Abstract
We carried out activation tagging screen to isolate genes regulating abscisic acid (ABA) response. From the screen of approximately 10,000 plants, we isolated ca 100 ABA response mutants. We characterized one of the mutants, designated ahs1, in this study. The mutant is ABAhypersensitive, and AtMYB52 was found to be activated in the mutant. Overexpression analysis to recapitulate the mutant phenotypes demonstrated that ATMYB confers ABA-hypersensitivity during postgermination growth. Additionally, AtMYB52 overexpression lines were droughttolerant and their seedlings were salt-sensitive. Changes in the expression levels of a few genes involved in ABA response or cell wall biosynthesis were also observed. Together, our data suggest that AtMYB52 is involved in ABA response. Others previously demonstrated that At- MYB52 regulates cell wall biosynthesis; thus, our results imply a possible connec-tion between ABA response and cell wall biosynthesis.
Keywords: abscisic acid (ABA), activation tagging, cell wall, MYB protein, stress response
INTRODUCTION
The plant hormone abscisic acid (ABA) affects plant growth and development throughout their life cycle from seed germination to seed maturation (Finkelstein, 2002). Although high concentrations of exogenous ABA inhibit both seed germination and postgermination growth, ABA is necessary for normal plant growth, and ABA-deficient mutants grow poorly under normal growth conditions. During vegetative growth, the endogenous ABA level increases under unfavorable environmental conditions, thereby helping plants cope with the adverse conditions (Xiong et al., 2002). In particular, ABA plays an essential role in adaptive responses to water deficit conditions by controlling stomatal movement to prevent water loss through transpiration (Schroeder et al., 2001). ABA also regulates the expression of numerous stress-responsive genes that are involved in protective response (Shinozaki and Yamaguchi-Shinozaki, 2007). During seed development, ABA regulates the accumulation of storage components and prevents the embryo from precocious germination.
Numerous proteins involved in various aspects of ABA response have been identified to date (Cutler et al., 2010). These include transcription factors, kinases, phosphatases and other signaling intermediates. Recently, several types of ABA receptors were also reported (Ma et al., 2009; Pandey et al., 2009; Park et al., 2009; Shen et al., 2006), and a core of the ABA signaling network was delineated. The ABA signaling pathway has been reconstituted in vitro using several key components that include the ABA receptor PYR1, the type 2C protein phosphatase ABI1, the protein kinase SnRK2.6 and the bZIP class transcription factor ABF2/AREB1 (Fujii et al., 2009).
A variety of transcription factors are involved in the regulation of ABA-responsive gene expression (Yamaguchi-Shinozaki and Shinozaki, 2005). A small subfamily of bZIP proteins named ABFs/AREBs regulates ABA-responsive genes via the G boxtype ABA response element (ABRE) (i.e. PyACGTGGC), which is present in numerous ABA-regulated genes (Kim, 2006). CBF/DREB and other AP2 domain proteins are known as positive or negative regulators of ABA and/or abiotic stress responses. MYB proteins such as AtMYB2, AtMYB96, AtMYB15, and AtMYB44 also regulate ABA and abiotic stress responses. Additionally, HD-ZIP, NAC, WRKY, or ZFHD proteins are known to mediate ABA and/or stress responses (Berri et al., 2009; Yamaguchi-Shinozaki and Shinozaki, 2005).
MYB proteins are involved in ABA and stress responses as well as many other cellular processes (Yanhui et al., 2006). A series of recent studies showed that a number of MYB genes, including MYB52, are involved in the regulation of secondary cell wall biosynthesis. For instance, MYB58 and MYB63 are regulators of lignin biosynthesis (Zhou et al., 2009), and MYB103, MYB85, MYB52 and MYB54 control secondary wall thickening (Zhong et al., 2008). Among the MYB transcription factors, hierarchical relationships exist, and it has been demonstrated that MYB52 is a downstream target of MY46, which is a master switch for secondary cell wall formation in Arabidopsis (Ko et al., 2009).
In the present study, we isolated an ABA response mutant by activation tagging screen. The mutant, referred to as ahs1, is hypersensitive to ABA, and determination of T-DNA insertion site and subsequent expression analysis revealed that MYB52 was activated in the mutant. Recapitulation experiments to confirm its role in ABA response showed that MYB52 is involved in ABA and stress responses. Taken together, the results presented herein suggest a possible connection between ABA response and cell wall biosynthesis.
MATERIALS AND METHODS
Plant growth and generation of activation-tagged lines
Arabidopsis thaliana ecotype Columbia (Col-0) and Landsberg erecta (Ler) were used in this study. The plants were grown under long day condition (16-h-light/8-h-dark cycle) at 22℃ aseptically or on soil. For aseptic growth, seeds were surfacesterilized by treatment with 70% ethanol for 1 min and with 30% bleach for 5 min, after which they were washed with sterile water five times before planting. For soil growth, seeds were sown on a mixture of vermiculite, perlite and peat moss (1:1:1 by weight), irrigated with 0.1% Hyponex (Hyponex Co., USA), placed at 4℃ for 3-5 days in the dark to break residual dormancy, and then transferred to normal growth conditions. Plants were watered once a week.
To generate activation-tagged transgenic plants, Arabidopsis plants (Col-0) were transformed with A. tumefaciens strain GV3101 harboring the vector pSKI015 (Weigel et al., 2000) according to Bechtold and Pelletier (Bechtold and Pelletier, 1998). Approximately 25,000 basta-resistant plants were recovered, and seeds were collected in pools of ca 100 transgenic plants. To screen for ABA-hypersensitive mutants, seeds from each pool were plated and germinated in the medium containing 0.3 μM ABA, and seedlings exhibiting abnormal germination and/or postgermination growth were selected and transferred to ABA-free medium. The plants were subsequently transferred to soil and their seeds were harvested. A total of ca 100 mutants were isolated from the primary screen of 100 pools, which is equivalent to 10,000 transgenic plants. The seeds from individual plants were then germinated and their phenotypes were confirmed. For analysis of the ahs1 mutant phenotypes shown in Fig. 1, we employed one of the heterozygous sublines (#8-3) because we could not recover homozygous lines. The T-DNA insertion site in the ahs1 mutant was determined by sequencing the left border flanking sequence after rescuing the plasmid, which was obtained by ligation of the ahs1 genomic DNA digested with Spe I according to Weigel et al. (2000).
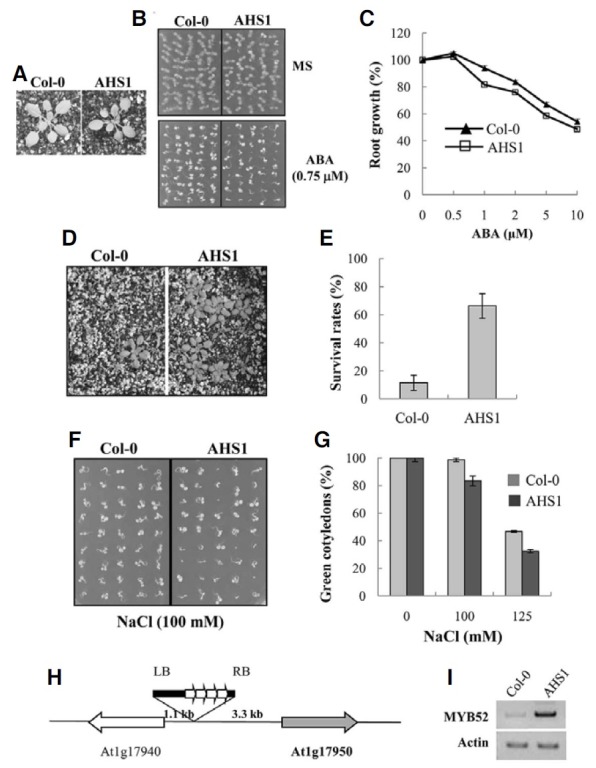
Fig. 1. The phenotypes of the tagging mutant ahs1. (A) Growth of ahs1 in soil. Plants were grown in soil for three weeks. (B, C) ABA sensitivity of ahs1. Seeds were germinated and grown in a medium containing 0.75 μM ABA for seven days (B). For root elongation assay in (C), seeds were germinated and grown in an ABA-free medium for three days, the seedlings were transferred to media containing various concentrations of ABA, and primary root growth was measured five days after the transfer. Experiments were carried out in triplicates (n = 6), and the small bars indicate the standard errors. (D, E) Drought tolerance of ahs1. Water was withheld from 11 day-old seedlings for 11 days, after which they were re-watered. The picture in (D) was taken two days after re-watering. The survival rates in (E) represent the means of three independent experiments (n = 20 each), and the small bars indicate the standard errors. (F, G) Salt tolerance of ahs1. Seeds were germinated and grown in media containing 100 mM or 150 mM NaCl for four days, and seedlings with green cotyledons were then counted. All experiments were conducted in triplicates (n = 50), and standard errors are indicated by the small bars. (H) A diagram showing the position of T-DNA insertion in the AHS mutant. (I) The expression level of AtMYB52 (At1g17950) (MYB52) was determined by RT-PCR. Actin-1 was used as an internal control.
Generation of transgenic lines and phenotype analysis
To generate the AHS1 overexpression (OX) lines, the coding region of AtMYB52 was amplified using the primer set 5′-TGC TCT AGA GTA TTA AAA AAT GAT GTG TAG TCG A-3′ and 5′-GAC AAA TTA ACA TAA ACC CTG AGA G-3′. After XbaI digestion, the coding region was cloned into pBI121 (Jefferson, 1987), which was prepared by removing the GUS coding after the XbaI-EcoICR1 digestion. To prepare the AtMYB52 promoter- GUS reporter construct, 2.4 kb of the 5′ flanking sequence was amplified employing the primer set 5′-TAG AAG CTT GTG GTT TGA TG G TAT TGA TTA AGT T-3′ and 5′-TTT TTA ATA CCT CTC TCC TTT TGA TC-3′ and then cloned into the HindIII-SmaI sites of pBI101.2 (Jefferson, 1987) after HindIII digestion. The constructs were introduced into A. tumefaciens strain GV3101, and Arabidosis plants (Col-0 for the promoter- GUS lines and Ler for the OX lines) were transformed according to the method described by Bechtold and Pelletier (1998). For the analysis of OX lines, we recovered seven T3 generation homozygous lines, and the T4 generation seeds from these lines were used for phenotype analysis.
Phenotype analysis of transgenic plants was conducted as described before (Kang et al., 2002; Kim et al., 2004). For aseptic growth, seeds were treated as described above and plated on MS medium (Murashige and Skoog, 1962) solidified with 0.8% Phytoagar after cold treatment at 4℃ for 3-5 days. The MS medium was supplemented with 1% sucrose, and various concentrations of ABA or NaCl was supplemented as indicated for ABA and salt sensitivity tests. For the drought test, water was withheld from ten to eleven day-old soil-grown plants until they lost turgor completely, at which time they were re-watered and their survival rates were determined. The same number of wild type and transgenic plants were grown on the same tray to minimize experimental variations.
Histochemical GUS assay was conducted as described by Jefferson et al. (1987). Briefly, T3 homozygous plants were immersed in a staining solution containing 1 mM X-gluc (5- bromo-4-chloro-3-indolyl-β-glucuronic acid) in the buffer (100 mM sodium phosphate, pH 7.0, 0.1 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 0.1% Triton X-100). Staining was conducted at 37℃ for the indicated times. At the end of staining, chlorophyll was removed from the plant tissues by immersing them in 95% ethanol.
RNA isolation and expression analysis
RNA was isolated employing a Qiagen RNeasy plant mini kit. Northern blot analysis was performed as described previously (Kang et al., 2002). For RT-PCR analysis, possible contaminating DNA was removed from RNA samples by DNase I treatment. The first strand cDNA was synthesized using Superscript III (Invitrogen) according to the supplier’s instructions. For semiquantitative RT-PCR, cDNA amplification was carried out within a linear range using gene-specific primers. For Real-Time RTPCR, the cDNA amplification was conducted using SsoFast EvaGreen supermix in conjunction with a Bio-Rad CFX96 Real- Time PCR System (Bio-Rad). Quantitation was conducted using the CFX96 Real-Time PCR Systems software. Actin-1 was employed as a reference gene. Primer sequences are available upon request.
Determination of subcellular localization of AtMYB52
The coding region of AtMYB52 was amplified using the primers 5′-CGG AGC TCA TGA TGT GTA GTC GAG GCC ATT G-3′ and 5′-ACA TAA ACC CTG AGA GGC AGA GTT-3′ and then digested with Sacl. The amplified fragment was subsequently cloned into the SacI-SmaI sites of p35S-FAST/EYFP in frame with the EYFP coding region. Tobacco (Nicotiana benthamiana) leaves were co-infiltrated with Agrobacterium strains (C58C1) containing the fusion construct and p19, respectively, according to the method described by Voinnet et al. (Voinnet et al., 2003). The tobacco epidermal cells were observed with a fluorescence microscope (Olympus BX51) 40 h after infiltration.
RESULTS
Activation tagging screen
We performed activation tagging screen to isolate genes involved in ABA response. A library of activation-tagged transgenic plants was generated using the vector pSKI015 (Weigel et al., 2000), and mutants exhibiting altered ABA response were isolated. One of the mutants, which is designated ahs1 (ABA-hypersenstive1), is shown in Fig. 1A. Under the normal growth condition, ahs1 did not exhibit distinct phenotypes other than smaller plant size. However, seedling establishment (i.e, cotyledon greening) of the mutant plants was inhibited more severely by ABA than wild type plants (Fig. 1B), suggesting that it may be ABA-hypersensitive. Enhanced ABA sensitivity was also observed in the later growth stage, i.e., inhibition of primary root elongation by ABA was more pronounced in ahs1 mutant than in wild type plants (Fig. 1C). Additionally, the mutant seedlings displayed higher survival rates under water-stress condition (Figs. 1D and 1E) and were hypersensitive to salt (Figs. 1F and 1G). Thus, our analysis of ahs1 mutant phenotypes indicated that it is ABA-hypersensitive and that it exhibits altered drought and salt tolerance.
To determine the T-DNA insertion site in the ahs1 mutant, the sequence flanking the left border was recovered by plasmid rescue (Weigel et al., 2000). Sequencing of the recovered genomic DNA fragment revealed that T-DNA was inserted in the intergenic region between the two genes, At1g17940 and At1g17950, which encode an unknown gene and a MYB gene, AtMYB52, respectively (Fig. 1H). Subsequently, RT-PCR was conducted to measure changes in the transcript levels of the two genes. As shown in Fig. 1I, the MYB gene transcript level was increased in the ahs1 mutant, whereas the transcript level of the unknown gene was not altered (data not shown). The results suggest that the ABA hypersensitivity of ahs1 mutant may result from the activation of the AtMYB52 gene.
Expression pattern and subcellular localization of AtMYB52
We determined the tissue-specific expression pattern of AtMYB52 by coupled reverse transcription and polymerase chain reaction (RT-PCR). As shown in Fig. 2A, AtMYB52 was more abundantly expressed in roots and siliques than in flowers and leaves. Similar RT-PCR analysis indicated that the expression of AtMYB52 in seedlings was slightly induced by ABA and high salt treatments (Fig. 2B). The expression patterns are in good agreement with those available in the public database (www. arabidopsis.org). To explore the expression pattern of AtMYB52 in detail, we prepared transgenic plants harboring an AtMYB52 promoter-GUS fusion construct and investigated the promoter activity by histochemical GUS staining of the transgenic plants. Figure 2C shows that the AtMYB52 promoter is active in roots of seedlings. In mature plants, promoter activity was observed in stems, flowers (i.e., receptacle, stigma, style and anthers) and siliques. Consistent with the RT-PCR result, stronger GUS staining was observed following ABA and salt treatments in seedlings, especially in the basal part and the maturation zone of lateral roots (Fig. 2D).
Fig. 2. Expression patterns of AtMYB52. (A) The expression of AtMYB52 (MYB52) in various tissues was determined by RT-PCR. (B) Induction patterns of AtMYB52 were determined by RT-PCR. ABA (100 μM), salt (150 mM NaCl) and mannitol (600 mM) treatments were conducted for 4 h. For cold treatment, plants were placed at 4 C for 24 h before RNA isolation. Untreated, control plants without any treatments. (C) GUS staining pattern of transgenic plants harboring the AtMYB52 promoter- GUS reporter gene construct. a, immature embryo; b, mature embryo; c, 11 day-old-seedling; d, five week-old whole plant; e, flower; f, silique. GUS staining was for 24 h. (D) GUS induction patterns. Plants were treated with 100 μM ABA (ABA) or 250 mM NaCl (NaCl) before GUS staining. GUS staining was conducted for 6 h. (E) Subcellular localization of AtMYB52 was investigated by Agroinfiltraion of tobacco leaves using an At- MYB52-YFP fusion construct. Tobacco leaves were observed with a fluorescence microscope 40 h after infiltration.
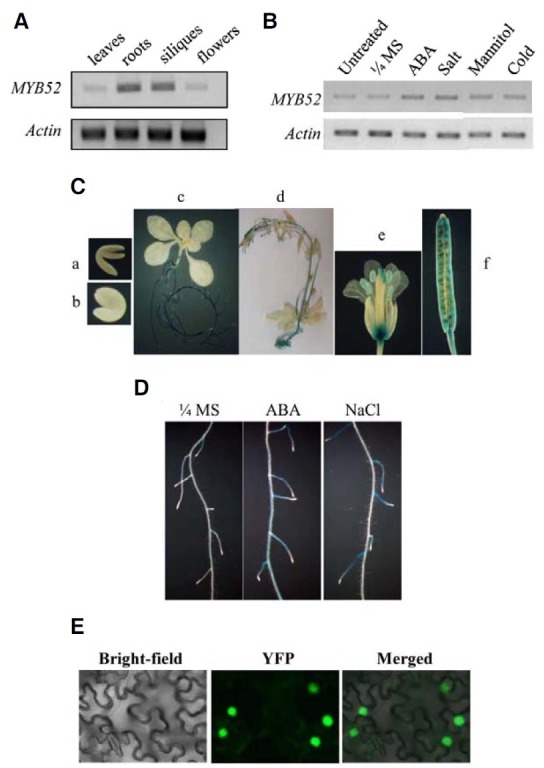
To examine subcellular localization of AtMYB52, an At- MYB52-YFP fusion protein construct was prepared and introduced into tobacco leaf cells by Agroinfiltration (Voinnet et al., 2003). The AtMYB52 localization was then investigated by fluorescence microscopy. As shown in Fig. 2E, the YFP signal was localized in the nucleus. Thus, our results indicated that AtMYB52 was nuclear-localized.
Generation and phenotype analysis of AtMYB52 overexpression lines
To confirm that the ahs1 phenotype resulted from the overexpression of AtMYB52, we generated AtMYB52 overexpression (OX) lines by expressing the AtMYB52 gene under control of the strong CaMV 35S promoter. Although Col-0 was used in the construction of the activation tagged lines, we used Ler for the construction of AtMYB52 overexpression lines because most of the genetic studies associated with ABA signaling were conducted in a Ler background. We recovered seven T3 generation homozygous lines and, after preliminary analysis, two representative lines were selected for further phenotype analyses.
As shown in Fig. 3A, the AtMYB52 OX lines exhibited dwarfism that was dependent on the AtMYB52 expression levels. However, similar to ahs1, the germination rates and overall growth of the OX lines were normal other than the dwarfism.
Fig. 3. ABA sensitivity of AtMYB52 overexpression lines. (A) Growth of the transgenic plants in soil. Plants were grown for three weeks. The expression levels of AtMYB52 determined by Northern analysis are shown in the left panel. Ler, wild type plants. #58 and #112 denote two transgenic lines. (B) Plants were grown in media containing ABA. (C) Cotyledon greening efficiency was determined seven days after seed sowing. Experiments were conducted in triplicate (n = 50 each) and the small bars represent the standard errors. (D) Root elongation assay. Seedlings were germinated and grown in a medium lacking ABA for three days, after which the seedlings were transferred to a medium containing various concentrations of ABA. Root elongation was then measured after five days. All experiments were conducted in triplicates (n = 6 each) and the small bars represent the standard errors.
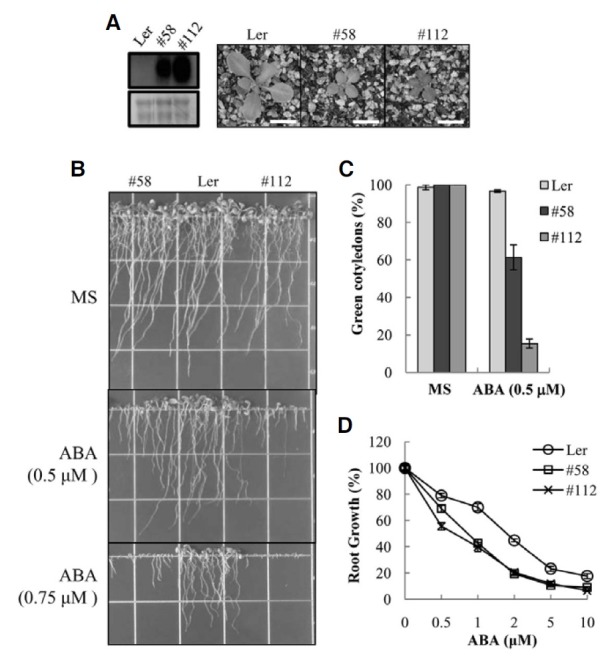
Because ahs1 was hypersensitive to ABA, we examined whether AtMYB52 OX lines were hypersensitive to ABA. We first scored the ABA effect on overall plant growth by growing the plants in the continual presence of ABA. The result (Fig. 3B) showed that growth of the OX lines was more severely inhibited by ABA than wild type plants. For example, at 0.5 μM ABA, root growth of the transgenic seedlings was severely inhibited and shoot development of many seedlings was arrested. By contrast, growth of wild type plants was marginally affected by the low concentration of ABA. In the presence of 0.75 μM ABA, the shoot and root growth of the transgenic seedlings was almost completely arrested, whereas wild type plants still grew, albeit at a lower rate (Fig. 3B). We next investigated shoot development at the seedling establishment stage by examining the efficiency of cotyledon greening. As shown in Fig. 3C, the cotyledons of most (~95%) of the wild type seedlings turned green at 0.5 μM ABA. With the AtMYB52 OX lines, green cotyledons were observed in 60% (#58) or 10% (#112) of the seedlings, respectively, indicating that transgenic shoot development was more sensitive to ABA inhibition. As another indicator of ABA sensitivity, we conducted a root elongation assay. The result (Fig. 3D) showed that root growth of the transgenic lines was more severely inhibited by ABA than that of the wild type plants at all doses of ABA. Thus, overexpression of AtMYB52 conferred ABA hypersensitivity during postgermination growth.
Drought and salt tolerance of AtMYB52 OX lines
The results presented above suggested that AtMYB52 affects ABA sensitivity. Because ABA mediates various abiotic stress responses (Xiong et al., 2002), especially drought response, we investigated whether AtMYB52 overexpression affected drought tolerance. Three-week-old plants were subjected to waterdeficit conditions by withholding water for two weeks. Subsequently, the plants were re-watered and their survival rates were determined. Figures 4A and 4B show that 75% of the AtMYB52 OX line #58 survived the treatment, whereas the wild type survival rate was less than 3%. Similarly, the survival rate of another AtMYB52 OX line, #112, was 81%, whereas that of the wild type was 10%. Thus, both ATMYB52 OX lines exhibited higher survival rates than wild type plants, indicating that they are drought-tolerant.
Fig. 4. Drought and salt tolerance of AtMYB52 overexpression lines. (A, B) Drought tolerance of the transgenic plants. Water was withheld from ten day-old plants for 16 days, after which they were re-watered. Experiments were performed in duplicates (n = 20 each) and the small bars represent the standard errors. (C, D) Salt tolerance of the transgenic plants. (C) shows representative plants, whereas (D) shows cotyledon greening efficiency. Experiments were conducted in triplicates (n = 55 each), and the small bars in (D) indicate the standard errors.
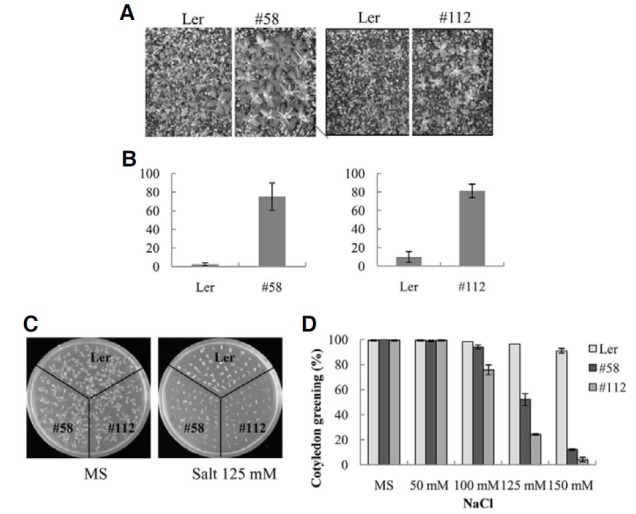
It has been well established that ABA mediates salt response (Xiong and Zhu, 2002). Hence, we next examined the salt sensitivity of the AtMYB52 OX lines. Figure 4C shows that shoot development of the transgenic plants was more extensively inhibited by the salt. For example, green cotyledons were observed in more than 95% of the wild type seedlings in the presence of 125 mM NaCl. In contrast, green cotyledons were developed in only 50% (#58) and 20% (#112), respectively, of the transgenic seedlings (Fig. 4D). At 150 mM NaCl, shoot development of the AtMYB52 OX lines was almost completely inhibited, whereas green cotyledons were observed in 95% of the wild type pants. Thus, shoot development of the AtMYB52 OX lines was hypersensitive to salt. We also tested the effect of mannitol on seedling growth, but no difference between the OX lines and wild type plants was detected (not shown). The results imply that AtMYB52 overexpression did not affect osmotic response and that the salt hypersensitivity of the transgenic lines may have been caused by altered ionic response rather than from changes in general osmotic response.
Target genes of AtMYB52
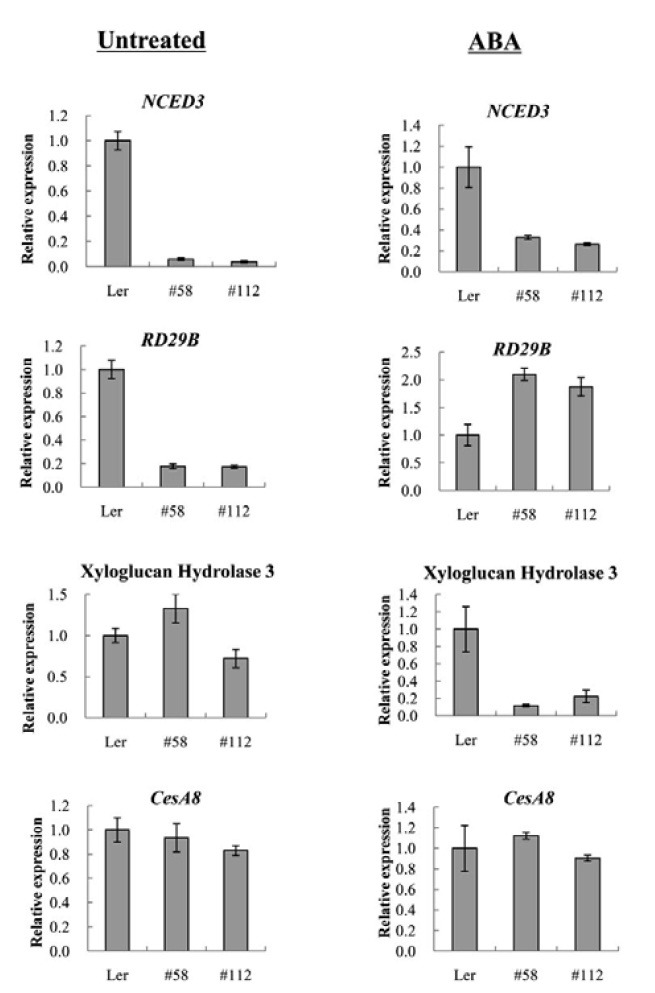
Because AtMYB52 OX affected ABA and stress responses, we compared the expression levels of a number of ABA-responsive or ABA biosynthetic genes in the transgenic and wild type plants by Real-Time RT-PCR (Fig. 5). Among the genes examined, the expression levels of NCED3 and RD29B were reduced in the AtMYB52 OX lines under the normal growth condition. Because AtMYB52 activity might be modulated posttranslationally by ABA, we also carried out similar experiments using RNA isolated from ABA-treated samples. Figure 5 shows that the relative RD29B expression levels in the transgenic lines were enhanced after ABA treatment, although NCED3 expression was still lower than the wild type level. As mentioned earlier, AtMYB52 is known to regulate cell wall biosynthesis (Zhong et al., 2008). Therefore, we determined the expression levels of two cell wall biosynthetic genes, CesA8 and the xyloglucan hydrolase 3 gene. We did not observe any significant changes in their expression levels under the normal condition. However, after ABA treatment, the expression levels of the xyloglucan hydrolase 3 gene in the transgenic lines were lower than that in the wild type.
Fig. 5. Expression analysis of putative AtMYB52 target genes. Expression of ABA-responsive or cell wall biosynthesis genes in the transgenic lines was determined by Real-Time RT-PCR. RNA was prepared from plants treated with 100 μM ABA for 4 h (ABA) or from untreated plants (Untreated). Reactions were performed in duplicate and the small bars indicate the standard errors.
DISCUSSION
Several MYB class transcription factors are known to be involved in ABA and stress responses in Arabidopsis. AtMYB2 regulates a subset of ABA-responsive genes, and its overexpression enhances ABA sensitivity (Abe et al., 2003). Similarly, AtMYB15, AtMYB44 and AtMYB96 are involved in ABA and drought responses (Ding et al., 2009; Jung et al., 2008; Seo et al., 2009), whereas AtMYB41 regulates osmotic response (Lippold et al., 2009). In this study, we showed that overexpression of AtMYB52 conferred ABA hypersensitivity, drought tolerance and salt sensitivity, which indicates that AtMYB52 also is involved in ABA and stress responses.
As mentioned before, other studies have shown that At- MYB52 is one of the downstream regulators of cell wall biosynthesis. Specifically, its overexpression affects the expression of several cell wall biosynthesis genes, although it does not influence secondary wall thickness, and dominant repression of its expression results in a reduction in secondary cell wall thickening (Ko et al., 2009; Zhong et al., 2008). Here, we demonstrated that AtMYB52 is involved in ABA response during postgermination growth. Thus, our observations raise an interesting possibility that ABA hypersensitivity of the AtMYB52 OX lines may be associated with changes in cell wall architecture. It has been reported that ABA inhibits seed germination by inhibiting cell-wall loosening and expansion (Gimeno-Gilles et al., 2009). On the other hand, cell wall arabinan is essential for guard cell function (Jones et al., 2003; 2005) and seedling growth (Gomez et al., 2009). Therefore, it can be speculated that AtMYB52 overexpression disturbed the biosynthesis of secondary cell wall components, which, in turn, caused a decrease in cell expansion, cell division/growth or stomatal movement. The decrease in the expression of the xyloglucan hydrolase3 gene in the presence of ABA (Fig. 5) that was observed herein supports this hypothesis. Alternatively, it is possible that AtMYB52 affects the expression of genes involved in ABA metabolism or ABA response. The results shown in Fig. 5 indicate that expression of the ABA biosynthetic gene NCED3 and the ABA-regulated gene RD29B was affected in the AtMYB52 OX lines. More extensive expression and physiological analyses will be necessary to elucidate the molecular mechanism underlying the ABAhypersensitivity of AtMYB52 OX lines.
In summary, we isolated an ABA-hypersensitive mutant ahs1 by activation tagging. AtMYB52 was activated in the mutant, and a recapitulation experiment showed that AtMYB52 overexpression conferred ABA-hypersensitivity during postgermination growth and enhanced drought tolerance of seedlings. Others have demonstrated that AtMYB52 is involved in cell wall biosynthesis. Thus, our results suggest a possible connection between cell wall biosynthesis and ABA-dependent growth regulation of seedlings.
Acknowledgments
This work was supported in part by grants from the Crop Functional Genomics Center of the 21C Frontier Program (CG2112) funded by the Ministry of Education, Science and Technology (MEST) and by the Mid-career Researcher Program through an NRF grant funded by the MEST (2008-0059137). The authors are grateful to the Kumho Life Science Laboratory of Chonnam National University for providing equipment and plant growth facilities.
References
- 1.Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi- Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. (2003);15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechtold N., Pelletier G. In planta Agrobacteriummediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. (1998);82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 3.Berri S., Abbruscato P., Faivre-Rampant O., Brasileiro A.C., Fumasoni I., Satoh K., Kikuchi S., Mizzi L., Morandini P., Pe M.E., et al. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. (2009);9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. (2010);61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 5.Ding Z., Li S., An X., Liu X., Qin H., Wang D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics. (2009);36:17–29. doi: 10.1016/S1673-8527(09)60003-5. [DOI] [PubMed] [Google Scholar]
- 6.Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell. (2002);14:S15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature. (2009);462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimeno-Gilles C., Lelievre E., Viau L., Malik-Ghulam M., Ricoult C., Niebel A., Leduc N., Limami A.M. ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture: modifying enzymes and structural proteins in Medicago truncatula embryo axis. Mol. Plant. (2009);2:108–119. doi: 10.1093/mp/ssn092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez L.D., Steele-King C.G., Jones L., Foster J.M., Vuttipongchaikij S., McQueen-Mason S.J. Arabinan metabolism during seed development and germination in Arabidopsis. Mol. Plant. (2009);2:966–976. doi: 10.1093/mp/ssp050. [DOI] [PubMed] [Google Scholar]
- 10.Jefferson R.A., Kavanagh T.A., Bevan M.W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. (1987);20:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones L., Milne J.L., Ashford D., McQueen-Mason S.J. Cell wall arabinan is essential for guard cell function. Proc. Natl. Acad. Sci. USA. (2003);100:11783–11788. doi: 10.1073/pnas.1832434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones L., Milne J.L., Ashford D., McCann M.C., McQueen- Mason S.J. A conserved functional role of pectic polymers in stomatal guard cells from a range of plant species. Planta. (2005);221:255–264. doi: 10.1007/s00425-004-1432-1. [DOI] [PubMed] [Google Scholar]
- 13.Jung C., Seo J.S., Han S.W., Koo Y.J., Kim C.H., Song S.I., Nahm B.H., Choi Y.D., Cheong J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. (2008);146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang J., Choi H., Im M., Kim S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. (2002);14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S.Y. The role of ABF family bZIP class transcription factors in stress response. Physiol. Plant. (2006);126:519–527. [Google Scholar]
- 16.Kim S., Kang J.Y., Cho D.I., Park J.H., Kim S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. (2004);40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 17.Ko J.H., Kim W.C., Han K.H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. (2009);60:649–665. doi: 10.1111/j.1365-313X.2009.03989.x. [DOI] [PubMed] [Google Scholar]
- 18.Lippold F., Sanchez D.H., Musialak M., Schlereth A., Scheible W.R., Hincha D.K., Udvardi M.K. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. (2009);149:1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. (2009);324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 20.Murashige T., Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plant. (1962);15:473–497. [Google Scholar]
- 21.Pandey S., Nelson D.C., Assmann S.M. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. (2009);136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 22.Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. (2009);324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder J.I., Kwak J.M., Allen G.J. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. (2001);410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- 24.Seo P.J., Xiang F., Qiao M., Park J.Y., Lee Y.N., Kim S.G., Lee Y.H., Park W.J., Park C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. (2009);151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y.Y., Wang X.F., Wu F.Q., Du S.Y., Cao Z., Shang Y., Wang X.L., Peng C.C., Yu X.C., Zhu S.Y., et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. (2006);443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 26.Shinozaki K., Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. (2007);58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 27.Voinnet O., Rivas S., Mestre P., Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. (2003);33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 28.Weigel D., Ahn J.H., Blazquez M.A., Borevitz J.O., Christensen S.K., Fankhauser C., Ferrandiz C., Kardailsky I., Malancharuvil E.J., Neff M.M., et al. Activation tagging in Arabidopsis. Plant Physiol. (2000);122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong L., Zhu J.-K. Salt tolerance. In the Arabidopsis Book, (American Society of Plant Biologists) (2002):1–24. doi: 10.1199/tab.0048. [DOI] [PMC free article] [PubMed]
- 30.Xiong L., Schumaker K.S., Zhu J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell. (2002);14(Suppl.):S165–183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi-Shinozaki K., Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stressresponsive promoters. Trends Plant Sci. (2005);10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Yanhui C., Xiaoyuan Y., Kun H., Meihua L., Jigang L., Zhaofeng G., Zhiqiang L., Yunfei Z., Xiaoxiao W., Xiaoming Q., et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. (2006);60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 33.Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. (2008);20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J., Lee C., Zhong R., Ye Z.H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. (2009);21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]


