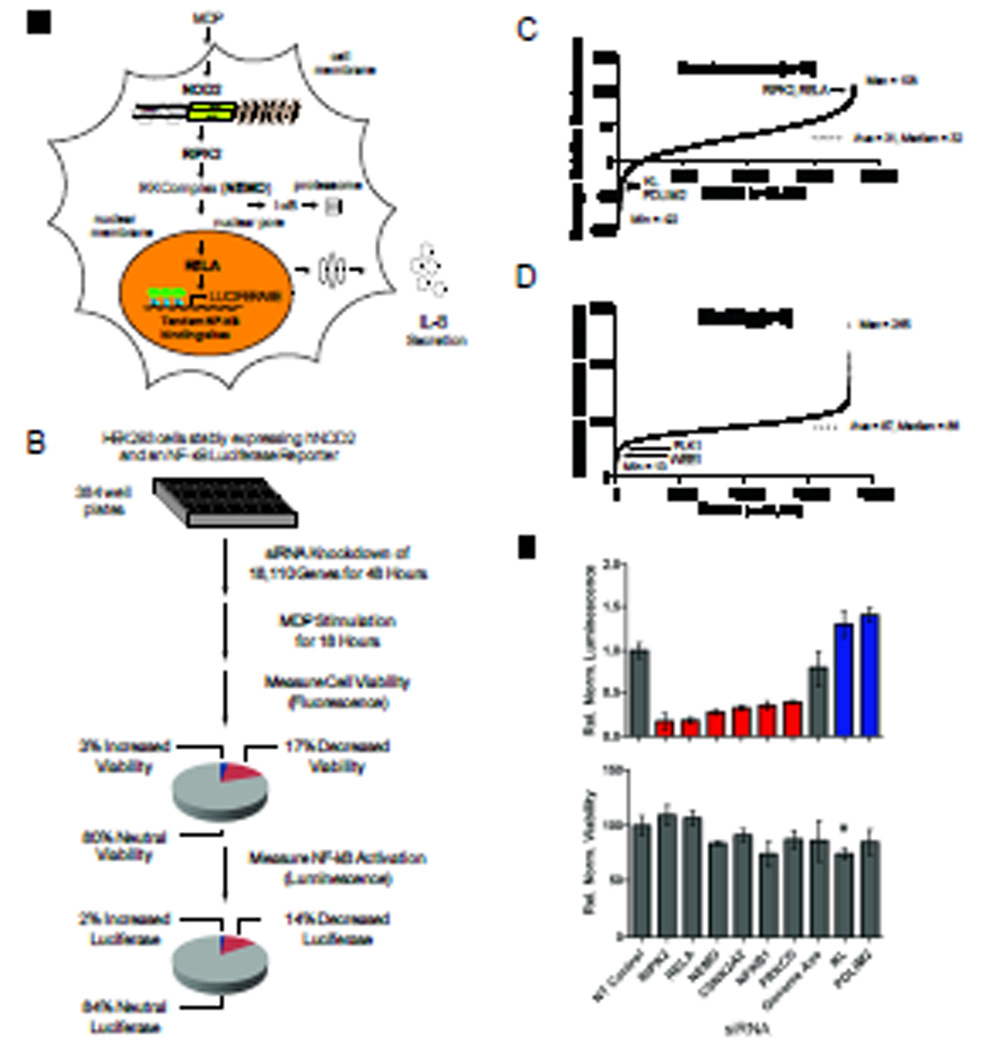
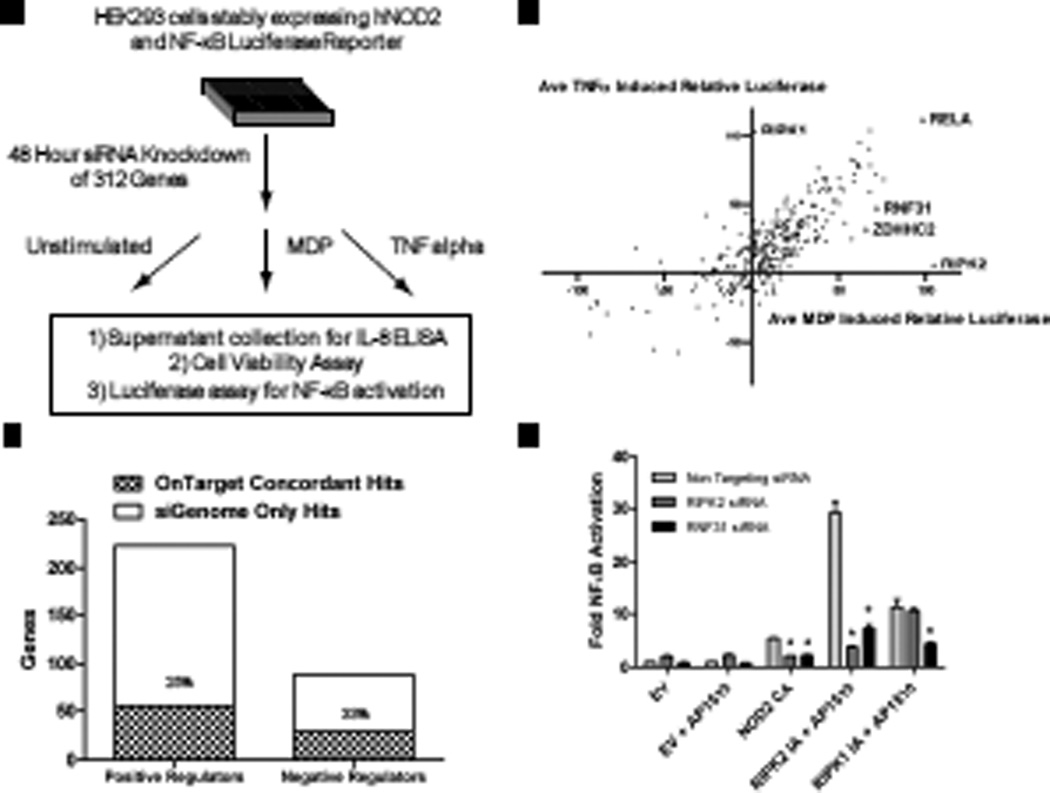
To identify regulators of the NOD2 signaling pathway, we performed a genome-wide siRNA screen. We engineered a luciferase-based reporter cell line derived from highly transfectable human embryonic kidney (HEK) 293 cells, and used this line to quantitatively measure NOD2-dependent signaling (Fig. 1A). HEK 293 cells are a well-established model for interrogating NOD2 signaling because they do not naturally have detectable amounts of NOD2 and they are normally unresponsive to stimulation with MDP (2). However, stimulation of HEK 293 cells stably expressing NOD2 with MDP resulted in NF-κB activation, phosphorylation of p38 MAPK, and secretion of the pro-inflammatory chemokine interleukin-8 (IL-8) (fig. S1). We developed a bioluminescence assay to monitor NF-κB activation with a stably incorporated luciferase reporter gene downstream of tandem copies of a consensus NF-κB–binding site. The screen was performed in triplicate with a commercially available siRNA library targeting 18,110 genes (Fig. 1B). The extent of luciferase activity after stimulation of cells with MDP was measured and displayed relative to that of cells transfected with control siRNAs present on each plate (table S1). On average, across all of the assay plates used in the screen, cells transfected with non-targeting (NT) siRNA and stimulated with MDP resulted in ~50-fold induction in luciferase activity. In comparison, the extent of luciferase induction upon silencing RIPK2, a serine and threonine protein kinase required for NOD2-dependent NF-κB activation (4) was on average ~7 fold, representing an 86% reduction in the extent of NOD2 signaling.
Fig. 1.
A genome-wide siRNA screen identifies regulators of NOD2 signaling. (A) Schematic representation of the reporter cell line engineered to quantitatively assess MDP-induced NOD2 signaling together with known components of the NOD2 signaling pathway. (B) The screen was carried out in triplicate with pools of four distinct siRNAs specific for each gene. Pie charts summarize the percentages of genes whose silencing affected cell viability and the percentages of genes not affecting viability that either decreased (red) or increased (blue) luminescence. (C) Rank order plot of the average normalized NF-κB luciferase reporter activity displayed relative to positive (RIPK2) and negative (non-targeting, NT) siRNA controls present on each assay plate. (D) Rank order plot of the average percentage cell viability displayed relative to NT siRNA and lysed cell controls present on each assay plate. (E) siRNA-mediated silencing of known components of the NOD2 signaling pathway led to decreased (red) or increased (blue) NF-κB luciferase activity relative to that of NT control siRNA cells (upper panel) without major effects on cell viability (lower panel). All luminescence values were statistically significant with I < 0.001. The asterisk indicates viability measurements with P < 0.001.
To enable comparisons to be made between plates, we normalized both luciferase and viability data sets with multiple positive and negative control wells present on each assay plate (figs. S2 and S3). For both the luciferase and viability assays, the data gave a normal distribution (fig. S4). Linear correlation analysis of the viability and luciferase assays indicated a high degree of reproducibility across replicas (fig. S5). Most of the Z’ factor scores, a measure of assay quality (13), for each assay plate were above 0.5 for both the luciferase and viability data sets, indicating a signal-to-noise ratio sufficient for performing robust high-throughput screens (fig. S6).
We classified genes as NOD2 regulators if their silencing reproducibly decreased the induction of the NF-κB luciferase reporter (positive regulators) or increased luciferase activity (negative regulators) relative to that of controls treated with NT siRNA (Fig. 1C and fig. S4). To avoid nonspecific effects of gene silencing on cell number, we used a fluorescence-based viability assay (14). Control wells present on each assay plate were used to calculate the percentage of viable cells (Fig. 1D and table S1). The luciferase signal weakly correlated with cell viability, especially for genes whose knockdown resulted in reduced viability, which most likely reflected a lack of sufficient cell numbers to produce luciferase (fig. S7). Therefore, we considered those genes whose silencing resulted in either (i) enhanced luciferase signal and increased viability (>120%) or (ii) decreased luciferase signal and decreased cell viability (<70%) to be inconclusive with respect to their effect on NOD2 signaling because of their nonspecific effects on cell viability. For example, knockdown of genes involved in core house-keeping functions, such as transcription and translation (fig. S8A) and cell survival (fig. S8B), resulted in decreases in both cell viability and luciferase activity. Together, these results demonstrate the importance of using a cell viability control to avoid false positives and to provide increased confidence in the ability of our screen to properly identify genes involved in the regulation of the NOD2–NF-κB signaling pathway.
Numerous NOD2 regulators interact with core components of the NF-κB pathway
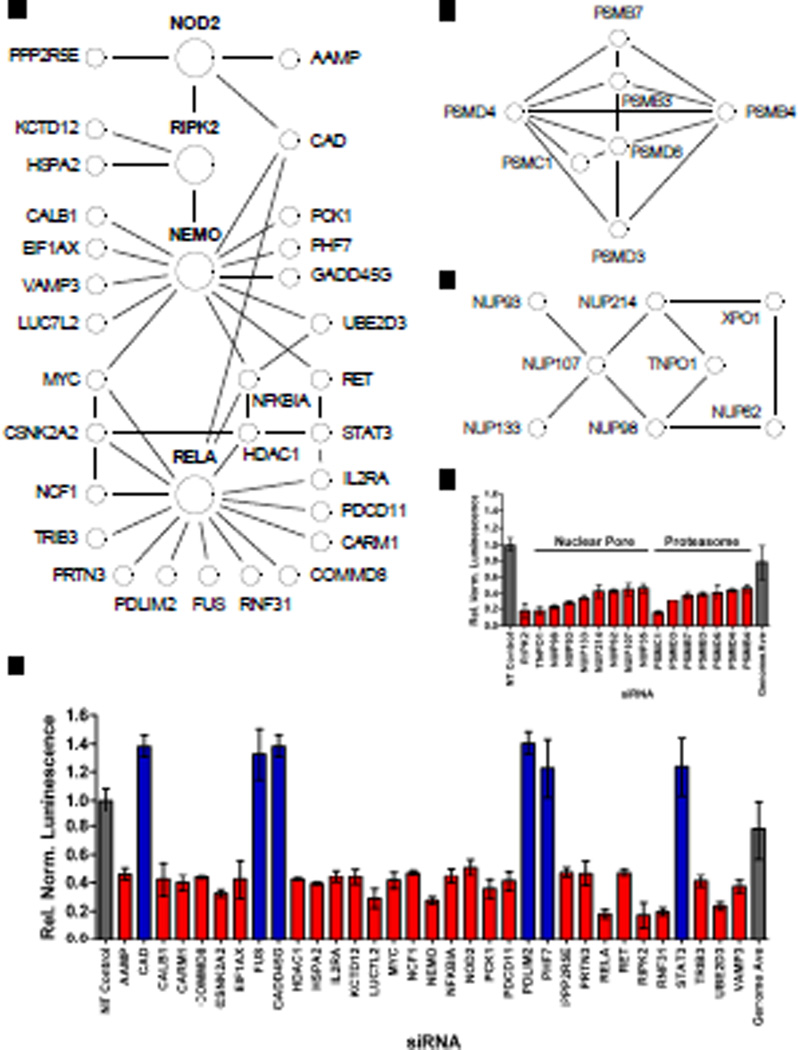
To confirm the validity of our experimental approach, we examined how known components of the NOD2 signaling pathway behaved in the screen. Indeed, knockdown of RIPK2 and other core components of the NF-κB signaling pathway, such as NEMO, NF-κB1, and avian reticuloendotheliosis viral oncogene homolog A (RelA) substantially diminished NOD2-dependent luciferase activity (Fig. 1E), thereby validating our approach. In addition, two serine and threonine kinases involved in NF-κB activation, protein kinase C δ (PRKCD) (15) and casein kinase 2 alpha’ (CSNK2A2) (16), were both classified as positive regulators in the screen (Fig. 1E). Notably, Klotho (KL), an anti-inflammatory protein that suppresses NF-κB signaling (17), and PDZ and LIM domain protein 2 (PDLIM2), a RelA-binding protein that inhibits NF-κB signaling through its E3 ubiquitin ligase activity against RelA (18), were each classified as negative regulators in the screen (Fig. 1E). Consistently, many proteins known to interact with core components of the NOD2 and NF-κB signaling pathways were categorized as positive and negative regulators in the screen (Fig. 2 and table S2). For example, the NOD2-interacting protein carbamoyl phosphate synthetase/aspartate transcarbamylase/dihydroorotase (CAD) was revealed as a negative regulator in the screen, consistent with previous reports (19, 20). Furthermore, PPP2R5E (protein phosphatase 2, regulatory subunit B', epsilon isoform), another NOD2-interacting protein (21), was categorized as a positive regulator of NOD2 signaling. Similarly, numerous RIPK2-, NEMO-, and RelA-interacting proteins were identified as either positive or negative regulators (Fig. 2, A and B). Furthermore, enrichment analysis of the top 700 positive regulators from the screen (table S3) revealed components of the proteasome (Fig. 2C) and nuclear pore complex (Fig. 2D) present significantly more often than what would be expected by chance consistent with the respective roles of these complexes in mediating NF-κB signaling via inhibitor of κB (IκB) degradation and translocation of NF-κB to the nucleus. Overall, the ability of our screen to identify many proteins already implicated in the regulation of the NOD2 and NF-κB signaling pathways validated our screening approach and provided increased confidence in our discovery of the many putative regulators not previously connected with NOD2 signaling.
Fig. 2.
Genome-wide screen identifies known regulators of NOD2 and NF-κB signaling pathways. (A) Numerous positive (red) and negative (blue) regulators identified in the siRNA screen interact with core components of the NOD2 and NF-κB signaling pathways (larger circles labeled in bold). Protein-protein interactions depicted by black lines were taken from publically available databases, such as STRING, with a confidence score of ≥ 0.4. (B) siRNA-mediated gene silencing led to decreased (red) or increased (blue) NF-κB luciferase activity upon stimulation with MDP relative to that of cells treated with NT control siRNA, without major effects on cell viability. (C to E) In addition, several components of protein complexes known to affect NF-κB signaling by mediating the proteasomal degradation of (C) IκB or (D) the nuclear translocation of RelA were significantly enriched (table S3) among (E) positive regulators in the screen. All luminescence values were statistically significant with P < 0.001.
Analysis of intersecting data sets reveals networks of putative NOD2 regulators
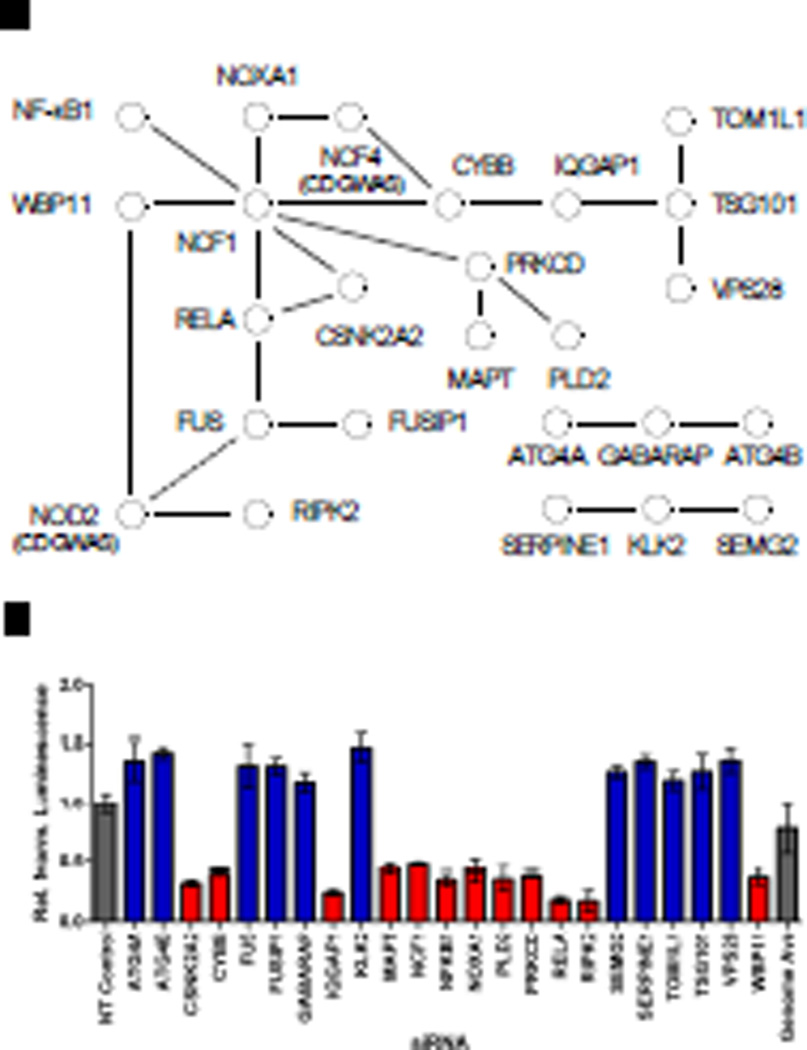
Next, we used additional lines of biochemical and functional data from the literature to identify experimental evidence supporting connections among hits from the screen. In one approach, we used the Search Tool for the Retrieval of Interacting Genes (STRING) database (22) to reveal numerous protein-protein interactions (PPIs) among hits from the screen (Fig. 3). For example, PPIs between two members of the endosomal sorting complex required for transport (ESCRT)-1 complex, tumor susceptibility gene 101 (TSG101) and vacuolar protein sorting 28 (VPS28), and three proteins involved in autophagy, ATG4A, ATG4B, and γ-aminobutyric acid receptorassociated protein (GABARAP), were identified among putative NOD2 negative regulators. In addition, multiple components of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex, including neutrophil cytosolic factor (NCF1), cytochrome b-245 beta (CYBB), and NADPH oxidase activator 1 (NOXA1), were each classified as positive regulators in the screen. Furthermore, two NOD2-interacting proteins identified by mass spectrometry, WW domain binding protein 11 (WBP11) and fused in sarcoma (FUS) (fig. S9), were identified as regulators of NOD2 signaling. We went on to identify numerous genes classified as hits in our screen with those potentially implicated in the NOD2 signaling pathway by microarray (table S4) or proteomic (table S5) studies. Similarly, we identified numerous hits from our screen that regulate NF-κB signaling (table S6). Together, these types of analysis with orthologonal data sets provide additional experimental support for a number of putative regulators and offer potential mechanistic insight into the level at which these regulators may act to regulate NOD2 signaling.
Fig. 3.
Genome-wide screen identifies networks of positive and negative regulators of the NOD2 and NF-κB signaling pathways. (A) Schematic depiction of selected protein-protein interactions among hits from the siRNA screen identified with a confidence score of ≥ 0.4 from the STRING database. Published interactions are depicted by black lines, whereas dashed lines indicate interactions with NOD2 from a NOD2 pull-down (fig. S9). (B) The relative effect on MDP-induced NF-κB signaling is shown for each gene in the network with positive and negative regulators shown in red or blue, respectively. All luminescence values were statistically significant; P < 0.001.
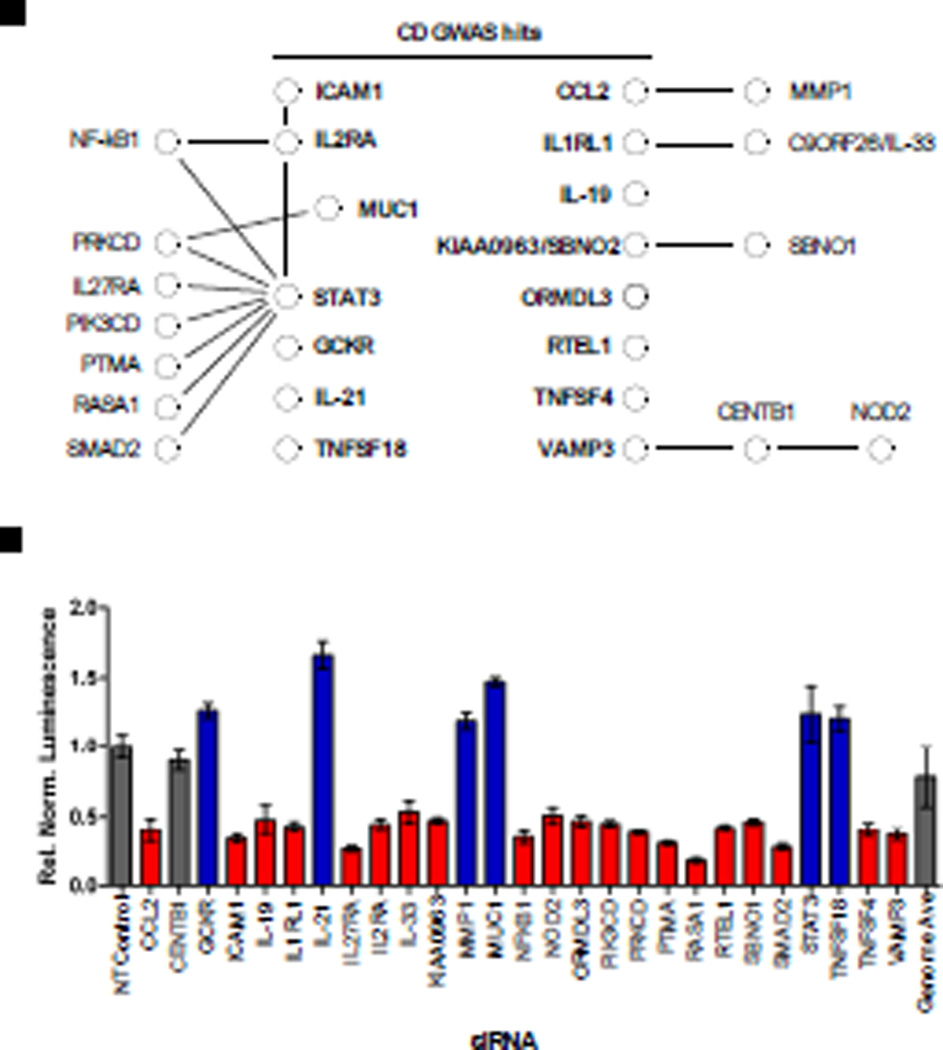
Given the link between NOD2 and Crohn’s disease, we compared genes categorized as hits in our screen with those associated with Crohn’s disease risk by genome-wide association studies (GWAS). Fifteen genes associated with Crohn’s disease risk were identified as hits in our screen (Fig. 4 and table S7). In addition, multiple genes whose products interact with these Crohn’s disease risk factors were also categorized as hits in our screen. These results not only support a role for these gene products in NOD2 signaling, but they also suggest that these regulators of the NOD2 and NF-κB signaling pathways may play a role in the pathogenesis of Crohn’s disease.
Fig. 4.
Many genes associated with Crohn’s disease risk affect NOD2 signaling. (A) Fifteen genes (labeled in bold) whose silencing either diminished (red) or enhanced (blue) NOD2 signaling in our siRNA screen have been identified as risk factors in the development of Crohn’s disease by GWAS (table S6). Black lines indicate published protein-protein interactions, taken from the STRING database, with other putative NOD2 regulators identified in the primary siRNA screen. A putative link between the Crohn’s disease risk factors VAMP3 and NOD2 is provided through their mutual interacting protein CENTB1 (62), which was not identified as a hit in our screen (gray). (B) siRNA-mediated silencing led to decreased (red) or increased (blue) NOD2-dependent NF-κB luciferase activity relative to that of cells treated with NT control siRNA, without major effects on cell viability. All luminescence values were statistically significant; P < 0.001.
Multiple NOD2 regulators were confirmed in secondary validation assays
To verify that the hits identified in our primary screen were bona fide regulators of NOD2 signaling, we carried out a series of secondary validation experiments with alternative siRNA pools that targeted 313 genes from our primary screen (Fig. 5A and table S8). In addition to measuring the induction of NF-κB–dependent luciferase activity, we used enzyme-linked immunosorbent assay (ELISA) analysis to measure IL-8 secretion, whose production is partially dependent on NF-κB activation (23). We found that 25% of the putative positive regulators and 33% of the putative negative regulators tested were concordant between the primary and secondary screens, a confirmation rate similar to those reported for other genome-wide siRNA screens (Fig. 5B and table S9) (19, 24). For example, siRNA-mediated knockdown of NCF1, a critical component of the NADPH complex that regulates the generation of reactive oxygen species, diminished NOD2-dependent NF-κB activity in both the primary and secondary screens (fig. S10, A and B). Given that many molecules that interact with NCF1 were identified as positive regulators in the primary screen (Fig. 3A and table S3), we continued to validate these results with primary bone marrow–derived macrophages (BMDMs) from Ncf1-deficient (Ncf1−/−) mice stimulated with MDP and analyzed for activation of the NOD2 pathway by Western blotting. BMDMs from Ncf1−/− mice exhibited impaired degradation of IκB compared to those from wild-type control mice (fig. S10C); however, p38 MAPK activation was not affected (fig. S10D). Moreover, the accumulation of phosphorylated IκB (pIκB) in Ncf1-deficient cells (fig. S10C) further supports an inability to properly degrade IκB, which is consistent with a role for NCF1 in mediating MDP-induced NF-κB activation.
Fig. 5.
Secondary validation of hits with independent siRNA pools. (A) Workflow summarizing the secondary validation screen with alternative sets of ON-TARGETplus siRNA pools consisting of four distinct siRNA sequences for each gene. (B) Summary of the secondary validation rate of positive and negative regulators tested for MDP-induced NF-κB luciferase activity. (C) Comparison of relative NF-κB luciferase activity after stimulation with MDP or TNF-α. Positive controls (RIPK2, RIPK1, and RelA) are highlighted along with two selected validated hits. (D) Epistasis analysis reveals that RNF31 is a general regulator of NF-κB signaling. HEK 293 cells were transfected with the indicated siRNA pools. After 48 hours, a second transfection with an NF-κB luciferase reporter and a Renilla reporter (for normalization) was used to monitor NF-κB pathway activation with (i) a constitutively active NOD2 construct (NOD2 CA), (ii) an inducibly active (IA) RIPK2 construct (RIPK2-IA), and (iii) an inducibly active RIPK1 construct (RIPK1-IA) relative to a pcDNA3 empty vector (EV) control. The dimerization agent AP1510 (ARIAD) was added as indicated to induce signaling. Experiments were performed in triplicate, of which one representative example is shown. *, P < 0.05 by a one way ANOVA and Tukey multiple comparison test performed using GraphPad Prism software.
Stimulation of cells with TNF-α identifies general regulators of NF-κB signaling
Because NOD2 signals through NF-κB, we were interested in separating candidate regulators that specifically regulate NOD2 signaling from those that are more general regulators of NF-κB signaling. To distinguish between these two possibilities, we measured NF-κB activation in response to stimulation with either MDP or TNF-α. As predicted, knockdown of RIPK2 inhibited MDP-induced, but not TNF-α–induced, NF-κB–dependent luciferase activity, whereas knockdown of RIPK1 inhibited TNF-α–induced, but not MDP-induced, NF-κB–dependent luciferase activity (Fig. 5C and table S9). Comparison of the responses to MDP and TNFα for each gene revealed that most of the tested genes affected both MDP- and TNF-α–induced NF-κB signaling, suggesting that most of the validated hits are likely downstream of RIPK2 and act as general regulators of the NF-κB pathway.
Apart from RIPK1 and RIPK2, we did not observe genes exhibiting robust specificity for either MDP or TNF-α. However, knockdown of ring finger protein 31 (RNF31) and zinc finger, DHHC-type containing 2 (ZDHHC2) exhibited more substantial inhibition of MDP-induced luciferase activity than of TNF-α–induced luciferase activity (Fig. 5C and table S9). To explore the level at which ZDHHC2 and RNF31 regulated NOD2 signaling, we performed epistasis analysis with various constructs that stimulate NF-κB signaling from different points in the pathway (Fig. 5D and fig. S11). We used a constitutively active (CA) version of NOD2 that lacks the inhibitory C-terminal leucine-rich repeats, as well as the inducibly active (IA) fusion proteins RIPK2-ΔCARD-Fpk3 (RIPK2 IA) and RIPK1-ΔCARD-Fpk3 (RIPK1 IA) whose CARD domains are replaced by tandem FK506 binding protein (FΚBP)-related dimerization domains. Previous studies showed that enforced oligomerization of RIPK2 or RIPK1 induces NF-κB activation that is dependent on the FΚBP-specific dimerization agent AP1510 (25). Using this approach, we showed that siRNA-mediated knockdown of RIPK2 inhibited activation of NF-κB signaling when stimulated with CA NOD2 or IA RIPK2, but not with IA RIPK1 alone, consistent with a specific requirement for RIPK2 in NOD2 signaling (Fig. 5D). However, knockdown of RNF31 (Fig. 5D) and ZDHHC2 (fig. S11) inhibited NF-κB signaling induced by all three constructs, indicating that both RNF31 and ZDHHC2 are capable of affecting NF-κB signaling downstream of RIPK1 and RIPK2.
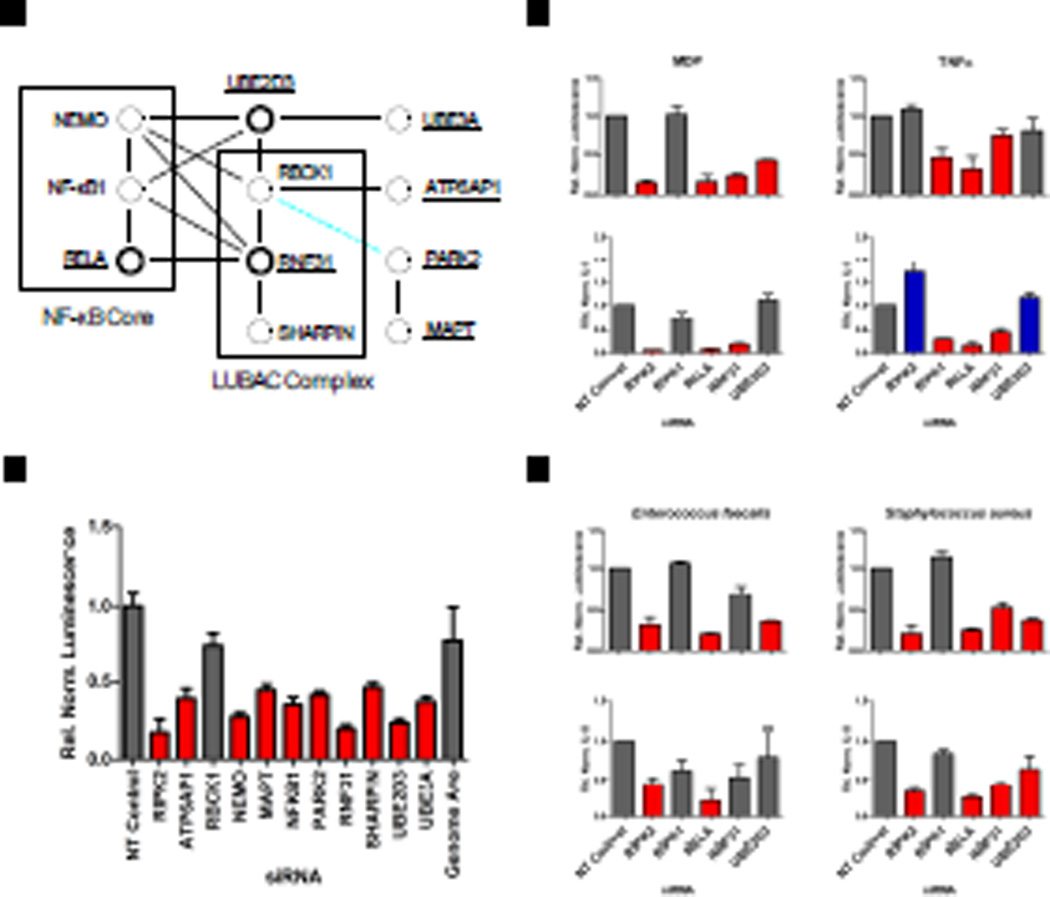
Members of LUBAC positively mediate NOD2 signaling
The ubiquitin conjugation machinery plays a critical role in the regulation of NF-κB signaling (as reviewed by 26). Linear ubiquitination mediated by the linear ubiquitin chain assembly complex (LUBAC) plays a role in the regulation of canonical NF-κB signaling in response to various stimuli, such as TNF-α, interleukin-1β (IL-1β), and lipopolysaccharide (LPS) (reviewed by 27). LUBAC is composed of three components: RanBP-type and C3HC4-type zinc finger containing 1 (RBCK1), SHANK-associated RH domain–interacting protein (SHARPIN), and RNF31, the latter two of which were identified as positive regulators of NF-κB in our screen. Analysis of protein-protein interaction databases identified a network of LUBAC-interacting proteins among hits from our primary screen linking LUBAC to core components of the NF-κB signaling pathway (Fig. 6, A and B). For example, two of the proteins in this network, RNF31 and ubiquitin-conjugating enzyme E2 D3 (UBE2D3), are known to be involved in the degradation of IκB (28) and the ubiquitination of NEMO (29), and both were validated in our secondary assays (Fig. 6C). Knockdown of RNF31 resulted in diminished MDP-induced NF-κB luciferase activity and IL-8 secretion but had only a weak effect on TNF-α–induced NF-κB luciferase activity and IL-8 secretion, whereas UBE2D3 was a positive regulator of MDP-induced NF-κB luciferase activity, but had no effect on MDP-induced IL-8 secretion or TNF-α–induced signaling.
Fig. 6.
Components of the LUBAC complex positively regulate NOD2 signaling. Two components of the LUBAC complex (RNF31 and SHARPIN) and many of their associated proteins were revealed as positive components of MDP-induced NOD2 signaling in the primary siRNA screen. (A) Protein-protein interactions are depicted by black lines, whereas a dashed blue line shows that PARK2 is a protein homolog of RBCK1 and that they share a similar domain structure. Factors required for MDP-induced activation of the NF-κB pathway are shaded in red, whereas those genes having no statistically significant effect are gray. Underlined names indicate genes tested by secondary screening, and bold circles indicate genes that were validated. (B) Induction of MDP-induced NF-κB luciferase activity relative to that of cells treated with NT siRNA from the primary screen is shown for each component of the protein-protein interaction network. All luminescence values were statistically significant; P < 0.001. (C) Induction of NF-κB luciferase reporter activity (upper panel) or IL-8 secretion (lower panel) after knockdown with ON-TARGETplus siRNA pools in response to stimulation with MDP (left) or TNF-α (right) or (D) infection with two gram positive bacteria, Enterococcus faecalis (left) or Staphylococcus aureus (right). Bars shaded in red and blue indicate P < 0.05.
To further examine roles for RNF31 and UBE2D3 in stimulating NF-κB activation, we treated our reporter cells with two different Gram positive bacteria that activate NOD2 signaling. We chose the common intestinal commensal bacterium Entercoccus faecalis, given the required role of NOD2 in mediating a cytokine response (30), and Staphylococcus aureus, another commensal organism that triggers a NOD2-mediated cytokine response both in vitro (31) and in vivo (32). We found that RNF31 was a positive regulator of bacterially-induced NF-κB luciferase activity and IL-8 secretion in response to S. aureus, whereas UBE2D3 positively regulated S. aureusinduced NF-κB luciferase activity and IL-8 secretion, as well as E. faecalis-induced NF-κB luciferase activity, but not IL-8 secretion (Fig. 6D). Together, these results support a critical role for LUBAC as a general regulator of NF-κB signaling and specifically in the modulation of NF-κB signaling downstream of NOD2.
DISCUSSION
Coordination of the inflammatory response is mediated by a wide range of effectors, and proper balance of responses is required for the maintenance of health (33). NOD2 signaling is capable of activating the NF-κB pathway, leading to an inflammatory response to bacteria that is dysregulated in human diseases such as Crohn’s disease (34). In the quiescent state, NF-κB family members reside within the cytoplasm where they are kept inactive because of physical associations with IκB family members. Degradation of IκB by the proteasome exposes a nuclear localization signal in NF-κB, which mediates its relocalization to the nucleus where it can bind to DNA and regulate the transcription of various target genes. Multiple core components of NOD2 and NF-κB signaling pathways including RIPK2, NEMO, NF-κB1, RelA, and subunits of the proteasome and the nuclear pore complex were classified as hits in our screen, providing evidence that our screen accurately detected bona fide NOD2 regulators. In addition, our genome-wide siRNA screen expanded the number of genes implicated in the regulation of the NOD2 and NF-κB signaling pathways.
Many steps in the NF-κB signaling pathway are regulated by protein ubiquitination (26). For example, Lys63-linked poly-ubiquitination of NEMO, an IKK scaffolding and regulatory protein, mediates the recruitment of the IKK-activating kinase TAK1. Consistent with this, our screen identified protein phosphatase magnesium-dependent 1 like (PPM1L), a phosphatase that binds to and dephosphorylates TAK1 (35), as an inhibitor of NOD2 signaling analogous to its previously reported role as an inhibitor of NF-κB activation downstream of TNF-α (36). Furthermore, we observed decreased NOD2 signaling upon knockdown of two components of LUBAC (RNF31 and SHARPIN), which is consistent with reports showing a role for LUBAC in the regulation of NOD2 signaling (37). Our results extend these observations by showing a role for RNF31 in the activation of the NOD2 pathway in response to bacterial stimulation. In vivo, sharpin deficient mice exhibit excessive inflammation (38), impaired secretion of pro-inflammatory cytokines, and a reduction in NF-κB activation (39–41), features that are somewhat analogous to the symptoms of Crohn’s disease. Biochemically, SHARPIN interacts with RNF31, and together they play a role in NF-κB activation by acting as an E3 ubiquitin ligase to catalyze the linear ubiquitination of NEMO (42–45), which agreeswith our secondary validation data showing a positive role for RNF31 in TNF-α–induced NF-κB activation. Although RBCK1, the third known component of the LUBAC complex, was not individually identified as a hit in our screen, Parkinson protein 2 (PARK2), a protein with the same domain structure as RBCK1 and that was previously implicated in NF-κB signaling (46), was classified as a positive regulator of NOD2 signaling. Notably, PARK2, like NOD2 and RIPK2, is linked to susceptibility to leprosy (47, 48). Furthermore, UBE2D3, an E2 ubiquitin conjugating enzyme that interacts with RBCK1 (49) was validated as a stimulator of NOD2 signaling in our screen. In addition, two ubiquitin-specific proteases (USP), USP2 and USP36, previously associated with TNF-α–induced NF-κB signaling (50), were classified as positive regulators of NOD2 signaling. Together, these data show a role for protein ubiquitination and, in particular, LUBAC, in NOD2 signaling.
Given that protein-protein interactions mediate the assembly and regulation of signal transduction pathways, we were pleased to identify numerous proteins known to interact with NOD2 and other core components of the NF-κB signaling pathway as hits in our screen. In addition, our screen implicated several additional protein complexes in the regulation of NOD2 signaling. For example, our identification of ESCRT-1 as having a role in the inhibition of NOD2 signaling is supported by the enhanced IL-8 secretion in response to NOD1 signaling that was observed upon knockdown of the ESCRT-I complex subunit tumor susceptibility gene 101 (TSG101) (51). In addition, several genes associated with Crohn’s disease appear to regulate oxidative stress (52), consistent with our identification of several components of the NADPH oxidase complex as regulators of NOD2 signaling. Finally, multiple interacting components of autophagy, an evolutionarily conserved process required for the degradation and recycling of cellular contents that can limit infection by certain intracellular pathogens, were identified as inhibitors of NOD2 signaling in our screen, the relevance of which is increased given evidence of their biochemical links to NOD2 (53, 54) coupled with the observation that multiple components of the autophagy network are associated with an increased risk of Crohn’s disease (reviewed by 55). Together, these examples illustrate the use of overlaying siRNA screening data with protein-protein interaction networks to provide insight into the regulatory mechanisms by which these proteins may influence NOD2 signaling, and to generate models worth pursuing by more traditional hypothesis-based research.
Biologically, activation of the NOD2 and NF-κB signaling pathways is critical for host defense against pathogens (reviewed by 56). Tight control of both the strength and duration of this response is necessary to achieve a balance between uncontrolled infection and tissue damage associated with chronic inflammation (57). Our screen identified many inhibitors of the NF-κB pathway. For example, MDP-induced activation of NF-κB was enhanced upon silencing PDLIM2, a protein that interacts with RelA and mediates its degradation (18). Furthermore, we validated the RNA-binding protein FUS and its interacting partner FUSIP1 as inhibitors of NOD2 signaling in our siRNA screen, the relevance of which is further supported by the identification of FUS as a NOD2-interacting protein by mass spectrometry. The systematic identification of genes whose products inhibit NF-κB signaling will improve our understanding of how this critical pathway is inhibited and may provide new avenues for therapeutic intervention.
Systems-level approaches are rapidly increasing our understanding of the complex feedback loops and crosstalk mechanisms that regulate NF-κB signaling. By comparing data from different approaches, we can get additional information to help comprehend these processes. To this end, we searched the literature for regulators of NF-κB signaling, including three published siRNA screens that examined (i) the translocation of RelA to the nucleus in response to TNF-α (19); (ii) the induction of NF-κB activation in response to Toll-like receptor (TLR) stimulation (24); and (iii) genes whose products are involved in NOD1-induced secretion of IL-8 (51), each of which identified largely distinct sets of genes whose products influence NF-κB signaling. In total, we found 96 hits from our primary screen that overlapped with genes identified in these previous studies. For example, eighteen hits from our screen were consistent with the NOD1 siRNA screen. NOD1 shares a similar domain structure with NOD2, it acts as a cytosolic receptor for bacterial peptidoglycan (PGN), and it is thought to share many downstream signaling components with NOD2, including RIPK2 (reviewed by 58). The siRNA library used in the NOD1 screen was much smaller than our library, and it used a transfection-based method to deliver NOD1 agonist into the cells. In contrast, our screen used a whole-genome approach and relied upon endogenous routes of entry of a more physiologically relevant amount of PGN agonist. Additionally, our use of a reporter gene enabled us to make a more direct measurement of NF-κB activity rather than having to rely on the induction of IL-8 production. As the number of published siRNA screens grows, key regulatory factors will emerge from the comparison of these types of data sets.
The NOD2 signaling pathway is just one of many upstream inputs that converge on the IKK complex and regulate the evolutionarily conserved NF-κB stress response pathway. These branches are unlikely to function in isolation in vivo. Therefore, we suggest that the relatively large number of genes identified to influence NOD2 signaling by our screen likely includes direct regulators as well as gene products in parallel pathways that indirectly affect NF-κB signaling. Indeed, our TNF-α counter screen revealed that apart from RIPK2, most of the hits identified are general regulators of NF-κB signaling. This lack of NOD2-specific components supports the idea that different danger-sensing receptors of the innate immune system likely use a small number of specific components before converging on a more general core NF-κB response.
Given the genetic link between NOD2 and Crohn’s disease, the identification by our screen of numerous genes associated with Crohn’s disease risk by GWAS as being putative NOD2 regulators supports a role for members of the NOD2 signaling pathway, beyond just NOD2, in the pathogenesis of Crohn’s disease. This idea is further supported by the identification of Crohn’s disease patients with impaired MDP responses in the absence of mutations in NOD2 (59). One potential application of our data set is in the analysis of large-scale GWAS and next-generation DNA sequencing efforts currently underway to explore the genetic basis of Crohn’s disease. For example, having a list of genes whose products influence NOD2 signaling provides candidates to explore among Crohn’s disease patients. In the absence of such a list, the search space for potentially causative variants is extremely large given the extent of sequence variation across individuals, which makes it very difficult to identify rare variants of modest effect that may underlie a given disease. Similarly, our data set should assist in the selection of candidate genes within gene-dense Crohn’s disease risk loci identified by GWAS.
In conclusion, we have identified numerous genes that regulate the NOD2 and NF-κB signaling pathways, providing a resource for future hypothesis-based studies aimed at understanding the detailed molecular mechanisms that regulate these pathways. It is becoming increasingly clear that complex networks of interacting proteins, and not simple linear pathways, are involved in the regulation of innate immunity. Our study offers a systems-level glimpse of the NOD2 signaling pathway, which will increase our understanding of how this pathway regulates inflammatory homeostasis and provides insight into the genetic basis for Crohn’s disease.
MATERIALS AND METHODS
Primary siRNA screen
The screen was carried out using a Bio-Tek ELx405 plate aspirator and Thermo Labsystems Multidrop liquid dispensers at the University of Michigan Center for Chemical Genomics.
HEK 293 cells stably expressing human NOD2 (HEK293-NOD2) and an NF-κB luciferase reporter were cultured on 384-well plates (Greiner Bio-One) in Dulbecco's modified eagle medium with 10% fetal bovine serum, 2 mM glutamine, 1 mM sodium pyruvate, and 1X Pen Strep (Gibco). Pools of four distinct siGENOME siRNAs (Thermo Scientific) were reverse transfected at a concentration of 20 nM with Lipofectamine 2000 (Invitrogen) diluted in Opti-MEM (Invitrogen) for 48 hours followed by stimulation with MDP (20 ng/ml, Bachem) for 18 hours. A PHERAstar plate reader (BMG Labs) was used to quantify both the cell viability assays (Cell Titre Fluor, Promega) and the NF-κB luciferase assays (Steady Glo, Promega).
Normalization
All cell viability values were calculated relative to multiple control wells of cells that received either non-targeting (NT) siRNA or Passive Lysis Buffer (PLB, Promega); such controls were present on each assay plate. The average value of the wells receiving NT siRNA was considered as 100%, and the average value of wells receiving PLB was considered as 0%. Therefore, wells with increased cell number had values greater than 100%, whereas those with decreased cell numbers relative to those of controls treated with NT siRNA had values less than 100%. Similarly, the average relative luminescence values for each well of the NF-κB luciferase reporter assay were calculated relative to multiple control wells present on each plate. RIPK2-specific siRNA was used as a positive control and NT siRNA was used as a negative control. For each plate, the average value of wells receiving RIPK2-specific siRNA was considered as 100% inhibition, and the average value of wells receiving NT siRNA was considered as 0%. Therefore, test siRNA that resulted in decreased luciferase (relative to that of wells receiving NT siRNA) resulted in values greater than 0, whereas test siRNA that resulted in increased luciferase (relative to that of wells receiving NT siRNA) resulted in values less than 0.
Secondary validation assays
Reporter cells were reverse transfected with 40 nM ON-TARGETplus siRNA pools (Thermo Scientific) for 48 hours, followed by stimulation with either MDP (20 ng/ml) or human TNF-α (10 ng/ml, R&D Biosystems). Culture supernatants were collected using a Biomek FX (Beckman) liquid handling robot and frozen at −20°C for subsequent ELISA analysis. Luciferase and cell viability assay on the remaining cells were analyzed as described above. For assay plates stimulated with TNF-α, wells transfected with RelA-specific siRNA were used as the positive control.
Measurement of IL-8 by ELISA
The concentration of human IL-8 in frozen culture supernatant was measured by Joel Whitfield at the University of Michigan immunology core facility using a sandwich ELISA (R&D Systems) according to the manufacturer’s recommended protocol adapted to a 384-well plate format. Cell supernatants were diluted 1:1 in assay buffer (Tris-buffered casein, Surmodics) and developed with TMB substrate (Surmodics). Concentrations of IL-8 (in pg/ml) were determined by comparison to an eight-point standard curve (in quadruplicate) prepared on each plate. Normalized amounts of IL-8 were calculated using the average amount of IL-8 in wells receiving RIPK2-specific siRNA set at 100% inhibition, whereas the average amount of IL-8 in wells that received NT siRNA was considered as 0% inhibition. Therefore, test siRNA that resulted in decreased IL-8 secretion (relative to that of cells that received NT siRNA) resulted in values greater than 0, whereas test siRNA that resulted in increased IL-8 secretion (relative to that of cells treated with NT siRNA) resulted in values less than 0. For assay plates stimulated with TNF-α, the average value of IL-8 secreted by cells that received RelA-specific siRNA was considered as 100% inhibition.
Bacterial Inoculation
E. faecalis (ATCC 47077) or S. aureus (strain 8325-4, a gift from Timothy Foster, Trinity College, Dublin, Ireland) were inoculated into Brain Heart Infusion media and incubated overnight at 37°C (for E. faecalis) or 30°C (for S. aureus) with shaking. Overnight cultures were sub-cultured into fresh media and incubated at 37°C with shaking for approximately 2 hours until reaching an OD600 of 0.6. The bacteria were washed and diluted in sterile phosphatebuffered saline (PBS). 40,000 bacteria were added to each well giving a bacterium to HEK293-NOD2 cell ratio of approximately 1:1. After a 1 hour infection, gentamicin (50 ng/µl, Gibco) was added to the media. Supernatants were collected after 17 hours. Luciferase and viability assays were carried as described for the primary screen.
Epistasis
HEK 293 cells were reverse transfected with 20 nM siRNA (siGENOME) as described earlier. After 48 hours, cells were transfected with pcDNA3, pcDNA3-NOD2ΔLRR, pcDNA3-RIPK2ΔCT-FPK3, or pcDNA3-RIPK1ΔCT-FPK3 together with NF-κB luciferase and Renilla luciferase reporter plasmids. After 6 hours, some wells received 100 nM AP1510 (ARIAD) to induce dimerization. Eighteen hours later, cells were lysed in passive lysis buffer (Promega) and analyzed with the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Preparation of BMDMs
B6(Cg)-Ncf1m1J/J mice (ncf1 mutant mice) were obtained from the Jackson Laboratory. Mice were housed in a pathogen-free facility, and all animal studies were conducted under protocols approved by the University of Michigan Committee on Use and Care of Animals. BMDMs were isolated as previously described (60). Briefly, femurs and tibia were removed from euthanized mice after sterilization with 70% ethanol. Bone marrow cells were resuspended in L-cellconditioned complete Iscove's Modified Dulbecco’s media (Gibco) and cultured for 5 to 6 days before overnight stimulation with MDP (10 µg/m) and subsequent analysis of IκB and MAPK proteins by Western blotting (Cell Signaling Technology Inc.).
Bioinformatics and statistical analysis
A custom-built relational database (M-screen) was developed and used for data storage, display, analysis, and queries (available at: http://mscreen.lsi.umich.edu/index.php). Assay quality was determined by calculating mean, standard deviation, coefficient of variation, and Z′ factor (13) values for each plate. After initial normalization to positive and negative controls on each plate, an additional normalization step was performed to normalize the percentage knockdown luciferase values to those for viability. Because the normalized knockdown luciferase values ranged from negative values to >100%, to convert to the luciferase scale, we first subtracted the values from their maximum rather than from 100. After normalization with the percentage viability, we then converted back to the percentage knockdown scale. The specific formula used was: max(L) – (100(max(L)- L) / V), where L = the normalized percentage knockdown of luciferase activity, and V = the percentage viability. Values were then normalized to the median of each plate, because substantial shifts in plate-to-plate medians were observed, and then global median normalization was used as a final step to make the samples comparable. To calculate P values, we tested for normalized knockdown luciferase values that were statistically significantly higher or lower than the overall median. We used an empirical Bayes method (a moderated t-test) that takes into account the relationship between the percentage knockdown of luciferase activity and variance to calculate P values (61), enabling more accurate sample variation estimates for each gene. P values were adjusted for multiple testing with the Bonferroni correction. Secondary validation analyses were performed in triplicate on 313 hits from the primary screen spread over two separate assay plates. For each of these plates, we analyzed both luciferase and IL-8 data by following the same steps as described earlier, except that no plate-to-plate normalization was performed.
Supplementary Material
Acknowledgments
We thank ARIAD Pharmaceuticals Inc. for providing AP1510. We are grateful to R. Jacob, M. Larsen, and S. Swaney at the Center for Chemical Genomics, University of Michigan (U of M), for assistance in carrying out the screen; J. Whitfield (U of M) for performing the IL-8 ELISA assay; A.I. Nesvizhskii (U of M), A. Sreekumar (Baylor College of Medicine), and A.M. Chinnaiyan (U of M) for assistance with the mass spectrometry; and P. Kuffa and S. Koonse for technical support. We would also like to thank J. Warner, M. Shaw, and N. Inohara for helpful advice and comments.
Funding: N.W. was supported by a Postdoctoral Fellowship from the U of M Center for Genetics in Health and Medicine. This work was supported by grants from the NIH (R01DK082437 (to C.M.) and 2R01DK61707 (to G.N.), the Michigan Institute for Clinical and Health Research (UL1RR024986), the Herman and Dorothy Miller Award for innovative research in immunology, career development awards from the Crohn’s & Colitis Foundation of America to C.M. and L.F., and the Eli and Edythe Broad Foundation Medical Research Program for Inflammatory Bowel Disease.
Footnotes
Author contributions:
N.W., A.B., and G.N. designed the research and wrote the manuscript; N.W. and A.B. carried out the siRNA screen and validation experiments; M.A.S. performed statistical analyses; C.M. assisted with creation of the NOD2 reporter cell line and did the NOD2 IP experiments; L.F. and Y.K. assisted with bacterial stimulations and experimental interpretation.
Competing interests:
The authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
Fig. S1. Reporter cell line characterization.
Fig. S2. Assay plate layout.
Fig. S3. Data normalization.
Fig. S4. Data distribution.
Fig. S5. Data reproducibility.
Fig. S6. Assay robustness.
Fig. S7. Luciferase activity correlates with cell viability.
Fig. S8. Reduced cell viability.
Fig. S9. FUS and WBP11 mass spectra data.
Fig. S10. NCF1 validation data.
Fig. S11. ZDHHC2 secondary validation.
Table S1. Primary screen results.
Table S2. Published NOD2-interacting proteins.
Table S3. Conceptgen enrichment of top 700 positive regulators.
Table S4. Hits supported by microarray studies.
Table S5. Hits supported by proteomic studies.
Table S6. Hits supported in the literature.
Table S7. References for Crohn’s disease GWAS hits.
Table S8. 313 secondary validation hits list – annotation.
Table S9. 313 secondary validation results.
REFERENCES AND NOTES
- 1.Chen G, Shaw MH, Kim YG, Nunez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–398. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 2.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 3.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 5.Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, Kelliher MA. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem. 2007;282:36223–36229. doi: 10.1074/jbc.M703079200. [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, Inohara N. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 9.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 10.Kanazawa N, Okafuji I, Kambe N, Nishikomori R, Nakata-Hizume M, Nagai S, Fuji A, Yuasa T, Manki A, Sakurai Y, Nakajima M, Kobayashi H, Fujiwara I, Tsutsumi H, Utani A, Nishigori C, Heike T, Nakahata T, Miyachi Y. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood. 2005;105:1195–1197. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- 11.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 12.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 14.Niles AL, Moravec RA, Eric Hesselberth P, Scurria MA, Daily WJ, Riss TL. A homogeneous assay to measure live and dead cells in the same sample by detecting different protease markers. Anal Biochem. 2007;366:197–206. doi: 10.1016/j.ab.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Lallena MJ, Diaz-Meco MT, Bren G, Paya CV, Moscat J. Activation of IkappaB kinase beta by protein kinase C isoforms. Mol Cell Biol. 1999;19:2180–2188. doi: 10.1128/mcb.19.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird TA, Schooley K, Dower SK, Hagen H, Virca GD. Activation of nuclear transcription factor NF-kappaB by interleukin-1 is accompanied by casein kinase II-mediated phosphorylation of the p65 subunit. J Biol Chem. 1997;272:32606–32612. doi: 10.1074/jbc.272.51.32606. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Banerjee S, Dey N, LeJeune WS, Sarkar PS, Brobey R, Rosenblatt KP, Tilton RG, Choudhary S. Klotho depletion contributes to increased inflammation in kidney of the db/db mouse model of diabetes via RelA (serine)536 phosphorylation. Diabetes. 2011;60:1907–1916. doi: 10.2337/db10-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 19.Gewurz BE, Towfic F, Mar JC, Shinners NP, Takasaki K, Zhao B, Cahir-McFarland ED, Quackenbush J, Xavier RJ, Kieff E. Genome-wide siRNA screen for mediators of NF-kappaB activation. Proc Natl Acad Sci U S A. 2012;109:2467–2472. doi: 10.1073/pnas.1120542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richmond AL, Kabi A, Homer CR, Marina-Garcia N, Nickerson KP, Nesvizhskii AI, Sreekumar A, Chinnaiyan AM, Nunez G, McDonald C. The Nucleotide Synthesis Enzyme CAD Inhibits NOD2 Antibacterial Function in Human Intestinal Epithelial Cells. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimmo ER, Stevens C, Phillips AM, Smith A, Drummond HE, Noble CL, Quail M, Davies G, Aldhous MC, Wilson DC, Satsangi J. TLE1 modifies the effects of NOD2 in the pathogenesis of Crohn's disease. Gastroenterology. 2011;141:972–981. doi: 10.1053/j.gastro.2011.05.043. e971–972. [DOI] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasumoto K, Okamoto S, Mukaida N, Murakami S, Mai M, Matsushima K. Tumor necrosis factor alpha and interferon gamma synergistically induce interleukin 8 production in a human gastric cancer cell line through acting concurrently on AP-1 and NF-kB-like binding sites of the interleukin 8 gene. J Biol Chem. 1992;267:22506–22511. [PubMed] [Google Scholar]
- 24.Chiang CY, Engel A, Opaluch AM, Ramos I, Maestre AM, Secundino I, De Jesus PD, Nguyen QT, Welch G, Bonamy GM, Miraglia LJ, Orth AP, Nizet V, Fernandez-Sesma A, Zhou Y, Barton GM, Chanda SK. Cofactors Required for TLR7- and TLR9-Dependent Innate Immune Responses. Cell Host Microbe. 2012;11:306–318. doi: 10.1016/j.chom.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inohara N, Koseki T, Lin J, del Peso L, Lucas PC, Chen FF, Ogura Y, Nunez G. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 26.Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emmerich CH, Schmukle AC, Walczak H. The emerging role of linear ubiquitination in cell signaling. Sci Signal. 2011;4:re5. doi: 10.1126/scisignal.2002187. [DOI] [PubMed] [Google Scholar]
- 28.Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai K, Ciechanover A. Identification of the ubiquitin carrier proteins E2s involved in signal-induced conjugation and subsequent degradation of IkappaBalpha. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- 29.Tang ED, Wang CY, Xiong Y, Guan KL. A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumor necrosis factor-alpha. J Biol Chem. 2003;278:37297–37305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 30.Kim YG, Shaw MH, Warner N, Park JH, Chen F, Ogura Y, Nunez G. Cutting edge: Crohn's disease-associated Nod2 mutation limits production of proinflammatory cytokines to protect the host from Enterococcus faecalis-induced lethality. J Immunol. 2011;187:2849–2852. doi: 10.4049/jimmunol.1001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hruz P, Zinkernagel AS, Jenikova G, Botwin GJ, Hugot JP, Karin M, Nizet V, Eckmann L. NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha-toxin-dependent innate immune activation. Proc Natl Acad Sci U S A. 2009;106:12873–12878. doi: 10.1073/pnas.0904958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deshmukh HS, Hamburger JB, Ahn SH, McCafferty DG, Yang SR, Fowler VG., Jr. Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect Immun. 2009;77:1376–1382. doi: 10.1128/IAI.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 34.Wullaert A, Bonnet MC, Pasparakis M. NF-kappaB in the regulation of epithelial homeostasis and inflammation. Cell Res. 2011;21:146–158. doi: 10.1038/cr.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li MG, Katsura K, Nomiyama H, Komaki K, Ninomiya-Tsuji J, Matsumoto K, Kobayashi T, Tamura S. Regulation of the interleukin-1-induced signaling pathways by a novel member of the protein phosphatase 2C family (PP2Cepsilon) J Biol Chem. 2003;278:12013–12021. doi: 10.1074/jbc.M211474200. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Wang L, Berman MA, Zhang Y, Dorf ME. RNAi screen in mouse astrocytes identifies phosphatases that regulate NF-kappaB signaling. Mol Cell. 2006;24:497–509. doi: 10.1016/j.molcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, Rieser E, Rickard JA, Bankovacki A, Peschel C, Ruland J, Bekker-Jensen S, Mailand N, Kaufmann T, Strasser A, Walczak H, Silke J, Jost PJ, Gyrd-Hansen M. The Ubiquitin Ligase XIAP Recruits LUBAC for NOD2 Signaling in Inflammation and Innate Immunity. Mol Cell. 2012;46:746–758. doi: 10.1016/j.molcel.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Seymour RE, Hasham MG, Cox GA, Shultz LD, Hogenesch H, Roopenian DC, Sundberg JP. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation immune system dysregulation and dermatitis. Genes Immun. 2007;8:416–421. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Sokolovska A, Seymour R, Sundberg JP, Hogenesch H. SHARPIN is essential for cytokine production, NF-kappaB signaling, and induction of Th1 differentiation by dendritic cells. PLoS One. 2012;7:e31809. doi: 10.1371/journal.pone.0031809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 41.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 42.Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 43.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Niu J, Shi Y, Iwai K, Wu ZH. LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 2011;30:3741–3753. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, Wegener E, Nakaso K, Culmsee C, Berninger B, Krappmann D, Tatzelt J, Winklhofer KF. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci. 2007;27:1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, Cui Y, Yan XX, Yang HT, Yang RD, Chu TS, Zhang C, Zhang L, Han JW, Yu GQ, Quan C, Yu YX, Zhang Z, Shi BQ, Zhang LH, Cheng H, Wang CY, Lin Y, Zheng HF, Fu XA, Zuo XB, Wang Q, Long H, Sun YP, Cheng YL, Tian HQ, Zhou FS, Liu HX, Lu WS, He SM, Du WL, Shen M, Jin QY, Wang Y, Low HQ, Erwin T, Yang NH, Li JY, Zhao X, Jiao YL, Mao LG, Yin G, Jiang ZX, Wang XD, Yu JP, Hu ZH, Gong CH, Liu YQ, Liu RY, Wang DM, Wei D, Liu JX, Cao WK, Cao HZ, Li YP, Yan WG, Wei SY, Wang KJ, Hibberd ML, Yang S, Zhang XJ, Liu JJ. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 48.Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, Sarno EN, Alter A, Montpetit A, Moraes ME, Moraes JR, Dore C, Gallant CJ, Lepage P, Verner A, Van De Vosse E, Hudson TJ, Abel L, Schurr E. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 49.Zenke-Kawasaki Y, Dohi Y, Katoh Y, Ikura T, Ikura M, Asahara T, Tokunaga F, Iwai K, Igarashi K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol. 2007;27:6962–6971. doi: 10.1128/MCB.02415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metzig M, Nickles D, Falschlehner C, Lehmann-Koch J, Straub BK, Roth W, Boutros M. An RNAi screen identifies USP2 as a factor required for TNF-alpha-induced NF-kappaB signaling. Int J Cancer. 2011;129:607–618. doi: 10.1002/ijc.26124. [DOI] [PubMed] [Google Scholar]
- 51.Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, Reed JC, Saleh M. Non-apoptotic role of BID in inflammation and innate immunity. Nature. 2011;474:96–99. doi: 10.1038/nature09982. [DOI] [PubMed] [Google Scholar]
- 52.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. 1641 e1631–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 55.Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn's disease: NOD2 autophagy and ER stress converge. Gut. 2011;60:1580–1588. doi: 10.1136/gut.2009.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruland J. Return to homeostasis: downregulation of NF-kappaB responses. Nat Immunol. 2011;12:709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 58.Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6:9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- 59.Seidelin JB, Broom OJ, Olsen J, Nielsen OH. Evidence for impaired CARD15 signalling in Crohn's disease without disease linked variants. PLoS One. 2009;4:e7794. doi: 10.1371/journal.pone.0007794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celada A, Gray PW, Rinderknecht E, Schreiber RD. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J Exp Med. 1984;160:55–74. doi: 10.1084/jem.160.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto-Furusho JK, Barnich N, Xavier R, Hisamatsu T, Podolsky DK. Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation. J Biol Chem. 2006;281:36060–36070. doi: 10.1074/jbc.M602383200. [DOI] [PubMed] [Google Scholar]
- 63.Zhonghong L, Lianjie L, Changqing Z, Ying H, Yu J, Yan L. The influence of survivin shRNA on the cell cycle and the invasion of SW480 cells of colorectal carcinoma. J Exp Clin Cancer Res. 2008;27:20. doi: 10.1186/1756-9966-27-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20 a potential cancer therapeutic target is negatively regulated by p53. Oncogene. 2008;27:1562–1571. doi: 10.1038/sj.onc.1210799. [DOI] [PubMed] [Google Scholar]
- 65.Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc Natl Acad Sci U S A. 2006;103:11964–11969. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu Q, Gou Y, Wang Q, Jin H, Cui L, Zhang Y, He L, Wang J, Nie Y, Shi Y, Fan D. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS One. 2011;6:e26401. doi: 10.1371/journal.pone.0026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heidel JD, Liu JY, Yen Y, Zhou B, Heale BS, Rossi JJ, Bartlett DW, Davis ME. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res. 2007;13:2207–2215. doi: 10.1158/1078-0432.CCR-06-2218. [DOI] [PubMed] [Google Scholar]
- 68.Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, Igarashi H, Nakamura T, Suzuki H, Watanabe Y, Hishida A, Ikuma M, Sugimura H. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–260. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.