Abstract
Scratch (scrt) genes are neural-specific in mammals, but their homologues have not been well studied in nonmammalian vertebrates. In this report, we isolated three zebrafish scrt genes, scratch1a (scrt1a), scratch1b (scrt1b), and scratch2 (scrt2), which belong to the Snail superfamily of zinc finger transcription factors. Spatiotemporal expression analysis revealed that scrt1a and scrt2 were initially detected in the central nervous system (CNS) during early somitogenesis while scrt1b was first detectable in neuronal clusters in the brain during late somitogenesis. Interestingly, scrt-expressing cells largely overlapped with huC-positive differentiating neurons and partially with neurogenin1-positive neuronal precursor cells. In addition, scrt-expressing cells were dramatically increased in mind bomb, a neurogenic mutant. Taken together, these results suggest that each zebrafish scrt gene is specifically expressed in neuronal cells and may be involved in differentiation of distinct neuronal populations in the vertebrate nervous system.
Keywords: central nervous system, neuronal differentiation, scratch, zebrafish
INTRODUCTION
The Scratch (Scrt) family is a member of the Snail superfamily of zinc finger transcription factors which are involved in regulating cell movement processes both during embryonic development and tumor progression (Barrallo-Gimeno and Nieto, 2005; Nieto, 2002). Like the Snail family proteins, the Scrt family contains one N-terminal basic amino acid-rich domain (SNAG), which is present only in vertebrates, and five zinc finger motifs acting as sequence-specific DNA-binding motifs. However, the Scrt family proteins are distinguished from other Snail family proteins by the presence of a Scratch domain. The SNAG domain of SCRT proteins is required for transcriptional repression and nuclear localization activities of the rat Gfi1 proto-oncoprotein (Grimes et al., 1996), although the SNAG domain of human SCRT is not necessary for such activities (Nakakura et al., 2001a). Currently, the exact function of the Scratch domain remains unclear.
Among Scratch homologues, the nematode scrt homologue ces-1 is important in controlling the death of specific neurons (Metzstein and Horvitz, 1999). In flies, scrt is expressed in most neuronal precursors and is involved in promoting neuronal cell fates (Roark et al., 1995). However, the mouse and human SCRT are expressed in newly differentiating, post-mitotic neurons and the mouse Scrt induces neuronal differentiation in P19 embryonic carcinoma cells (Nakakura et al., 2001b), implying it plays a role in neuronal differentiation. In particular, the human SCRT acts as a neural-specific transcriptional repressor in vitro by antagonizing the function of the basic helix-loop-helix (bHLH) proteins MASH-1 and E12 (Nakakura et al., 2001a; 2001b).
In this study, we characterized the expression of three zebrafish scratch paralogs during development. These genes were specifically expressed in distinct sets of neuronal populations in the central nervous system (CNS) during early embryogenesis. Interestingly, the scrt-expressing cells overlapped with huC-positive neurons and the number of scrt-expressing cells was significantly increased in the mind bomb mutant embryos, suggesting that scrt genes are expressed in differentiating neurons, similar to their mammalian counterparts.
MATERIALS AND METHODS
Zebrafish maintenance
Adult fish were maintained at 28.5℃ in a 14 h light/10 h dark cycle. Wild-type and mibta52b embryos (Itoh et al., 2003) were raised as described previously (Westerfield, 1995). Embryonic stages were determined by the hours post-fertilization (hpf) and microscopic observation. Embryos were treated with 0.2 mM PTU (1-phenyl-2-thiourea) to prevent pigment formation.
Cloning of the zebrafish scratch genes
To isolate the zebrafish scratch genes, total RNA was isolated from wild-type zebrafish embryos at 24 hpf using TRIZOL (MRC, Inc.) and reverse-transcribed. Full-length scratch1a (Genbank Acc. No. BC154637), scratch1b (BC091823), and scratch2(BC135018) were generated by PCR amplification using the following primers: scrt1a forward primer, 5′-GTTCTCCAGC GTTGCACTTGCATG-3′; scrt1a reverse primer, 5′-CACCCCA AAACTCAGAGGACATTG-3′; scrt1b forward primer, 5′-GCA GGATCAACTCTTCTATTGTGCGG-3′; scrt1b reverse primer, 5′-CTCACTTCCCTGTCACGTGTTAGGATGA-3′; scrt2 forward primer, 5′-GCGGGTCAGTCTGCGACCGATG-3′; scrt2 reverse primer, 5′-GCTCGCAAAAATCCCCACAAGACT-3′. The amplified PCR products were inserted into pEasy-T3 cloning vectors (BioPrince), and sequenced using an automated sequencer.
Whole-mount in situ hybridization
To synthesize RNA probes, pEasy-T3 plasmids containing scrt1a, scrt1b, and scrt2 full-length cDNAs were linearized with PstI, SalI, and HindIII, respectively. Anti-sense RNA probes were transcribed in vitro using T7 RNA polymerase and digoxigenin or fluorescein-labeled UTP. Antisense digoxigeninlabeled RNA probes for ngn1 (Kim et al., 1997) and huC/elavl3 (Kim et al., 1996) were produced using a digoxigenin-RNA labeling Kit (Roche, Germany) according to the manufacturer’s instructions. Whole-mount in situ hybridization was performed as previously described (Gwak et al., 2010).
RESULTS AND DISCUSSION
Isolation of zebrafish scratch genes
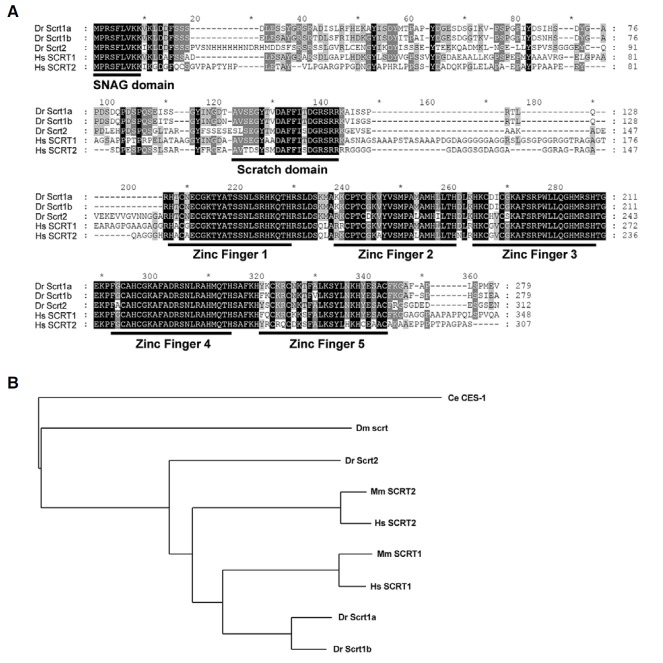
To analyze the expression patterns of the zebrafish scrt family genes in developing embryos, we isolated three scratch genes based on the similarity of amino acid sequences to human SCRATCH. The amino acid alignment of zebrafish and human SCRT revealed highly conserved regions, such as the Nterminal SNAG domain, which is not present in invertebrates, the Scratch domain in the central region, and five zinc finger motifs in the C-terminus (Fig. 1A). Based on comparison of the putative amino acid sequences and phylogenetic relationships, we designated these genes scratch1a, scratch1b, and scratch2 (Fig. 1B).
Fig. 1. Comparison of the amino acid sequences of Scratch homologues. (A) Multiple sequence alignments were performed using Clustal X. Scratch homologues contain one SNAG domain in the N-terminus, one Scratch domain in the central region, and five C2H2 zinc finger motifs in the C-terminus. Identical residues are shown in black. (B) The phylogenetic tree of zebrafish Scrts and their putative orthologues from other species. Human (Hs) SCRT1 (Genbank Acc. No. NP_112599), human SCRT2 (NP_149120), mouse (Mm) SCRT1 (NP_570963), mouse SCRT2 (NP_001153882), zebrafish (Dr) Scrt1a (NP_001107073), zebrafish Scrt1b (NP_001014369), zebrafish Scrt2 (NP_998802), Drosophila (Dm) scrt (AAA91035), and C. elegans (Ce) CES-1 (AAF01678).
Expression profiles of scratch mRNA in developing embryos
In mammals, Scrt expression appears in newly differentiating, post-mitotic neurons in the brain and spinal cord, as well as in the retina and in lung neuroendocrine cells (Marín and Nieto, 2006; Nakakura et al., 2001a). To determine scrt expression profiles during early development of zebrafish, we performed RT-PCR and whole-mount in situ hybridization. Expression of scrt1a and scrt1b was first detected at 24 hpf and of scrt2 at the 3-somite stage, coinciding with the onset of primary neurogenesis which begins at 10.5 hpf and is complete before 24 hpf (Figs. 2A, B, B′, and B′′) (Mueller and Wullimann, 2003; Park et al., 2003). At the 2-cell and shield stages, no transcripts were detected, indicating that the three scratch genes are only expressed zygotically.
Fig. 2. Expression profile of scratch genes during early development. (A) The three scratch genes are expressed from somite-stages to the early larval stage but not the cleavage- and gastrula-stages. Actin was used as a loading control. (B) At the 12-somite stage (15 hpf), scrt1a expression was detected in subsets of neurons in forebrain, hindbrain, and spinal cord. (C) At the 22-somite stage (20 hpf), scrt1b was expressed in subsets neurons in the forebrain, hindbrain and spinal cord. (D) Expression of scrt2 was first detectable in the trigeminal ganglia at the 3-somite stage (11 hpf). (E) At 24 hpf, scrt1a was expressed in the dorsal telencephalon, diencephalon, tegmentum, hindbrain and spinal cord neurons. (F) At 24 hpf, expression of scrt1b was detected in the dorsal telelecephalon, tegmentum, and hindbrain. (G) At 24 hpf, scrt2 was specifically expressed in the dorsal telencephalon, diencephalon, tegmentum, hindbrain and spinal cord neurons. (H) At 72 hpf, scrt1a was detected in the ganglion cell layer in the retina, and the tectum, midbrain-hindbrain boundary, hindbrain, and spinal cord neurons. (I) At 72 hpf, scrt1b was expressed in the tectum and hindbrain. (J) At 72 hpf, expression of scrt2 was strongly detected in the retina, forebrain, midbrain, midbrain-hindbrain boundary, and hindbrain. Abbreviations: d, diencephalon; dt, dorsal telencephalon; f, forebrain; gcl, ganglion cell layer; h, hindbrain; m, midbrain; mhb, mid-hindbrain boundary; ret, retina; sp, spinal cord; tc, tectum; teg, tegmentum; tg, trigeminal ganglia. Scale bars = 200 μm.
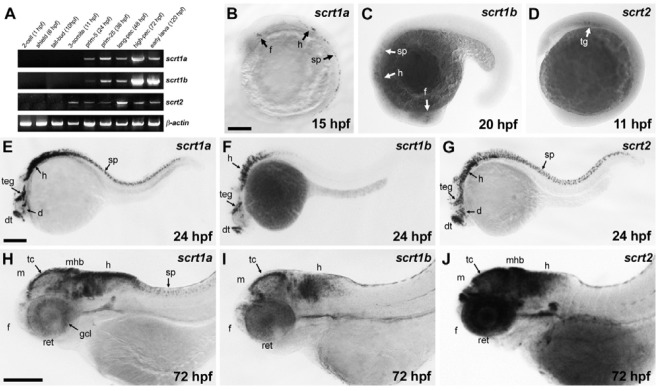
To further examine temporal and spatial expression of scratch genes during early development, whole-mount in situ hybridization was employed. Consistent with the RT-PCR results, transcripts of the three scratch genes were not detected in the cleavage and gastrula stages (data not shown). Expression of scrt1a was first detected in clusters of neurons in the forebrain, hindbrain and spinal cord at 15 hpf (Fig. 2B). At 24 hpf, scrt1a was expressed in the dorsal telencephalon, diencephalon, tegmentum, hindbrain and spinal cord neurons (Fig. 2E). As development proceeded, strong expression of scrt1a was maintained in the CNS, including the forebrain, tectum, midbrainhindbrain boundary, hindbrain, and spinal cord at 72 hpf; scrt1a was also specifically expressed in the ganglion cell layer of the retina (Fig. 2H). Expression of scrt1b was first detected in clusters of neurons in the forebrain, hindbrain, and spinal cord at 20 hpf (Fig. 2C). At 24 hpf, scrt1b was expressed in the dorsal telencephalon, tegmentum, and hindbrain, as well as weakly in spinal cord neurons (Fig. 2F). At 72 hpf, scrt1b was detectable in the forebrain, tectum, and hindbrain, but not at the midbrainhindbrain boundary, or in the retina or spinal cord (Fig. 2I).
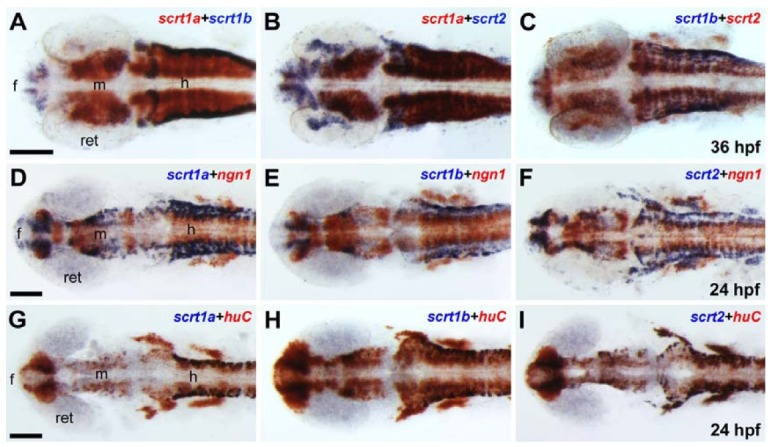
Expression of scrt2 was first detected in the trigeminal ganglia at 11 hpf (Fig. 2D). At 24 hpf, scrt2 was expressed in the dorsal telencephalon, diencephalon, tegmentum, hindbrain, and spinal cord neurons (Fig. 2G). Expression of scrt2 was detected in the forebrain, tectum, midbrain-hindbrain boundary, and hindbrain, as well as in the retina, at 72 hpf. This expression pattern partially overlapped with scrt1a and scrt1a (Fig. 2J). To further investigate the expression patterns of the three scrt paralogs, we performed double-labeling whole-mount in situ hybridization. At 36 hpf, the expression domains of scrt1a-scrt1b, scrt1a-scrt2, and scrt1b-scrt2 partially overlapped with each other at clusters of neurons in the forebrain, midbrain, and hindbrain (Figs. 3A-3C). In summary, the expression of the three scrt genes was restricted to the CNS, including the brain, spinal cord, and retina, and was similar in terms of temporal and spatial expression, implying that these genes might play similar roles during early neurogenesis.
Fig. 3. Expression of scratch genes during early neurogenesis. Images are dorsal view with anterior to the left for all panels. (A-C) 36 hpf, (D-I) 24 hpf. (A, B) Expression of scrt1a partially overlapped with that of scrt1b (A) and scrt2 (B) in the CNS at 36 hpf. (C) Expression of scrt1b overlapped predominantly with that of scrt2 at 36 hpf. (D-F) scrt1a- (D), scrt1b- (E), and scrt2-expressing cells (F) partially overlapped with ngn1- positive neuronal precursor cells at 24 hpf. (G-I) scrt1a- (G), scrt1b- (H), and scrt2-expressing cells (I) are all huC-positive, differentiating neurons in the CNS at 24 hpf. Abbreviations: f, forebrain; h, hindbrain; m, midbrain; ret, retina. Scale bars = 100 μm.
Scratch genes are expressed in differentiating neurons
To identify whether scrt-expressing cells are neuronal precursors or differentiating neurons, double-labeling whole-mount in situ hybridization was carried out with neuronal markers. neurogenin1/ ngn1, a marker for neuronal precursors, is required for the development of cranial sensory ganglia precursors (Andermann et al., 2002; Kim et al., 1997). The scrt-expressing cells partially overlapped with ngn1-positive neuronal precursors at 24 hpf (Figs. 3D-3F). However, scrt-expressing cells completely overlapped with huC/elavl3-positive differentiating neurons in the brain as well as cranial ganglia, indicating that scrt is expressed in most differentiating neurons (3G-I) (Kim et al., 1996). These results suggest that the expression of zebrafish scrt genes is specifically restricted to differentiating neurons during early neurogenesis.
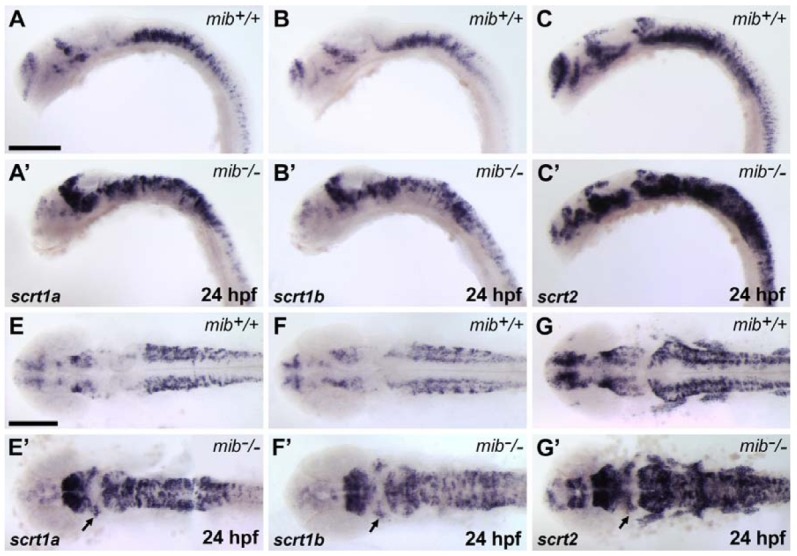
We next examined scrt expression in the mind bomb mutant (mib), which has increased levels of differentiating neurons and reduced levels of neuronal precursors in the CNS (Itoh et al., 2003). Since scrt-expressing cells completely overlap with huC-positive differentiating neurons, we expected that scrt-expressing cells would be increased in mib mutant embryos. We found that the number of scrt-expressing cells was dramatically increased and the bilateral expression domains of scrt were fused across the midline in mib mutants at 24 hpf (Figs. 4A-4G′). Furthermore, ectopic scrt-positive cells were detected in the midbrain of the mib mutant embryos (Figs. 4E′-4G′). This ectopic expression indicated that neuronal precursor cells in the mib mutant undergo premature differentiation.
Fig. 4. Increase in scratch-expressing cells in mind bomb/mib, a neurogenic mutant. (A-C′) Lateral view with anterior to the left. (E-G′) Dorsal view with anterior to the left. The numbers of scrt1a (A, A′, E, and E′), scrt1b (B, B′, F, and F′), and scrt2 (C, C′, G, and G′)-expressing cells increased markedly in mib mutant embryos at 24 hpf. In mib mutant embryos, ectopic scrt-positive cells in the midbrain were detected compared to wild-type embryos (arrows in E′, F′, and G′). Scale bars = 200 μm.
In conclusion, zebrafish scratch genes are specifically expressed in a subset of differentiating neurons in the CNS during early development, similar to their mammalian counterparts. The analysis of zebrafish scrt expression provides a first step in understanding the functional roles of these genes during vertebrate neurogenesis.
Acknowledgments
This study was supported by research funds from the Chungnam National University in 2010.
References
- 1.Andermann P., Ungos J., Raible D.W. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev. Biol. (2002);251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- 2.Barrallo-Gimeno A., Nieto M.A. The snail genes as inducers of cell movement and survival: implications in development and cancer. Development. (2005);132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 3.Grimes H.L., Chan T.O., Zweidler-McKay P.A., Tong B., Tsichlis P.N. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. (1996);16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gwak J.W., Kong H.J., Bae Y.K., Kim M.J., Lee J., Park J.H., Yeo S.Y. Proliferating neural progenitors in the developing CNS of zebrafish require Jagged2 and Jagged1b. Mol. Cells. (2010);30:155–159. doi: 10.1007/s10059-010-0101-4. [DOI] [PubMed] [Google Scholar]
- 5.Itoh M., Kim C.H., Palardy G., Oda T., Jiang Y.J., Maust D., Yeo S.Y., Lorick K., Wright G.T., Ariza-McNaughton L., et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of notch signaling by delta. Dev. Cell. (2003);4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 6.Kim C.H., Ueshima E., Muraoka O., Tanaka H., Yeo S.Y., Huh T.L., Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. (1996);216:109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim C.H., Bae Y.K., Yamanaka Y., Yamashita S., Shimizu T., Fujii R., Park H.C., Yeo S.Y., Huh T.L., Hibi M., et al. Overexpression of neurogenin induces ectopic expression of HuC in zebrafish. Neurosci. Lett. (1997);239:113–116. doi: 10.1016/s0304-3940(97)00908-7. [DOI] [PubMed] [Google Scholar]
- 8.Marín F., Nieto M.A. The expression of Scratch genes in the developing and adult brain. Dev. Dyn. (2006);235:2586–2591. doi: 10.1002/dvdy.20869. [DOI] [PubMed] [Google Scholar]
- 9.Metzstein M.M., Horvitz H.R. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol. Cell. (1999);4:309–319. doi: 10.1016/s1097-2765(00)80333-0. [DOI] [PubMed] [Google Scholar]
- 10.Mueller T., Wullimann M.F. Anatomy of neurogenesis in the early zebrafish brain. Brain Res. Dev. Brain Res. (2003);140:137–155. doi: 10.1016/s0165-3806(02)00583-7. [DOI] [PubMed] [Google Scholar]
- 11.Nakakura E.K., Watkins D.N., Schuebel K.E., Sriuranpong V., Borges M.W., Nelkin B.D., Ball D.W. Mammalian Scratch: a neural-specific Snail family transcriptional repressor. Proc. Natl. Acad. Sci. USA. (2001a);98:4010–4015. doi: 10.1073/pnas.051014098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakakura E.K., Watkins D.N., Sriuranpong V., Borges M.W., Nelkin B.D., Ball D.W. Mammalian Scratch participates in neuronal differentiation in P19 embryonal carcinoma cells. Brain Res. Mol. Brain Res. (2001b);95:162–166. doi: 10.1016/s0169-328x(01)00246-7. [DOI] [PubMed] [Google Scholar]
- 13.Nieto M.A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell. Biol. (2002);3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 14.Park S.H., Yeo S.Y., Yoo K.W., Hong S.K., Lee S., Rhee M., Chitnis A.B., Kim C.H. Zath3, a neural basic helixloop- helix gene, regulates early neurogenesis in the zebrafish. Biochem. Biophys. Res. Commun. (2003);308:184–190. doi: 10.1016/s0006-291x(03)01353-6. [DOI] [PubMed] [Google Scholar]
- 15.Roark M., Sturtevant M.A., Emery J., Vaessin H., Grell E., Bier E. scratch, a pan-neural gene encoding a zinc finger protein related to snail, promotes neuronal development. Genes Dev. (1995);9:2384–2398. doi: 10.1101/gad.9.19.2384. [DOI] [PubMed] [Google Scholar]
- 16.Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press; (1995). [Google Scholar]