Abstract
Multiple cytosine guanine dinucleotides (CpG island) are found in the VIM promoter region. The levels of VIM promoter methylation and VIM gene expression were investigated in 7 cervical cancer cell lines and 50 human tissue samples with a distinctive degree of malignant trans-formation. While multiple CpG sites in the VIM promoter were highly methylated in CIN III and invasive carcinoma cells, they were rarely methylated in normal cells. Our result shows that methylation in the VIM promoter appears to start from CIN I and CIN II, relatively early stages of multistep carcinogenesis. This epigenetic alteration in VIM promoter suggests the availability as a biomarker for the early diagnosis and prevention of cervical cancer. We also show that hypermethylation in the VIM promoter is responsible for transcriptional silencing of the VIM gene in cervical cancer cells. In addition, our result shows that exogenous overexpression of the VIM gene in SiHa cervical cancer cells slightly activated cell proliferation and migration as shown in soft agar colony formation and migration assays.
Keywords: cervical cancer, epigenetic gene regulation, methylation biomarker, VIM
INTRODUCTION
Cervical cancer still remains a fatal disease worldwide in spite of various advanced treatments, (Pisani et al., 1999). Upon human papillomavirus (HPV) infection, cervical epithelial cells develop from premalignant cervical lesions to malignant invasive cancer via multistep processes (Bosch et al., 1995; Durst et al., 1992; Walboomers et al., 1999; Zur, 2002); Normal, CIN I (Cervical Intraepithelial Neoplasia I, mild dysplasia), CIN II (moderate dysplasia), CIN III (severe dysplasia), and invasive cervical carcinoma. Fortunately, most cases of CIN do not develop into cervical cancer and only a small percentage of cases becomes cervical cancer if they are not well treated (Agorastos et al., 2005). Moreover, this process is a relatively slow event with a long interval between infection and cancer. Therefore, the development of diagnostic systems for early detection is considered as very important to prevent cancer. Among diagnostic technologies, DNA methylation based biomarker has been considerered as a powerful tool for the diagnosis of many cancers (Laird, 2003).
In the effort of finding the sensitive and effective DNA methyaltion based biomarker, we initially focused on the VIM gene. Several studies have showed that VIM gene can be used as a biomarker for the early detection of several cancers. Shirahata et al. (2009) showed that aberrant methylation of the VIM gene is frequently found to be methylated in advanced colorectal carcinomas and is closely related with colorectal carcinomas. Chen et al. (2005) found that VIM gene is transcriptionally inactive in normal colon epithelial cells with very low level of methylation on CpG region near the first exon. However, this region is found to be highly methylated in primary tumor of colon cancer. Costa et al. (2010) suggested VIM gene as a biomarker that allows for early detection of bladder cancer using urine samples. The methylation levels in VIM gene promoter were significantly higher in bladder cancer tissues compared to normal bladder mucosa. Kitamura et al. (2009) showed that well-differentiated adenocarcinoma was signifycantly methyllated in gastric carcinomas when compared to poorly differentiated. These findings prompted us to investigate the correlationship between the level of VIM promoter methylation and cervical cancer development as well as the effect of promoter methylation on VIM gene expression during the development of cervical cancer. VIM gene encodes vimentin, a member of the intermediate filament family that is especially found in connective tissue. Intermediate filaments, along with microtubules and actin microfilaments, make up the cytoskeleton (Fuchs and Weber, 1994). Vimentin is known to be involved in various biological processes including maintaining cell shape and stabilizing cytoskeletal interactions. Viemntin also plays vital roles in cell adhesion, migration, and signaling (Ivaska et al., 2007). VIM gene expression is considered as a classic marker of mesenchymal cells, such as fibroblasts (Lodish et al., 1995).
In this report, we suggest that VIM promoter methylation can be used as an effective biomarker for the diagnosis of cervical cancer. In addition, we show that overexpression of VIM activates proliferation and migration in cervical cancer cells.
MATERIALS AND METHODS
Cervical cancer cell lines and human tissue samples
Seven cervical cancer cell lines were used for this study. C33A, CaSki, HeLa, and SiHa cells were purchased from the American Type Culture Collection (ATCC). The other cell lines, SNU- 17, -703, and -1299 were obtained from the Korean Cell Line Bank (KCLB, Korea). Each cell line was grown in one of the following different media: C33A, HeLa, and SiHa cells in DMEM medium (WelGENE Inc, Korea); CaSki, SNU-703, and SNU- 1299 cells in RPMI 1640 medium (GibcoBRL, Korea); SNU-17 in AR5 medium (KCLB, Korea). All media were supplemented with 10% fetal bovine serum (GibcoBRL, Korea) and 1% Antibiotic- Antimycotic (GibcoBRL, Korea). All of these cells were cultured at 37℃ in a humidified atmosphere composed of 95% air and 5% CO2.
Preparation of genomic DNA and total RNA from tissue samples of patients
A total of 50 human tissue samples were kindly provided by Dr. Chang-Jin Kim at Soonchunhyang University Hospital (Korea). These tissue samples originated from cervical cancer patients and their information is represented according to the histological tumor grade and age in Table 1. All patients provided with informed consent and the procedure of obtaining tissue samples was approved by the institutional review board of Soonchunhyang University Cheonan Hospital. The uterine cervical tissues were obtained either by punch biopsy or hysterectomy from patients having cervical intraepithelial neoplasm invasive carcinoma. The histopathologic diagnoses of tissues were made by a pathologist. The normal uterine epithelium and cervical epithelial lesions were manually dissected with a 26 gauge needle through a light microscope by a pathologist and collected in a lysis buffer (10 mM Tris-HCl, pH 8.0; 0.1 mM EDTA, pH 8.0; 2% SDS; proteinase K, 0.15 mg), following incubation at 60℃ until the samples were completely lysed. Genomic DNA was then extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol precipitated at -20℃ overnight. The DNA pellet was then dissolved and quantified using a Nano- Drop ND-1000 Spectrophotometer (Nanodrop Technologies, USA). After the same tissue samples were treated with the lysis buffer at 55℃ for 3 h, total RNA was extracted using TRIzol Reagent (Invitrogen, Korea) according to the manufacturer’s protocol.
Table 1.
Human cervix tissue samples used in MSP and BSP analysis
| Numbera | Diagnosis samplesb (Age) | ||||
|---|---|---|---|---|---|
| Normalc | CIN I | CIN II | CIN III | Carcinoma | |
| 1 | 08-3488-1d (48) | 07-692 (44) | 07-4215 (34) | 07-852 (51) | 7227 (75) |
| 2 | 08-7568-1b (49) | 07-949 (32) | 07-4406 (27) | 07-1631 (48) | 12593 (42) |
| 3 | 08-3782-1b (40) | 07-1573 (40) | 07-4556 (36) | 07-1858 (56) | 10931 (75) |
| 4 | 08-7275-1a (43) | 07-1888 (46) | 07-4751 (37) | 07-1854 (41) | 6956 (59) |
| 5 | 08-3665-1b (68) | 07-1899 (45) | 07-4926 (38) | 07-2346 (43) | 10919 (82) |
| 6 | 08-5386-1a (44) | 07-1857 (42) | 07-5660 (39) | 07-2914 (53) | 8026 (71) |
| 7 | 08-3513-1a (42) | 07-1855 (43) | 07-5881 (41) | 07-8302 (25) | 5739 (38) |
| 8 | 08-3513-1b (NA)d | 07-2687 (22) | 07-5908 (41) | 07-9619 (38) | 6851 (56) |
| 9 | 08-3488-1b (NA) | 07-2888 (25) | 07-5929 (22) | 07-10051 (72) | 4321 (81) |
| 10 | 08-5889-1a (45) | 07-3349 (23) | 07-6000 (27) | 07-10432 (65) | 5822 (46) |
aNumber of cases examined
bSamples were collected from patient in different histological type of cervical cancer (different tumor grade or clinical stage) in cervical carcinogenesis (See “Materials and Method”).
cNormal tissue samples are from adjacent tumor tissue.
dNot available
Reverse transcription (RT)-PCR
Following the manufacturer’s instructions, total RNA was extracted from 7 cervical cancer cell lines using a RNeasy Minikit (Qiagen, Korea). For reverse transcription, 1 μg RNA of each sample was subjected to cDNA synthesis using an oligo (dT) primer and RevertAid First Strand cDNA Synthesis Kit (Fermentas, Korea) in accordance with the manufacturer’s instructions. PCR amplification was performed using 10 ng cDNA, a primer pair of pRT-VIM-F and pRT-VIM-R, and AccuPower PCR PreMix (Bioneer, Korea). The nucleotide sequence of the primers and the conditions for gene amplification are listed in Table 2. Primers were synthesized by Bioneer (Bioneer, Korea). The amplification reaction was carried out using a GeneAmp PCR System 9700 from Applied Biosystems. The amplification products were electrophoresed on a 2% agarose gel stained with ethidium bromide and then visualized using a UV-illuminator.
Table 2.
Oligonucleotide sequences and conditions for PCR analysis
| Primer namesa | Primer sequencesb | Conditionsc | Amplicon size (bp) | Sources |
|---|---|---|---|---|
| pRT-VIM-F | AGCAGGAGTCCACTGAGTACCGGAG | 55℃, 30 s, 35 | 377 | This study |
| pRT-VIM-R | TGAGTGGGTATCAACCAGAGGGAGTG | |||
| pMSP-UM-VIM-F | ATTTTTTTGGTTTAGTTTTAGGTGG | 60℃, 30 s, 35 | 143 | This study |
| pMSP-UM-VIM-R | ACATAATCCCATTACTTCAACACT | |||
| pMSP-M-VIM-F | GGATTTTTTTGGTTTAGTTTTAGGC | 60℃, 30 s, 35 | 146 | This study |
| pMSP-M-VIM-R | AACATAATCCCGTTACTTCAACG | |||
| pBSP-VIM-F | CGTAAGCTTGGGTGAGTTTAGTTTAGATTATTAT | 56℃, 30 s, 35 | 209 | This study |
| pBSP-VIM-R | CTAGAATTCAAAAAAAATCCCCTCCCACTAC | |||
| pVIM-BamHI-F | CCGTGGATCCATGTCCACCAGGTCCGTGT | 60℃, 1 min, 35 | 1421 | This study |
| pVIM-EcoRI-R | CCTAGAATTCTTATTCAAGGTCATCGTGA | |||
aF, forward primer; R, reverse primer; M, methylated-specific primers; U, unmethylated-specific primers.
bAll sequences shown in the 5′ → 3′ direction. Restriction enzymes are underlined as italic.
cConditions are shown as the order of annealing temperature, elongation time, and number of cycles.
Methylation-specific PCR (MSP) and bisulfite sequencing PCR (BSP) analyses
Genomic DNA was extracted from 7 cervical cancer cell lines using DNeasy Blood & Tissue Kit (Qiagen, Korea) following the manufacturer’s instructions. Bisulfite treatment was performed using 1 μg of genomic DNA at 55℃ for 16 h according to the instructions for the EZ DNA Methylation Kit (Zymo Research, USA) and 1 unit of HotStart prime Taq (Qiagen, Korea). For MSP analysis, bisulfite-treated DNA was subjected to PCR amplification using two pairs of primer for unmethylated and methylated DNAs, pMSP-UM-VIM-F/pMSP-UM-VIM-R and pMSP-M-VIM-F/ pMSP-M-VIM-R. For BSP analysis, bisulfite-treated DNA was subjected to PCR reaction using the primers of pBSP-VIM-F and pBSP-VIM-R (Table 2). The amplified PCR products were cloned into a pBlueScript-SK (+) vector using HindIII and EcoRI restriction enzymes and transformed into DH5α competent cells. Plasmids purified from amphicillin-positive colonies were sequenced using an M13 forward or reverse primer by Solgent (Korea). The nucleotide sequence of each primer and the amplification conditions are given in Table 2. Primers used in this study were synthesized by Bioneer (Korea).
5′-aza-2′-deoxycytidine (5′-Aza-dC) and trichostain A (TSA) treatment
SNU-1299 cells were treated with a DNA methyltransferase inhibitor of 5′-Aza-dC (Sigma, Korea) and/or a histone deacetylase inhibitor of TSA (Sigma, Korea). Briefly, the cells were plated onto 100 mm plates for 24 h before treatment. The SNU- 1299 cells were treated with 1 μM of 5′-Aza-dC for 24 h, 48 h, 72 h or 96 h and/or 0.1 or 0.3 μM of TSA for 48 h.
Construction of pcDNA3-VIM
For the functional study of vimentin protein, VIM overexpressing vector, pcDNA3-VIM, was constructed as follows. The full length cDNA of human VIM was amplified using a primer set of pVIM-BamHI- F/pVIM-EcoRI-R (Table 2) and the pCMV-SPORTS6- VIM plasmid as a template. pCMV-SPORTS6 plasmid (Invitrogen Life Technology) containing the human VIM cDNA was obtained from the 21C Frontier Human Gene Bank (http:// genebank.kribb.re.kr). Amplified PCR products were cloned into pcDNA3 using BamHI and EcoRI restriction enzymes to generate the pcDNA3-VIM plasmid.
Soft agar colony forming assay and wound healing migration assay
SiHa cells were transiently transfected with 4 μg of pcDNA3- VIM plasmid using the LipofectAMINE™ reagent (Gibco) according to the manufacturer’s instructions. A pcDNA3 plasmid without the VIM gene was also transfected as a control. Overexpression of the VIM protein was verified by Western blotting using the VIM antibody (Epitomics Inc. Cat. #2707-1). For the soft agar colony forming assay, cells were then counted, diluted, and seeded in duplicate at 50 cells per culture dish (6-well plate). Cells were incubated for 26 h at 37℃. Colonies were allowed to grow for 22 days and counted after staining with 1% Gimmsa solution. For the wound healing assay, SiHa cells (1 × 105) were plated onto 60-mm tissue culture dishes and allowed to create a confluent monolayer. Cells were grown for 12 h after transfection with pcDNA3 or pcDNA3-VIM plasmid. The cell monolayer was then scratched in a straight line to make a “scratched wound” with a 0.2 ml pipette tip, and the cell debris was removed by washing the cells with phosphate-buffered saline. DMEM medium supplemented with 10% FBS was added and closure of the scratch was photographed at 0 h, 24 h, and 48 h.
Statistical analysis
Statistical analyses were carried out with the Statistical Package for the Social Sciences (SPSS) software. Associations of the VIM promoter methylation and cervical carcinogenesis were determined using a Chi-Square test (also chi-squared or χ2 test). Statistical significance was set at a P value of less than 0.05.
RESULTS
The methylation status of VIM promoter and its gene expression in 7 cervical cancer cell lines
Putative CpG islands in the VIM promoter were predicted using the Methprimer program [http://www.urogene.org//methprimer, (Li and Dahiya, 2002)] with the default setting (%GC > 50%, ObsCpG/ExpCpG > 0.6) (Fig. 1A). CpG sites are frequently found in the VIM promoter region (Fig. 1A) and the methylation level of the VIM promoter was investigated in 7 cervical cancer cell lines by using MSP and BSP assays. In the MSP assay, methylation specific bands were detected in all tested cervical cancer cell lines except CaSki, HeLa and SNU-17 (Fig. 1B). In the case of SiHa and SNU-1299 cell lines, very high levels of methylated PCR products were amplified from the VIM promoter (Fig. 1B). The accuracy of the MSP method in detecting methylated DNA was verified by using a BSP assay. Our data showed that VIM promoters were exclusively hypermethylated in SiHa and SNU-1299 cervical cancer cell lines (Fig. 1C). We obtained generally concordant results for the methylation status of the VIM promoter when compared with the MSP and BSP assays. It is well known that hypermethylation in the promoter region often repress gene expression in many cancer cells. This implies a possible connection between VIM promoter hypermethylation and gene expression in cervical cancer cells. Therefore, we measured the VIM gene expression level in 7 cervical cancer cell lines. The genomic structure of the VIM gene is shown in Fig. 2A, which is mapped to chromosome 18p11.32. The full-length transcript of the VIM gene is shown in Fig. 2B. From the RT-PCR results, a relatively high level of VIM gene expression was observed in the CaSki, HeLa and SNU- 17 cell lines, while low levels of VIM gene expression were found in C33A and SNU-703. However, no transcript was detected in the SiHa and SNU-1299 cell lines (Fig. 2C). These results reveal an inverse relationship between the VIM promoter methylation level and gene expression. For example, high levels of VIM promoter methylation were detected in the SiHa and SNU-1299 cervical cancer cell lines, wherein low levels of VIM gene expression were observed. Therefore, we subsequently tested whether the VIM promoter hypermethylation is responsible for transcriptional silencing of the VIM gene in cervical cancer cells.
Fig. 1. Methylation status of the VIM promoter in 7 cervical cancer cell lines. (A) Predicted CpG island in the genomic DNA containing the VIM gene. CpG dinucleotides in the VIM promoter are shown as vertical lines. Number, +1, indicates the translation start site of the VIM gene. Arrows indicate the position of primers used in the MSP or BSP assays (Table 2). The oligonucleotides used in this work are named as indicated. Abbreviations F and R denote forward and reverse, respectively. (B) MSP analysis of the VIM promoter in 7 cervical cancer cell lines. Methylation status is presented by the presence or absence of bands. UM or M represents PCR products amplified by oligonucleotide primers specific for unmethylated or methylated DNA, respectively. (C) Bisulfite sequencing of the CpG sites in the VIM promoters of 7 cervical cancer cell lines. Methylated or unmethylated cytosines are represented as closed or open circles, respectively. Their spacing reflects the CpG density of the region. Each row represents an individual cloned allele that was sequenced following sodium bisulfite DNA modification.
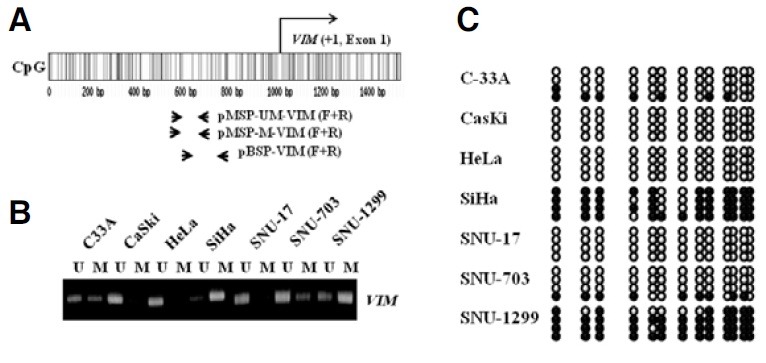
Fig. 2. VIM gene expression in 7 cervical cancer cell lines. (A) Genomic DNA for the VIM gene is represented as exons (black boxes) and introns (thin lines linking the black boxes). The putative minimal promoter of the VIM gene is indicated by a horizontal thick line. Numbers indicate the position of exons coding the VIM gene. (B) The full-length transcript of the VIM gene is shown, where exons are indicated as boxes and numbers. Arrows indicate the positions of pRT-VIM-F and -R primers used in the RT-PCR (See Table 2). Abbreviations F and R denote forward and reverse, respectively. Nucleotide sequences were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/ BLAST/). (C) The transcriptional level of the VIM gene was measured in 7 cervical cancer cell lines. β-actin served as an internal control for the integrity of the cDNA.
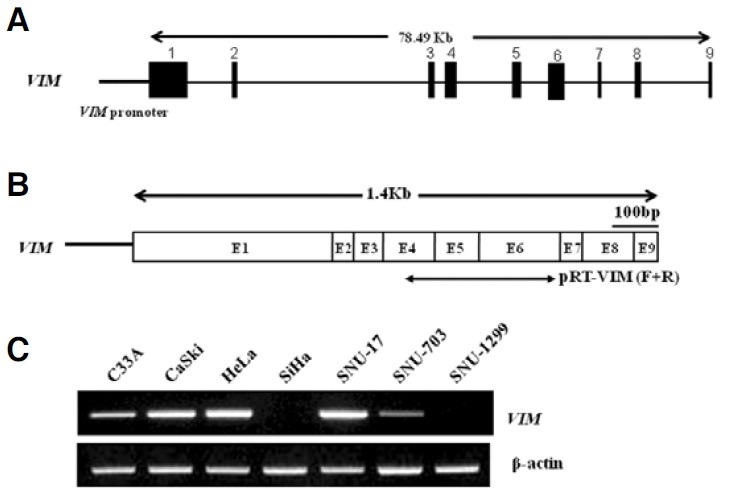
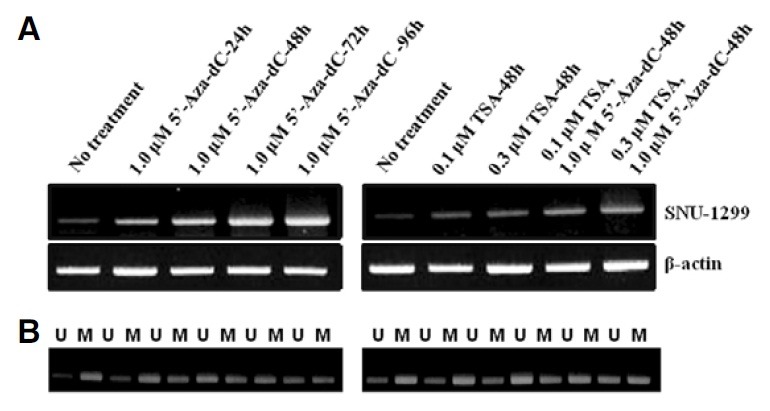
Reactivation of the VIM gene expression by the treatment of 5′-Aza-dC and TSA
To test the hypothesis that VIM promoter hypermethylation repress VIM gene expression in cervical cancer cell lines, the total RNA was isolated from the SNU-1299 cell line after treatment of 5′-Aza-dC and/or TSA as described in “Materials and Methods”. The expression level of the VIM gene was measured using RT-PCR. Consistent with our hypothesis, the VIM gene expression was reactivated by 1.0 μM 5′-Aza-dC treatment in SNU-1299 cell lines as shown in Fig. 3A. This result suggests that hypermethylation in the VIM promoter is associated with transcriptional silencing of VIM gene in SNU-12 99 cells. The VIM gene expression was also found to be reactivated by 0.1 or 0.3 μM TSA, confirming the negative effect of deacetylated DNA on VIM gene expression. Demethylation of the VIM promoter by 5′-Aza-dC and/or TSA.was confirmed with MSP analysis. The treatment of 5′-Aza-dC and/or TSA PCR obviously increased the products amplified by oligonucleotide primers specific for unmethylated DNA (Fig. 3B). Therefore, we concluded that VIM promoter hypermethylation is at least in part responsible for the transcriptional silencing of VIM gene expression in cervical cancer cells.
Fig. 3. RT-PCR analysis demonstrating reactivation of VIM gene expression in SNU-1299 cell line treated with demethylase 5′-Aza-dC and/or TSA. (A) RT-PCR analysis. RT-PCR was carried out using cDNA from cells subjected to different concentrations of the drug (See “Methods and Materials”). The β-actin served as an internal control for the integrity of the cDNA. (B) MSP analysis of the VIM promoter after treatment of 5′-Aza-dC and/or TSA. Methylation levels are presented by the thickness of bands. UM or M represents PCR products amplified by oligonucleotide primers specific for unmethylated or methylated DNA, respectively.
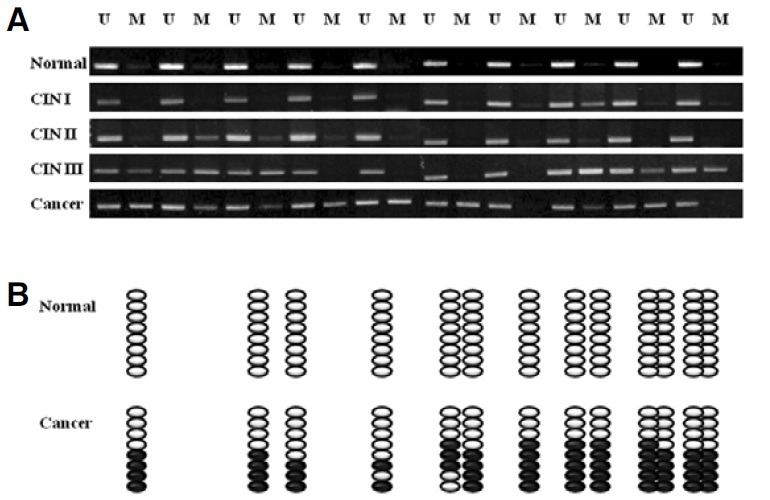
Methylation incidence in VIM promoter during multistep processes of cervical carcinogenesis
Discovery of the VIM promoter methylation in cervical cancer cell lines suggested the possibility of the same situation in vivo. Therefore, the present investigation was extended to human tissue samples from five different stages of cervical carcinogenesis; normal cervix, CIN I, CIN II, CIN III, and invasive carcinoma. We evaluated the methylation levels of the VIM promoter in a total of fifty cervical tissue samples. Briefly, genomic DNA extracted from tissue samples was modified by sodium bisulfite and then subjected to MSP and BSP analyses (See “Materials and Methods”). Our MSP analysis showed that nonor partial methylated VIM promoters were detected in normal, CIN I, and CIN II tissue samples, whereas a high degree of methylation was observed in CIN III and invasive carcinoma (Fig. 4A). A BSP assay was also performed to confirm the MSP results. The first normal and cancer tissue samples were chosen as a representative samples for BSP anaylysis. In other words, eight clones from each normal and cancer samples were sequenced and evaluated for the frequency of VIM methylation. None of the normal cells exhibited hypermethylation. In contrast, invasive carcinoma cells showed high levels of methylation in the VIM promoter regions (Fig. 4B). Our methylation analysis showed a correlation between VIM hypermethylation and cervical cancer development. The level of VIM hypermethylation was very low at the early stages but started to increase with tumor development. This implies that VIM promoter methylation is associated with cervical cancer development.
Fig. 4. Schematic representation of correlation between VIM promoter hypermethylation and cervical cancer development. (A) MSP analysis of VIM promoter in normal, CIN I, CIN II, CIN III, and invasive cervical carcinoma tissues. The symbols of U or M denote PCR products amplified by oligonucleotide primers specific for unmethylated or methylated DNA, respectively. (B) Bisulfite sequencing of CpG sites in the VIM promoters of normal and carcinoma tissue samples. Each row represents an individual cloned allele that was sequenced. Circles represent CpG sites and their spacing reflects the CpG density of the region. Unmethylated or methylated cytosines are represented as open or closed circles, respectively.
Functional study of vimentin by soft agar colony forming and migration assays
We have shown that methylation level of the VIM promoter increased as normal cells developed into cervical cancer. It is well known that the transcription of several key tumor suppressor genes is found to be inactivated by promoter hypermethylation in many cancer cells (Baylin and Ohm, 2006; Egger et al., 2004; Esteller et al., 2001; Issa, 2004; Ushijima, 2005). These facts imply that VIM may act as a tumor suppressor gene in the cervix. Therefore, a soft agar colony forming assay was used to examine the tumor suppression ability of the VIM gene (See “Materials and Methods”). Figure 5A shows the number of G418-resistant colonies arising from cells transfected with the control expression vector (pcDNA3) and VIM overexpressing vector (pcDNA3-VIM). The colony number slightly but not significantly increased in the VIM overexpressing SiHa cells compared to the control cells (Figs. 5A and 5B). The VIM overexpression increased the colony-forming ability about 20% (P < 0.01). This result implies that vimentin does not provide a tumor suppressor activity in cervical cancer cells. The expression of vimentin protein in the pcDNA3-VIM transfected SiHa cells was confirmed by Western blot analysis (Fig. 5C). We also performed a wound healing migration assay to investigate whether VIM expression is involved in cervical cancer cell migration. The pcDNA3-VIM transfected SiHa cells migrated and completely covered the scratch after 48 h, whereas the pcDNA3 transfected SiHa cells migrated slowly, remaining the the scratched area uncovered even after 48 h. This result suggests that the overexpression of the VIM gene activates wound healing of SiHa cells (Fig. 5D). Exogenous expression of VIM in SiHa cell line exhibiting hypermethylation in the VIM promoter slightly increased motility in vitro, indicating the involvement of VIM in cell migration.
Fig. 5. Functional study of VIM in SiHa cervical cancer cells. (A) Soft agar colony-forming assays. SiHa cervical calls were transfected with pcDNA3 or pcDNA3-VIM vectors and the transfected cells were plated with 50 cell numbers per 6-well plate for 22 days. (B) Cervical cancer colony formation by the pcDNA3-VIM expression vector. (C) Western blotting verifying the overexpression of the VIM gene. (D) Phase micrographs of SiHa cells at various times after monolayer wounding. The pcDNA3 or pcDNA3-VIM-transfected SiHa cells were scratched and the closure of the scratch was photographed at the indicated times. This experiment was repeated 3 times and the closest result average is presented.
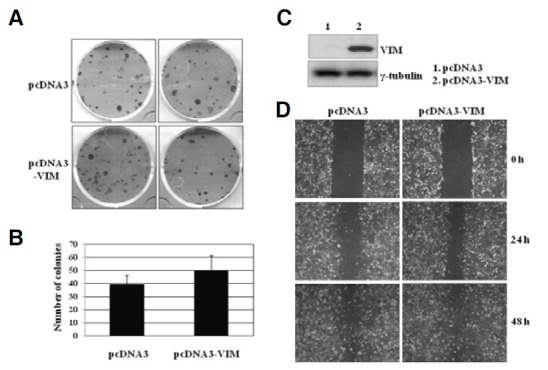
Taken together, these findings showed that VIM expression positively affects the proliferation and migration in cervical cancer cells.
DISCUSSION
In an effort to identify a methyaltion-based sensitive biomarker for the detection of cervical cancer, we evaluated the levels of VIM promoter methylation in human tissue samples as well as cervical cancer cell lines. Our study showed that methylation level of the VIM promoter increased as normal cells developed into cervical cancer. We also show that VIM promoter hypermethylation is at least in part responsible for the transcriptional silencing of VIM gene in cervical cancer cells.
It is well known that the transcription of several key tumor suppressor genes is found to be inactivated by promoter hypermethylation in many cancer cells. It was therefore expected that VIM may act as a tumor suppressor gene in the cervix. However, our result showed that vimentn appears to have an oncogenic characteristics rather than to suppress a tumorigenic phenotype. VIM expression appears to positively affect the proliferation and migration in cervical cancer cells. This result is consistent with previous work, in which down-regulation of VIM gene expression inhibits migration and invasion of colon and breast cancer cells (McInroy and Maatta, 2007). Many tumor suppressor genes have been identified that they are commonly unmethylated and expressed in normal colon mucosa but are methylated and silenced in cancer. In which the overexpression of the tumor suppressor genes in cancer cells suppress the tumorigenic phenotype. However, the overexpression of the VIM gene in cervical cancer cells did not show the same effect. In fact, it has been knwon that vimentin is usually transcriptionally silent in normal epithelium. Many studies involving vimentin have suggested that deregulation of VIM gene expression linked to cancer. For example, the VIM gene is known to be overexpressed in hepatocellular carcinoma (HCC) and is significantly associated with HCC metastasis, suggesting that overexpression of vimentin may plays a vital role in the metastasis of HCC (Hu et al., 2004). Present result showed that VIM promo-ter hypermethyaltion in SUN-1299 cervical cancer cell line is responsible for the transcriptional silencing of VIM gene. How-ever, the transcriptional level of VIM gene needs to be tested in more cervical cancer cell lines and also needs to be compared between normal and cervical cancer cells.
In conclusion, our study shows that the VIM promoter is frequently hypermethylated in cervical cancer cells and this epigenetic alteration of the VIM gene is associated with cancer cell development as well as transcriptional silencing of the VIM gene. This suggests that VIM promoter methylation can be used as a methylation-based tumor biomarker for the diagnosis of cervical cancer. In addition, our result shows the positive effect of vimentin on proliferation and migration in cervix.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (Ministry of Education, Science and Technology), Sookmyung Women’sUniversity (2010), and a grant (Project No. 10012714) from Ministry of Knowledge Economy, Republic of Korea.
References
- 1.Agorastos T., Miliaras D., Lambropoulos A.F., Chrisafi S., Kotsis A., Manthos A., Bontis J. Detection and typing of human papillomavirus DNA in uterine cervices with coexistent grade I and grade III intraepithelial neoplasia: biologic progression or independent lesions? Eur. J. Obstet. Gynecol. Reprod. (2005);121:99–103. doi: 10.1016/j.ejogrb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Baylin S.B., Ohm J.E. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. (2006);6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 3.Bosch F.X., Manos M.M., Munoz N., Sherman M., Jansen A.M., Peto J., Schiffman M.H., Moreno V., Kurman R., Shah K.V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J. Natl. Cancer Inst. (1995);87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 4.Chen W.D., Han Z.J., Skoletsky J., Olson J., Sah J., Myeroff L. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J. Natl. Cancer Inst. (2005);97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 5.Costa V.L., Henrique R., Danielsen SA., Duarte-Pereira S., Eknaes M., Skotheim R.I., Rodrigues A., Magalhães J.S., Oliveira J., Lothe R.A., et al. Three epigenetic biomarkers, GDF15, TMEFF2 and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin. Cancer Res. (2010);16:5842–5851. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 6.Durst M., Glitz D., Schneider A., Zur H.H. Human papillomavirus type 16 (HPV 16) gene expression and DNA replication in cervical neoplasia: analysis by in situ hybridization. Virology. (1992);189:132–140. doi: 10.1016/0042-6822(92)90688-l. [DOI] [PubMed] [Google Scholar]
- 7.Egger G., Liang G., Aparicio A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. (2004);429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M., Corn P.G., Baylin S.B., Herman J.G. A gene hypermethylation profile of human cancer. Cancer Res. (2001);61:3225–3229. [PubMed] [Google Scholar]
- 9.Fuchs E., Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. (1994);63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 10.Hu L., Lau S.H., Tzang C.H., Wen J.M., Wang W., Xie D., Huang M., Wang Y., Wu M.C., Huang J.F., et al. Association of Vimentin overexpression and hepatocellular car-cinoma metastasis. Oncogene. (2004);23:298–302. doi: 10.1038/sj.onc.1206483. [DOI] [PubMed] [Google Scholar]
- 11.Issa J.P. CpG island methylator phenotype in cancer. Nat. Rev. Cancer. (2004);4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 12.Ivaska J., Pallari H.M., Nevo J., Eriksson J.E. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp. Cell Res. (2007);313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 13.Jung S., Jeong D., Kim J., Yi L., Koo K., Kim C.H., Kim C.J., Lee M.S. The role of hLHX6-HMR as a methylation biomarker for early diagnosis of cervical cancer. Oncology Rep. (2010);23:1675–1682. doi: 10.3892/or_00000811. [DOI] [PubMed] [Google Scholar]
- 14.Jung S., Yi L., Jeong D., Kim J., Koo K., Lee J., An S., Oh T., Kim C.H., Kim C.J., et al. The role of ADCYAP1, Adenylate cyclase activating polypeptide 1, as a methylation biomarker for early diagnosis of cervical cancer. Oncology Rep. (2011);25:245–252. [PubMed] [Google Scholar]
- 15.Kitamura Y.H., Shirahata A., Sakata M., Goto T., Mizukami H., Saito M.M., Ishibashi K., Kigawa G., Nemoto H., Sanada Y., et al. Frequent methylation of Vimentin in well-differentiated gastric carcinoma. Anticancer Res. (2009);29:2227–2230. [PubMed] [Google Scholar]
- 16.Laird P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. (2003);3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 17.Li L.C., Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. (2002);18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 18.Lodish H., Baltimore D., Berk A., Zipursky S.L., Mastudaira P., Darnell J. Molecular cell biology. 3rd ed. Scientific American Books; New York, USA: (1995). [Google Scholar]
- 19.McInroy L., Maatta A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem. Biophys. Res. Commun. (2007);360:109–114. doi: 10.1016/j.bbrc.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 20.Pisani P., Parkin D.M., Bray F., Ferlay J. Erratum: Estimates of the worldwide mortality from 25 cancers in 1990. Int. J. Cancer. (1999);83:870–873. doi: 10.1002/(sici)1097-0215(19991210)83:6<870::aid-ijc35>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Shirahata A., Sakata M., Sakuraba K., Goto T., Mizukami H., Saito M., Ishibashi K., Kigawa G., Nemoto H., Sanada Y., et al. Vimentin methylation as a marker for advanced colorectal carcinoma. Anticancer Res. (2009);29:279–281. [PubMed] [Google Scholar]
- 22.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat. Rev. Cancer. (2005);5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 23.Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. (1999);189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 24.Zur H.H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. (2002);2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]


