Abstract
OBJECTIVES:
Intrahepatic cholestasis of pregnancy (ICP) has a complex etiology with a significant genetic component. Heterozygous mutations of canalicular transporters occur in a subset of ICP cases and a population susceptibility allele (p.444A) has been identified in ABCB11. We sought to expand our knowledge of the detailed genetic contribution to ICP by investigation of common variation around candidate loci with biological plausibility for a role in ICP (ABCB4, ABCB11, ABCC2, ATP8B1, NR1H4, and FGF19).
METHODS:
ICP patients (n=563) of white western European origin and controls (n=642) were analyzed in a case–control design. Single-nucleotide polymorphism (SNP) markers (n=83) were selected from the HapMap data set (Tagger, Haploview 4.1 (build 22)). Genotyping was performed by allelic discrimination assay on a robotic platform. Following quality control, SNP data were analyzed by Armitage's trend test.
RESULTS:
Cochran–Armitage trend testing identified six SNPs in ABCB11 together with six SNPs in ABCB4 that showed significant evidence of association. The minimum Bonferroni corrected P value for trend testing ABCB11 was 5.81×10−4 (rs3815676) and for ABCB4 it was 4.6×10−7(rs2109505). Conditional analysis of the two clusters of association signals suggested a single signal in ABCB4 but evidence for two independent signals in ABCB11. To confirm these findings, a second study was performed in a further 227 cases, which confirmed and strengthened the original findings.
CONCLUSIONS:
Our analysis of a large cohort of ICP cases has identified a key role for common variation around the ABCB4 and ABCB11 loci, identified the core associations, and expanded our knowledge of ICP susceptibility.
INTRODUCTION
Intrahepatic cholestasis of pregnancy (ICP), also known as obstetric cholestasis, occurs in ∼1 in 150 UK pregnancies. Maternal symptoms of the disease include pruritus, raised serum bile acids, and deranged liver function tests (1,2,3,4). Liver transaminases can be increased as much as 100-fold, but the bilirubin level is usually normal or mildly raised. Fetal consequences of ICP include spontaneous and iatrogenic preterm labor, meconium-stained amniotic fluid, fetal distress, and intrauterine death (5,6,7,8). Approximately 80% of patients present after 30 weeks of gestation and diagnosis is routinely confirmed by elevated serum bile acid concentrations and liver function tests (9). Higher levels of fasting and nonfasting serum bile acids (>40 μM) have been associated with increased risk of adverse pregnancy outcomes including spontaneous preterm labor, meconium passage, stillbirth, and prolonged admission to the neonatal unit (8,10). Severe ICP, defined by maternal serum bile acid levels of ≥40 μM, affects 1 in 1,000 pregnancies in the United Kingdom (10). Treatment is usually with ursodeoxycholic acid, to which ∼70% of patients respond with improvement in maternal liver function tests, bile acids, and pruritus (11,12). It is currently not known whether ursodeoxycholic acid treatment reduces the risk of adverse pregnancy outcome, although the results of two recent studies were encouraging (12,13). The impact of ICP for the offspring was further highlighted by a recent study that reported increased rates of obesity and dyslipidemia in the 16-year-old offspring of affected women (14).
ICP has a complex, multifactorial etiology with hormonal, environmental, and genetic influences. Considerable evidence for a genetic predisposition to this disease comes from significant familial clustering (15,16), population-specific risk differences (17), and increased risk with an affected first-degree relative (18). Genes mutated in progressive familial intrahepatic cholestasis (PFIC) and the related condition benign recurrent intrahepatic cholestasis (BRIC), namely ABCB4, ABCB11, and ATP8B1, have been implicated in the pathogenesis of ICP in a number of different studies. Initial studies identified heterozygous mutations of the phosphatidyl choline floppase ABCB4 (MDR3) in familial (19) and sporadic (20) cases. Homozygous mutations of this gene cause a severe childhood-onset liver disease, PFIC3 (21). These first studies of the genetics of ICP susceptibility were confirmed and expanded by a number of subsequent studies (22,23,24). Mutations in ABCB4 cause a number of other biliary disorders, including drug-induced cholestasis (25) and low phospholipid-associated cholelithiasis (26). A small single-nucleotide polymorphism (SNP)/haplotype study of common variation around this locus in 52 “severe” ICP cases with serum bile acid levels of >40 μM provided additional evidence for a role in ICP susceptibility (27). Hence, ABCB4 mutations and variation are linked to a spectrum of cholestatic disease of varying severity.
ABCB11 (the bile salt export pump), another member of the ABC transporter superfamily, is the high-affinity liver-specific transporter responsible for the export of conjugated bile acids into the canaliculus (28). Homozygous loss-of-function mutations of this gene cause the cholestatic diseases PFIC2 and BRIC2 (29). The role of genetic variation at this locus in ICP susceptibility has recently been explored in detail, with several recurrent mutations identified, together with the confirmation of the p.444A variant as a population susceptibility allele (30). Further analysis of an Italian cohort has further established ABCB11 variation as playing a role in ICP susceptibility (31).
Acting together with ABCB4, this transporter is responsible for bile salt-dependent bile flow. The phosphatidyl choline flopped by ABCB4 complexes with bile salts exported by ABCB11 and cholesterol transported by ABCG5/G8 to form mixed micelles in the canalicular tree and protect the ductal epithelium from the detergent action of the bile salts.
The involvement of another familial cholestasis gene, ATP8B1 (mutated in PFIC1/BRIC1), has not been established definitively. This protein is proposed to function as a phosphatidyl serine flippase in the canalicular membrane and has been studied to a limited extent in ICP cohorts (32,33). Furthermore, recent work has identified a functional interdependence with the ABCB4 protein (34).
Bile salt-independent bile flow is primarily the result of another transporter, ABCC2 (MRP2 (multidrug resistance-related protein 2)), that transports bilirubin and other organic anions (including some bile acids) across the canalicular membrane (35). Involvement of genetic variation around ABCC2 in ICP has been reported in South American populations (36).
The activity of the transporters responsible for bile formation is regulated by the principal bile acid sensor FXR (NR1H4), and functional variation of this receptor has been identified in ICP (37). FXR acts as the master regulator of bile acid homeostasis by sensing intracellular concentrations of bile acids and regulating their metabolism and transport via modulation of promoter activity in key genes (38,39). In addition, key feedback signaling from the gut is performed by fibroblast growth factor 19 (FGF19). This is a peptide hormone released by enterocytes via FXR-mediated transcriptional activation. It downregulates hepatocyte CYP7A1 activity (via FGFR4/β-klotho-mediated transduction) and hence bile acid synthesis (40) and is vital in controlling appropriate levels of hepatic FXR activity.
In addition to ABCB4 and ABCB11, these other loci represent biologically plausible candidates for a role in ICP susceptibility. Of note, a number of small studies in a variety of populations have examined other loci postulated to play a role in ICP susceptibility (reviewed in Dixon and Williamson (17) without providing definitive evidence of involvement. Although several of the genes have already been demonstrated to play a role in ICP in Caucasians (ABCB4, ABCB11, and NR1H4), there is still a paucity of information concerning the contribution of other key genes (ABCC2, ATP8B1, and FGF19) in the etiology of ICP.
In order to clarify and expand the genetic factors known to play a role in ICP, we sought to investigate the common variation around the six candidate loci described above in a large ICP cohort and further investigate these findings in a second cohort.
METHODS
Patients (initial cohort)
A cohort of 563 ICP patients of white western European origin together with 642 controls of the same origin from the Rotunda Thrombophilia study (41) were analyzed in a case–control design. This cohort is an extension of our previously described cases (27) and includes all previously studied women. For this study, fasting and nonfasting maternal serum bile acid levels were used to make the diagnosis of ICP as it was not possible to obtain fasting samples from all women attending antenatal clinics.
This study conformed to the guidelines outlined by the 1975 Declaration of Helsinki and permission was obtained from the Ethics Committees of the Hammersmith Hospitals NHS Trust, London (REC 97/5197), the Ethics Committee of the Faculty of Medicine at the University of Göteborg, University Hospital Aachen, University Hospital Dusseldorf, and the Ethics committee of the Rotunda Hospital, Dublin.
All ICP patients were diagnosed on the basis of clinical symptoms in combination with routine laboratory investigations, as described previously (27,33). ICP was diagnosed in pregnant women with pruritus without evidence of rash apart from dermatitis artefacta, and confirmation of the diagnosis was made with raised serum liver transaminases and/or bile acids. Women were excluded if another hepatic disorder was diagnosed following identification of abnormal hepatitis serology (hepatitis A, B, or C), or extrahepatic biliary obstruction following ultrasound examination. Genomic DNA was extracted from buffy coats prepared from EDTA blood samples using standard methods, principally with the Qiagen blood mini kit (Qiagen, Crawley, UK). DNA purity and concentration were determined with an ND-1000 Nanodrop spectrophotometer (Thermofisher Scientific, Loughborough, UK).
Polymorphisms around the six candidate loci (ABCB4, ABCB11, NR1H4, ABCC2, ATP8B1, and FGF19) drawn from the HapMap database (version 2, release 22) were analyzed with Haploview (v4.1, Broad Institute, Boston, MA). Markers were selected from the genomic region of each locus including 5 kb up- and downstream of the coding region. Following filtering of the marker set to exclude rare alleles (minor allele frequency <0.05) the tagger algorithm was used to select markers from the HapMap database. This identified a set of SNPs that efficiently captured genetic variation at each locus so that all untyped variants had high correlation (R2 >0.8) with one member of the typed set. In cases where markers had been previously reported to be associated with ICP, these markers were force-included in the selection algorithm.
In total, 83 markers were identified encompassing the six loci as follows: 23 around ABCB11, 14 around ABCB4, 11 around ABCC2, 22 around ATP8B1, 6 around NR1H4, and 5 around FGF19 (Supplementary Table S1 online).
Primers were designed for each selected SNP using “Primer Picker” (KBioscience, Hoddesdon, UK). Genotyping was undertaken using a competitive allele-specific PCR SNP genotyping system utilizing FRET quencher cassette oligonucleotides (KASPar, KBioscience) with DNA concentrations adjusted as appropriate.
Statistical analysis
Provided there was no evidence of departure from Hardy–Weinberg equilibrium (Pearson's χ2 test), SNP data were analyzed by Armitage's trend test (PLINK v1.07; see ref. (42). Subsequently, the data were tested for a difference in haplotype frequencies between cases and controls by χ2 tests (R v2.10.1, Haplostats V1.4, see ref. (43) at each locus. All P values were subjected to Bonferroni correction for multiple testing by multiplying uncorrected P values by the number of independent tests performed (78 for SNP testing and 6 for haplotype analysis). Following correction, P values of <0.05 were considered significant.
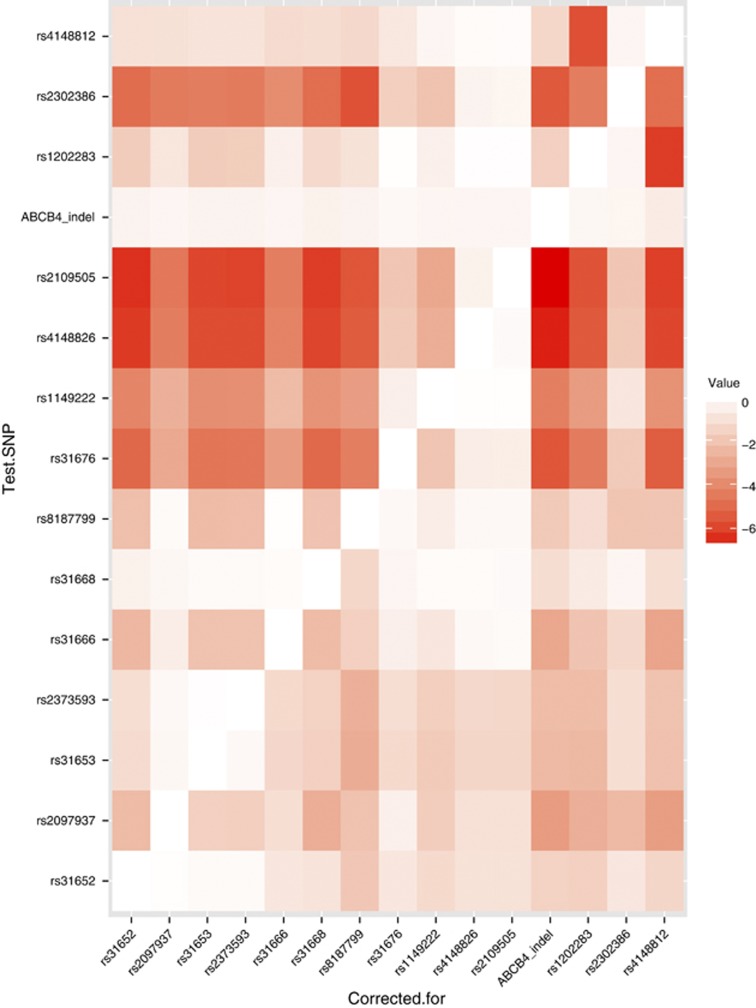
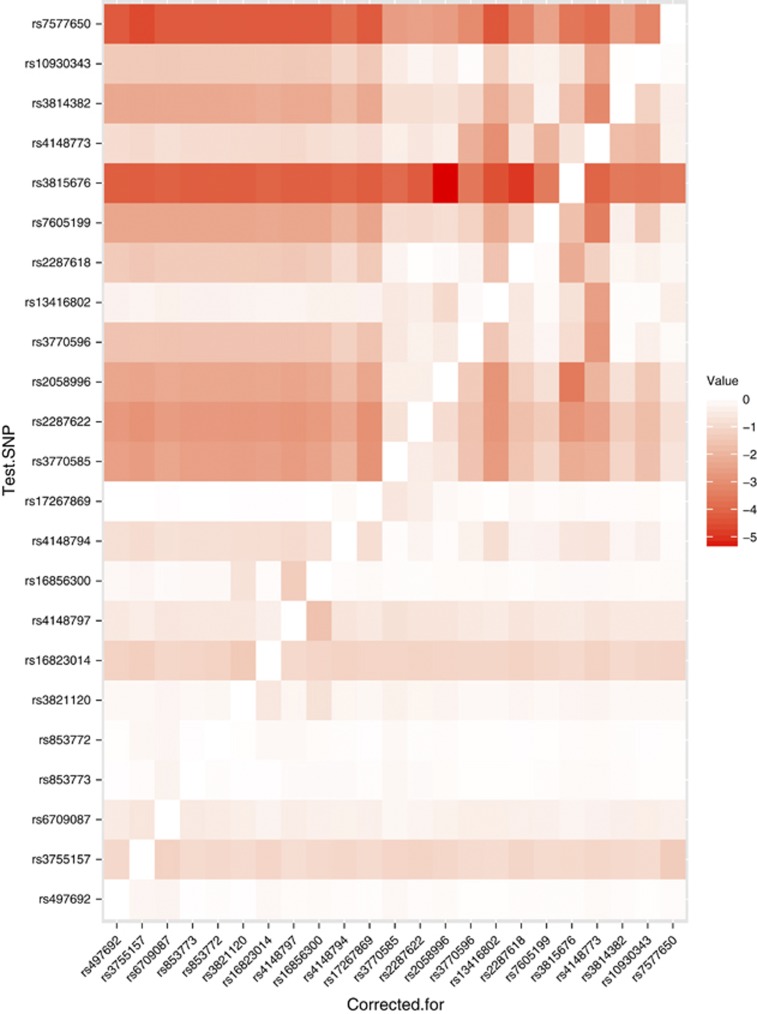
The association signals were further analyzed using logistic regression performed in R. In particular, we tested each SNP conditional on each other SNP using likelihood ratio tests to determine whether multiple signals were present at each associated loci. Heat maps, in which color indicates the strength of evidence for association, were used to visualize these results.
Second cohort analysis
The SNPs identified by this analysis, namely rs2109505 in ABCB4 together with rs7757650 and rs3815676 in ABCB11, were then tested in a second cohort. Thus, 227 further ICP cases, identified as part of on-going recruitment, were collected in the same way as the first cohort (see above), together with cases from Kings College Hospital (ethics REC 02/03/033), and DNA extracted and SNP genotyping performed as described above. None of these cases have been reported in our previous genotyping studies.
Association was tested using this cohort alone, and then in a combined analysis of the three SNPs in each cohort, with the statistical tests described above.
RESULTS
In total, 78 markers passed quality control and Hardy–Weinberg testing and were used in association analysis of the two cohorts. Association analysis with the Armitage trend test identified six SNPs in ABCB11 together with six SNPs in ABCB4 showing significant evidence for association ( Table 1). The strongest association signals were seen with rs2109505 in ABCB4 and with rs7577650 in ABCB11. No SNPs from the other four loci showed evidence of association (Supplementary Table S1).
Table 1. Single-nucleotide polymorphisms of ABCB11 and ABCB4 showing significant evidence of association with ICP (trend test).
| dbSNP | Location (bp) | MAF (ICP) | MAF (C) | OR (95% CI) | P value | P (corr) |
|---|---|---|---|---|---|---|
| ABCB11 | ||||||
| rs2287622 | 169,538,574 | 0.33 | 0.40 | 1.39 (1.17–1.64) | 0.000159 | 0.012363 |
| rs2058996 | 169,542,195 | 0.38 | 0.46 | 1.39 (1.18–1.65) | 0.000145 | 0.011287 |
| rs7605199 | 169,564,700 | 0.47 | 0.45 | 1.36 (1.16–1.6) | 0.000374 | 0.029172 |
| rs3815676 | 169,578,625 | 0.015 | 0.052 | 3.32 (1.93–5.71) | 7.45×10−6 | 0.000581 |
| rs3814382 | 169,597,234 | 0.45 | 0.38 | 1.35 (1.15–1.60) | 0.000511 | 0.039874 |
| rs7577650 | 169,599,456 | 0.31 | 0.40 | 1.52 (1.28–1.80) | 2.4×10−6 | 0.00018 |
| ABCB4 | ||||||
| rs2097937 | 86,868,839 | 0.16 | 0.22 | 1.50 (1.22–1.86) | 0.00018 | 0.014017 |
| rs31676 | 86,907,816 | 0.16 | 0.24 | 1.45 (1.15–1.83) | 1.72×10−6 | 0.000134 |
| rs1149222 | 86,911,711 | 0.14 | 0.27 | 1.63 (1.31–2.02) | 8.66×10−6 | 0.000676 |
| rs4148826 | 86,912,355 | 0.11 | 0.20 | 1.95 (1.54–2.45) | 1.56×10−8 | 1.22×10−6 |
| rs2109505 | 86,917,342 | 0.11 | 0.20 | 1.98 (1.57–2.49) | 5.9×10−9 | 4.6×10−7 |
| rs2302386 | 86,929,880 | 0.078 | 0.14 | 1.98 (1.51–2.60) | 2.95×10−7 | 2.3×10−5 |
bp, base pairs; C, control; CI, confidence interval; dbSNP, single-nucleotide polymorphism database; ICP, intrahepatic cholestasis of pregnancy; MAF, minor allele frequency; OR, odds ratio; P (corr), P value corrected for multiple testing.
The data set was investigated further by haplotype analysis across each of the six loci, which identified significant differences in frequencies between cases and controls for ABCB11 and ABCB4. No other significant differences in haplotype frequencies across the other loci (ABCC2, ATP8B1, NR1H4, and FGF19) were identified (Supplementary Table S2).
ABCB11
HapMap analysis identified 23 tagging polymorphisms that passed quality control. Association analysis of these markers subsequent to genotyping identified six markers significantly associated with altered risk for ICP: rs228762, rs2058996, rs7605199, rs3815676, rs3814382, and rs7577650 ( Table 1). The most strongly associated marker was rs7577650 (trend test corrected P=1.8×10−4).
ABCB4
HapMap analysis identified 14 polymorphisms that passed quality control. Association analysis of these markers subsequent to genotyping identified six markers significantly associated with altered risk for ICP: rs2097937, rs31676, rs1149222, rs4148826, rs2109505, and rs2302386 ( Table 1). The most strongly associated marker was rs2109505 (P=4.6×10−7).
ABCC2, ATP8B1, NR1H4, and FGF19
Around these loci, HapMap analysis identified 41 polymorphisms but none showed significant evidence for association in our cohort (Supplementary Table S1). The previously reported ABCC2 association (36) was not detected in this cohort.
Haplotype analysis
The data set was investigated further by haplotype analysis across all six loci (Supplementary Tables S2 and S3a and b). Significant differences were identified in haplotype distributions between cases and controls for ABCB11 (global corrected P value 0.02) and ABCB4 (global corrected P value 5.76×10−5).
Conditional analysis of association and second cohort analysis
Heat maps were generated using conditional analysis with logistic regression to visualize the patterns of evidence for association across the locus. This analysis showed that the association signal in ABCB4 was explained by the SNP rs2109505, with no evidence for further signals ((P>0.05; Figure 1). This is shown in the diagram by the red horizontal signal for this SNP (i.e., significant evidence for association regardless of which other SNPs are corrected for) and the white column (“corrected for”) for this SNP, indicating no other significant signal when the effect of this SNP is corrected for. In contrast, analysis of the associated SNPs in ABCB11 identified evidence for two independent signals, at the SNPs rs3815676 and 7577650: each remains significant (P<10−4) correcting for the other, or any other SNP at this locus (Figure 2). In the heat map, the strong red color again indicates evidence for association, but in this case the “corrected for” column shows evidence of the second signal, indicated by the darker red block. The key SNPs identified by this analysis were genotyped in the second cohort. Using the controls from the first cohort for comparison, the association with each SNP was confirmed and when analyzed together the combined cohort indicated strong evidence for association ( Table 2). The odds ratios calculated for the trend tests are allelic and, as ICP is rare, are approximately equivalent to the corresponding relative risk. Thus, for the association with rs3825676 (a rare SNP compared with the majority studied), the odds ratio indicate a risk of ICP that is 3.79 times higher in the homozygote than the heterozygote (with a corresponding reduction in risk in the other homozygote genotype.). For the other SNPs analyzed using this test, the same principle applies. Hence, for the much commoner SNP rs7577650, the homozygotes have a 1.4 times change in risk for ICP, and for rs2109505 in ABCB4 (again a much commoner SNP), the odds ratio shows a change in risk of 2.06-fold.
Figure 1.
Heat map showing conditional analysis of association signals at the ABCB4 locus. Each row plots the P value for the row single-nucleotide polymorphism (SNP), corrected for the column SNP, with color intensity indicating the size of the P value by order of magnitude units, hence 5 indicates a P value of <10−5. Thus, the map explores the independent effect of each SNP by removing the effects of each of the others in turn to determine if a single or multiple association signals are present.
Figure 2.
Heat map showing conditional analysis of association signals at ABCB11 locus using the same technique as Figure 1 to identify whether multiple associations are present.
Table 2. Second cohort analysis and combined analysis of ICP associations (trend test P values).
| dbSNP identifier | Original cohort | Second cohort | Combined cohort (n=790) | Combined MAF (ICP) | MAF (C) | OR (95% CI) |
|---|---|---|---|---|---|---|
| ABCB11 | ||||||
| rs3815676 | 5.8×10−4 | 4.6×10−4 | 4.6×10−8 | 0.013 | 0.049 | 3.79 (2.30–6.26) |
| rs7577650 | 1.8×10−4 | 1.9×10−2 | 2.9×10−6 | 0.32 | 0.40 | 1.46 (1.25–1.70) |
| ABCB4 | ||||||
| rs2109505 | 4.6×10−7 | 3.3×10−6 | 1.6×10−11 | 0.11 | 0.20 | 2.06 (1.67–2.54) |
C, control; CI, confidence interval; dbSNP, single-nucleotide polymorphism database; ICP, intrahepatic cholestasis of pregnancy; MAF, minor allele frequency; OR, odds ratio.
DISCUSSION
We present here the results of the analysis of six candidate loci for susceptibility to ICP. We have extended and expanded prior studies on these biologically plausible candidates and confirmed key roles for variation around two of the loci studied by identifying groups of strongly associated polymorphisms. We have demonstrated significant association of SNPs and haplotypes of the key transporters responsible for bile formation, the bile salt export pump ABCB11, and the phosphatidyl choline floppase ABCB4, with ICP. After determining the key association signals with conditional analysis, these findings were confirmed in a second cohort.
Heterozygous mutant alleles of ABCB4 have been described in a spectrum of cholestatic disease, including ICP (17). Our analysis has demonstrated strong association at this locus, confirming that common polymorphisms together with rare mutations (16,17,19,20) in this region are implicated in ICP. Of the six polymorphisms showing association, the synonymous variant rs2109505 (c.711 A>T, p.I237I) was the most significant, identified as the key signal by conditional analysis and confirmed in the second cohort. This association has been previously reported in smaller population-based studies in ICP (22,27). In silico analysis of splicing using predictive tools together with mini-gene construct mRNA analysis in COS-1 cells failed to identify an effect of this variant on ABCB4 splicing (data not shown).
The contribution of synonymous mutations to disease susceptibility via a number of different mechanisms is being increasingly realized (44); however, the possibility remains that this association is because of linkage disequilibrium between rs2109505 and an unknown causative variant.
The association seen at the ABCB11 locus (the bile salt export pump) expands the role of this gene in ICP. We and others previously identified rare heterozygous mutant alleles in ICP, together with an association with rs2287622 (the p.444A polymorphism) (30,45,46). In this study we have extended the analysis across the gene, and unraveled the genetic architecture underlying susceptibility at this locus. Importantly the association signal in the cases, confirmed in the expanded cohort with the second group of patients, is composed of two signals, the major one from rs7577650 but with rs3815676 contributing independently to risk, although at a relatively low frequency. The p.444A variant remains associated with disease but our comprehensive analysis suggests that a different marker drives this association, namely rs7577650. These findings have important implications as studies in other populations have postulated roles for the 444A polymorphism in drug-induced cholestasis (46) and in hepatitis C-related cirrhosis (47). Given that the SNP rs7577650 is intronic, and that the regression analysis shows it to be driving the association seen with 444A, it is possible that the underlying functional variation has yet to be identified. Deep resequencing of this region using next-generation platforms will be necessary to identify the catalog of variation around the identified associations and identify the underlying causative risk alleles, for both the rs7577650/444A signal and the new independent signal we have identified, rs3815676.
Genetic variation around the multidrug resistance-related protein ABCC2 represents an attractive candidate for ICP susceptibility because of the localization and function of the protein (35). A previous study of a South American population proposed an association with ICP (36), but this was not replicated in a European Caucasian population (46). By saturating the genomic region of ABCC2 with tagging SNPs in our larger cohort (n=563 vs. 70 in the initial report) we have shown that common variation of this transporter does not play a major role in ICP susceptibility in our Caucasian cohort (including the SNP identified in the initial report (rs3740066)).
The causative gene for PFIC1/BRIC1 was also included in our analysis. Previous small studies have reported heterozygous SNPs of ATP8B1 in ICP cases (32,33) but have not demonstrated conclusive functional effects of these variants in in vitro studies (48). Our analysis has excluded common variation around this locus as having a large role in the disease.
We previously identified genetic variation at the NR1H4 (farnesoid-X receptor) locus in our ICP cohort (37) and demonstrated functional effects for some of these variants. However, the population frequencies were very low in both cases and controls. In the expanded cohort used in this study, the frequency of these variants has not changed significantly in cases or controls. Thus, common genetic variation around FXR is unlikely to play a major role in the etiology of ICP. A weakness of this study is that the cohort is not sufficiently large to identify rare variants that confer susceptibility to ICP, despite being the largest available cohort to our knowledge. This is exemplified by the NR1H4 results that confirmed the presence of rare functional variants but did not establish these variants as common susceptibility alleles for ICP.
FGF19 secretion and signaling represents a key part of the enterohepatic circulation by regulating CYP7A1 expression (40), and hence this was a plausible candidate locus for ICP. However, common variation around this locus does not seem to play a major role in disease susceptibility in our cohort.
It is important to recognize however that weaker associations at these other candidate loci may be identified by future studies with much larger cohorts.
In addition to the genetic susceptibility to ICP, evidence is accumulating for the involvement of other factors in this complex condition. During the third trimester when the disease usually presents, circulating levels of reproductive hormones are at their highest and recent work has identified progesterone metabolites as capable of reducing bile acid uptake in hepatocytes, thereby having a potential role in ICP (49,50,51). Estrogen metabolites have also been implicated (52). Environmental factors have also been identified that may play a role, including seasonal variation and selenium levels (53). A proposed disease mechanism is that altered concentrations of specific hormone metabolites can unmask the disease in genetically susceptible individuals.
The identification of a role for common variation in ABCB4 and ABCB11 in the etiology of ICP is of relevance in a clinical context. The condition has a spectrum of severity. At present, the principal way the severity of maternal disease is classified is in terms of the maternal serum bile acid level. ICP typically presents in the third trimester of pregnancy and resolves after delivery of the baby. Approximately 20% of cases have early-onset disease (10) and a similar proportion of cases have associated biliary diseases when they are not pregnant, (54) e.g., cholelithiasis. Although there are studies that demonstrate an association between the level of serum bile acids and rates of adverse pregnancy outcome (8,10) the relationships are not straightforward. It will be of value to clinicians managing this condition for future studies to establish whether specific genotypes in the mother are associated with an increased risk of specific clinical features of ICP, including severity of disease and associated maternal and offspring diseases. Furthermore, patients with ABCB4 mutations respond to ursodeoxycholic acid, and hence it is feasible that the SNPs reported in this study will be associated with treatment response. This study was not designed to evaluate treatment response, and hence it was not possible to establish whether this is the case. At present, it is also not known whether mutation screening is justified in ICP. However, with the emerging use of next-generation sequencing technology in the clinic, it will be valuable for future studies to evaluate whether women with early-onset severe disease, particularly if they have a family history of ICP or related biliary disease, have mutations in ABCB4 or ABCB11.
We have identified population risk alleles for ICP in the two genes primarily responsible for bile formation; the phosphatidyl choline floppase ABCB4 (MDR3) and the bile salt export pump ABCB11. The identification of the functional variants that underlie these association signals will lead to a greater understanding of the mechanisms responsible for susceptibility to this cholestatic disease of pregnancy.
Acknowledgments
We thank all the patients who agreed to participate in this study, researchers responsible for collecting samples, and all the midwives and clinicians contributing to the care of patients in this study. We are grateful for the assistance of the Women's Health Research Centre staff at Imperial College London.
Guarantor of the article: Peter H. Dixon, PhD.
Specific author contributions: Study conception and design: P.H.D., C.A.W., J.W., and C.W.; patient recruitment and clinical data collection, sample collection, and processing: J.C., J.D., S.C., R.B., R.M., S.J., A.S., V.G., P.P., M.S., R.K., F.L., R.M.T., C.L.C., H.-U.M., A.G., K.N., and M.G.; data generation and analysis: P.H.D., C.A.W., R.B., P.P., M.S., and J.W.; manuscript drafting: P.H.D., J.W., and C.W. All authors reviewed, commented upon, and approved the final submission.
Financial support: Our research is supported by the British Liver Trust, the Lauren Page Trust, the Genesis Research Trust, the Biomedical Research Centre at Imperial College Healthcare NHS Trust, the German Research Council, and the Department of Research and Development (FoU), Västra Götaland, Sweden.
Potential competing interests: None.
Supplementary Material
References
- Lammert F, Marschall HU, Matern S. Intrahepatic cholestasis of pregnancy. Curr Treat Options Gastroenterol. 2003;6:123–132. doi: 10.1007/s11938-003-0013-x. [DOI] [PubMed] [Google Scholar]
- Geenes V, Williamson C. Intrahepatic cholestasis of pregnancy. World J Gastroenterol. 2009;15:2049–2066. doi: 10.3748/wjg.15.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon A, Tribe RM, Nelson-Piercy C, et al. Pruritus in pregnancy: a study of anatomical distribution and prevalence in relation to the development of obstetric cholestasis. Obstet Med. 2011;3:25–29. doi: 10.1258/om.2010.090055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribe RM, Dann AT, Kenyon AP, et al. Longitudinal profiles of 15 serum bile acids in patients with intrahepatic cholestasis of pregnancy. Am J Gastroenterol. 2010;105:585–595. doi: 10.1038/ajg.2009.633. [DOI] [PubMed] [Google Scholar]
- Rioseco AJ, Ivankovic MB, Manzur A, et al. Intrahepatic cholestasis of pregnancy: a retrospective case-control study of perinatal outcome. Am J Obstet Gynecol. 1994;170:890–895. doi: 10.1016/s0002-9378(94)70304-3. [DOI] [PubMed] [Google Scholar]
- Fisk NM, Storey GN. Fetal outcome in obstetric cholestasis. Br J Obstet Gynaecol. 1988;95:1137–1143. doi: 10.1111/j.1471-0528.1988.tb06791.x. [DOI] [PubMed] [Google Scholar]
- Williamson C, Hems LM, Goulis DG, et al. Clinical outcome in a series of cases of obstetric cholestasis identified via a patient support group. BJOG. 2004;111:676–681. doi: 10.1111/j.1471-0528.2004.00167.x. [DOI] [PubMed] [Google Scholar]
- Glantz A, Marschall HU, Mattsson LA. Intrahepatic cholestasis of pregnancy: relationships between bile acid levels and fetal complication rates. Hepatology. 2004;40:467–474. doi: 10.1002/hep.20336. [DOI] [PubMed] [Google Scholar]
- Dann AT, Kenyon AP, Wierzbicki AS, et al. Plasma lipid profiles of women with intrahepatic cholestasis of pregnancy. Obstet Gynecol. 2006;107:106–114. doi: 10.1097/01.AOG.0000189096.94874.9c. [DOI] [PubMed] [Google Scholar]
- Geenes V, Chappell LC, Seed PT, et al. Association of severe intrahepatic cholestasis of pregnancy with adverse pregnancy outcomes: a prospective population-based case-control study Hepatology 2013(in press). [DOI] [PMC free article] [PubMed]
- Glantz A, Marschall HU, Lammert F, et al. Intrahepatic cholestasis of pregnancy: a randomized controlled trial comparing dexamethasone and ursodeoxycholic acid. Hepatology. 2005;42:1399–1405. doi: 10.1002/hep.20952. [DOI] [PubMed] [Google Scholar]
- Chappell LC, Gurung V, Seed PT, et al. Ursodeoxycholic acid versus placebo, and early term delivery versus expectant management, in women with intrahepatic cholestasis of pregnancy: semifactorial randomised clinical trial. BMJ. 2012;344:e3799. doi: 10.1136/bmj.e3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacq Y, Sentilhes L, Reyes HB, et al. Efficacy of ursodeoxycholic acid in treating intrahepatic cholestasis of pregnancy: a meta-analysis. Gastroenterology. 2012;143:1492–1501. doi: 10.1053/j.gastro.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Papacleovoulou G, Abu-Hayyeh S, Nikolopoulou E, et al. Maternal cholestasis during pregnancy programs metabolic disease in offspring. J Clin Invest. 2013;123:3172–3181. doi: 10.1172/JCI68927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes H, Ribalta J, Gonzalez-Ceron M. Idiopathic cholestasis of pregnancy in a large kindred. Gut. 1976;17:709–713. doi: 10.1136/gut.17.9.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbach RT, Sivak DA, Braun WE. Familial recurrent intrahepatic cholestasis of pregnancy: a genetic study providing evidence for transmission of a sex-limited, dominant trait. Gastroenterology. 1983;85:175–179. [PubMed] [Google Scholar]
- Dixon PH, Williamson C. The molecular genetics of intrahepatic cholestasis of pregnancy. Obstet Med. 2008;1:65–71. doi: 10.1258/om.2008.080010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloranta ML, Heinonen S, Mononen T, et al. Risk of obstetric cholestasis in sisters of index patients. Clin Genet. 2001;60:42–45. doi: 10.1034/j.1399-0004.2001.600106.x. [DOI] [PubMed] [Google Scholar]
- Jacquemin E, Cresteil D, Manouvrier S, et al. Heterozygous non-sense mutation of the MDR3 gene in familial intrahepatic cholestasis of pregnancy. Lancet. 1999;353:210–211. doi: 10.1016/S0140-6736(05)77221-4. [DOI] [PubMed] [Google Scholar]
- Dixon PH, Weerasekera N, Linton KJ, et al. Heterozygous MDR3 missense mutation associated with intrahepatic cholestasis of pregnancy: evidence for a defect in protein trafficking. Hum Mol Genet. 2000;9:1209–1217. doi: 10.1093/hmg/9.8.1209. [DOI] [PubMed] [Google Scholar]
- de Vree JM, Jacquemin E, Sturm E, et al. Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc Natl Acad Sci USA. 1998;95:282–287. doi: 10.1073/pnas.95.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenbach R, Linton KJ, Wiltshire S, et al. ABCB4 gene sequence variation in women with intrahepatic cholestasis of pregnancy. J Med Genet. 2003;40:e70. doi: 10.1136/jmg.40.5.e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floreani A, Carderi I, Paternoster D, et al. Hepatobiliary phospholipid transporter ABCB4, MDR3 gene variants in a large cohort of Italian women with intrahepatic cholestasis of pregnancy. Dig Liver Dis. 2008;40:366–370. doi: 10.1016/j.dld.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Schneider G, Paus TC, Kullak-Ublick GA, et al. Linkage between a new splicing site mutation in the MDR3 alias ABCB4 gene and intrahepatic cholestasis of pregnancy. Hepatology. 2007;45:150–158. doi: 10.1002/hep.21500. [DOI] [PubMed] [Google Scholar]
- Lang C, Meier Y, Stieger B, et al. Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacogenet Genomics. 2007;17:47–60. doi: 10.1097/01.fpc.0000230418.28091.76. [DOI] [PubMed] [Google Scholar]
- Rosmorduc O, Poupon R. Low phospholipid associated cholelithiasis: association with mutation in the MDR3/ABCB4 gene. Orphanet J Rare Dis. 2007;2:29. doi: 10.1186/1750-1172-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth HE, Glantz A, Keppeler H, et al. Intrahepatic cholestasis of pregnancy: the severe form is associated with common variants of the hepatobiliary phospholipid transporter ABCB4 gene. Gut. 2007;56:265–270. doi: 10.1136/gut.2006.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe J, Stieger B, Meier PJ. Functional expression of the canalicular bile salt export pump of human liver. Gastroenterology. 2002;123:1659–1666. doi: 10.1053/gast.2002.36587. [DOI] [PubMed] [Google Scholar]
- Strautnieks SS, Bull LN, Knisely AS, et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet. 1998;20:233–238. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- Dixon PH, van Mil SW, Chambers J, et al. Contribution of variant alleles of ABCB11 to susceptibility to intrahepatic cholestasis of pregnancy. Gut. 2009;58:537–544. doi: 10.1136/gut.2008.159541. [DOI] [PubMed] [Google Scholar]
- Anzivino C, Odoardi MR, Meschiari E, et al. ABCB4 and ABCB11 mutations in intrahepatic cholestasis of pregnancy in an Italian population. Dig Liver Dis. 2013;45:226–232. doi: 10.1016/j.dld.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Painter JN, Savander M, Ropponen A, et al. Sequence variation in the ATP8B1 gene and intrahepatic cholestasis of pregnancy. Eur J Hum Genet. 2005;13:435–439. doi: 10.1038/sj.ejhg.5201355. [DOI] [PubMed] [Google Scholar]
- Mullenbach R, Bennett A, Tetlow N, et al. ATP8B1 mutations in British cases with intrahepatic cholestasis of pregnancy. Gut. 2005;54:829–834. doi: 10.1136/gut.2004.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A, Romero MR, Kunne C, et al. Complementary functions of the flippase ATP8B1 and the floppase ABCB4 in maintaining canalicular membrane integrity Gastroenterology 20111411927–1937.e1921–1924. [DOI] [PubMed] [Google Scholar]
- Jemnitz K, Heredi-Szabo K, Janossy J, et al. ABCC2/Abcc2: a multispecific transporter with dominant excretory functions. Drug Metab Rev. 2010;42:402–436. doi: 10.3109/03602530903491741. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Castano G, Burgueno A, et al. Association of the multidrug-resistance-associated protein gene (ABCC2) variants with intrahepatic cholestasis of pregnancy. J Hepatol. 2008;48:125–132. doi: 10.1016/j.jhep.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Van Mil SW, Milona A, Dixon PH, et al. Functional variants of the central bile acid sensor FXR identified in intrahepatic cholestasis of pregnancy. Gastroenterology. 2007;133:507–516. doi: 10.1053/j.gastro.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Papacleovoulou G, Abu-Hayyeh S, Williamson C. Nuclear receptor-driven alterations in bile acid and lipid metabolic pathways during gestation. Biochim Biophys Acta. 2011;1812:879–887. doi: 10.1016/j.bbadis.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Jones S. Mini-review: endocrine actions of fibroblast growth factor 19. Mol Pharm. 2008;5:42–48. doi: 10.1021/mp700105z. [DOI] [PubMed] [Google Scholar]
- Cooley SM, Donnelly JC, Walsh T, et al. The impact of positive acquired thrombophilia serology on ultrasound, obstetric outcome and the placenta in a low-risk primigravid population. Obstet Med. 2011;4:15–19. doi: 10.1258/om.2010.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, et al. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Pauli-Magnus C, Lang T, Meier Y, et al. Sequence analysis of bile salt export pump (ABCB11) and multidrug resistance p-glycoprotein 3 (ABCB4, MDR3) in patients with intrahepatic cholestasis of pregnancy. Pharmacogenetics. 2004;14:91–102. doi: 10.1097/00008571-200402000-00003. [DOI] [PubMed] [Google Scholar]
- Meier Y, Zodan T, Lang C, et al. Increased susceptibility for intrahepatic cholestasis of pregnancy and contraceptive-induced cholestasis in carriers of the 1331T>C polymorphism in the bile salt export pump. World J Gastroenterol. 2008;14:38–45. doi: 10.3748/wjg.14.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata R, Baur K, Stieger B, et al. A common polymorphism in the ABCB11 gene is associated with advanced fibrosis in hepatitis C but not in non-alcoholic fatty liver disease. Clin Sci. 2011;120:287–296. doi: 10.1042/CS20100246. [DOI] [PubMed] [Google Scholar]
- Folmer DE, van der Mark VA, Ho-Mok KS, et al. Differential effects of progressive familial intrahepatic cholestasis type 1 and benign recurrent intrahepatic cholestasis type 1 mutations on canalicular localization of ATP8B1. Hepatology. 2009;50:1597–1605. doi: 10.1002/hep.23158. [DOI] [PubMed] [Google Scholar]
- Glantz A, Reilly SJ, Benthin L, et al. Intrahepatic cholestasis of pregnancy: amelioration of pruritus by UDCA is associated with decreased progesterone disulphates in urine. Hepatology. 2008;47:544–551. doi: 10.1002/hep.21987. [DOI] [PubMed] [Google Scholar]
- Abu-Hayyeh S, Martinez-Becerra P, Sheikh Abdul Kadir SH, et al. Inhibition of Na+-taurocholate Co-transporting polypeptide-mediated bile acid transport by cholestatic sulfated progesterone metabolites. J Biol Chem. 2010;285:16504–16512. doi: 10.1074/jbc.M109.072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Hayyeh S, Papacleovoulou G, Lovgren-Sandblom A, et al. Intrahepatic cholestasis of pregnancy levels of sulfated progesterone metabolites inhibit FXR resulting in a pro-cholestatic phenotype. Hepatology. 2013;57:716–726. doi: 10.1002/hep.26055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milona A, Owen BM, Cobbold JF, et al. Raised hepatic bile acid concentrations during pregnancy in mice are associated with reduced farnesoid X receptor function. Hepatology. 2010;52:1341–1349. doi: 10.1002/hep.23849. [DOI] [PubMed] [Google Scholar]
- Kauppila A, Korpela H, Makila UM, et al. Low serum selenium concentration and glutathione peroxidase activity in intrahepatic cholestasis of pregnancy. Br Med J. 1987;294:150–152. doi: 10.1136/bmj.294.6565.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall HU, Shemer EW, Ludvigsson JF, et al. Intrahepatic cholestasis of pregnancy and associated hepatobiliary disease: a population-based cohort study. Hepatology. 2013;58:1385–1391. doi: 10.1002/hep.26444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.