Abstract
Autophagy and endocytic pathway are highly regulated catabolic processes. Both processes are crucial for cell growth, development, differentiation, disease and homeostasis and exhibit membrane rearrangement for their function. Autophagy and endocytic pathway represent branches of the lysosomal digestive system, autophagy being responsible for degradation of cytoplasmic components and endocytic pathway for degradation of exogenous substances. Here we report that autophagy is activated when endocytic pathway regulatory genes such as rab-5 and rabx-5 are disrupted. Defects in the ubiquitin binding domain of RABX-5 are critical in activating autophagy. We also observed that the elevated autophagy level does not contribute to lifespan extension of rabx-5 mutant. Our results suggest that autophagy may compensate for the endocytic pathway when regulatory genes for the endocytic pathway malfunction, providing a case of complementation between two functionally related cellular processes.
Keywords: autophagy, endocytic pathway, GFP::LGG-1, RAB-5, RABX-5
INTRODUCTION
Autophagy is a ubiquitous cellular process that involves a process of self-cannibalization through the lysosomal degradation pathway and is evolutionarily conserved in all eukaryotic organisms. Autophagy is a complex catabolic process that digests various cytosolic proteins and organelles through autophagosomal- lysosomal pathway (Klionsky, 2005). Autophagy allows cell survival under starvation conditions by the bulk degradation of cytoplasmic constituents generally by non-selective sequestration. In C. elegans autophagy is essential for lifespan extension, reproductive development, programmed cell death, necrotic cell death, neurodegenerative diseases, cell size regulation, neurotransmitter receptor trafficking and dauer development (Melendez and Levine, 2009). Earlier reports have suggested the positive correlation between the elevated autophagy and extended lifespan in various model organisms (Dwivedi and Ahnn, 2009).
Endocytic pathway resembles autophagy in a way as it is responsible for uptake of extracellular constituents and its degradation. Endocytic pathway is regulated by Rab GTPases and their associated factors. RAB-5, a small GTPase, plays a crucial role in regulating early endocytic pathway (Bucci et al., 1992; Chavrier et al., 1990; Christoforidis et al., 1999; De Reniz et al., 2002). RABX-5 is a guanine nucleotide exchange factor (GEF) for RAB-5 (Horiuch et al., 1997). RABX-5 contains an ubiquitin binding domain at its N-terminus followed by helical bundle and Vps9 domains functioning as catalytic core. Another GEF for RAB-5 is RME-6 that is conserved from worms to human and also includes Vps9 domain but lacks ubiquitin binding domain. RME-6 is distinct from other RAB-5-associated proteins in that RME-6 acts at the clathrin-coated pit (Sato et al., 2005).
Growing evidence suggests that there may be a crosstalk between autophagy and endocytic pathway. Inactivation of certain C. elegans Vps genes lead to the accumulation of enlarged endosomal structures and an increase in autophagy (Michelet et al., 2009; Roudier et al., 2005). In addition, certain regulators in autophagy and endocytic pathway are reportedly involved in both processes. BEC-1, the C. elegans ortholog of Atg6/Vps30/ Beclin1 and a key regulator of the autophagic machinery, also contributes to endosome function (Ruck et al., 2011). UVRAG, the human ortholog of Vps38 and a Beclin1-binding protein, has a role in endocytosis and autophagy (Liang et al., 2008).
To study inter-dependence of autophagy and endocytic pathway in C. elegans, we used GFP::LGG-1 as a reporter gene for autophagy to investigate the effects of RNAi for endocytic pathway genes such as rab-5, rabx-5, rme-6 and double mutant of rabx-5 and rme-6. We also used dyn-1 mutant to check the level of autophagy. Finally, we studied the lifespan of endocytic mutants showing elevated autophagy. Our data suggest that autophagy may compensate endocytic pathway at early endosomes and that the elevated autophagy does not contribute to extending the lifespan of endocytic mutants.
MATERIALS AND METHODS
C. elegans strains and maintenance
The following strains were obtained from the Caenorhabditis Genetics Center (CGC) at University of Minnesota, USA: Bristol N2, CX51 dyn-1(ky51) X, CB1370 daf-2(e1370) III, rabx-5 (tm1512) III, and rme-6(b1014). Worm breeding and handling were conducted as previously described (Brenner, 1974).
Analysis of autophagic events
The level of autophagy in various mutants was assessed using GFP::LGG-1 translational reporter characterized previously (Melendez et al., 2003). The wild type worms carrying the extrachromosomal array GFP::LGG-1 were kindly provided by Alicia Melendez. The extrachromosomal array GFP::LGG-1 was introduced into rabx-5(tm1512)and dyn-1(ky51) mutants by crossing. GFP-positive puncta were counted (using 1,000-fold magnification on a Zeiss Axioplan II microscope equipped with fluorescent optics) in the seam (lateral epidermal) cells of L3 transgenic animals. When performing RNAi experiments to count GFP::LGG-1 positive foci, young adults (P0) were fed with RNAi bacteria, and the L3 progeny of the F1 generation were examined. All RNAi clones used were obtained from gene service constructed by Julie Ahringer’s group at the The Wellcome CRC Institute, University of Cambridge, Cambridge, England. At least 3 to 10 seam cells were examined in each of 10 to 20 animals from at least two independent trials and averaged. Statistical data analysis was done using unpaired, two-tailed t-test. We have checked the level of GFP::LGG-1 in dyn-1 mutants as dynamin is the key component in clathrin-mediated endocytosis and is involved in pinching off the coated pit from the plasma membrane to form a clathrin-coated vesicles (CCVs) (Grant and Sato, 2006).
Sequence alignment and homology modeling
Pairwise protein sequence alignment was performed using ClustalW2 web server hosted by European Bioinformatics Institute (Chenna et al., 2003). Homology modelling was done using SWISS-MODEL web server (Arnold et al., 2006). The resulting homology structural model figure was generated using PyMOL (Schrödinger, LLC).
Lifespan analysis
The lifespan analysis after RNAi treatments at 20℃ was done in F1 and F3 generation as standardized previously in the laboratory (Dwivedi et al., 2009). In all experiments, the prefertile period of adulthood (L4 stage) was used as t = 0 for lifespan analysis. Animals that ruptured, bagged (exhibited internal progeny hatching), or crawled off the plates were censored from lifespan analysis data. Each lifespan experiment was repeated at least three times with n = 75 to 100 animals per experimental group. Kaplan-Meier survival analysis was used to compare the mean lifespan of different treatments, and p values were calculated using the logrank test (the Mantel-Cox test) to determine the relative effects on wild-type N2, rabx-5 (tm1512) and rme-6(b1014) mutant animals.
RESULTS AND DISCUSSION
Activation of autophagy by knockdown of RAB-5 and RABX-5
To study whether RABX-5 is required in autophagy, we employed GFP-tagged LGG-1 protein as a reporter for autophagy. LGG-1 is the worm ortholog of yeast Atg8p and localizes to preand autophagosomal membranes. During autophagy, GFP:: LGG-1 forms fluorescent puncta. To minimize endogenous auto-fluorescence in adult animals, we restricted our observations to the hypodermal seam cells of L3 larvae. GFP::LGG-1 in wild-type worms fed with bacteria expressing L4440 vector shows diffused expression pattern (Fig. 1A). However, worms treated with RNAi targeted against rab-5, rabx-5, rme-6 and rabx-5/rme-6 genes, GFP::LGG-1 puncta were observed (Fig. 1A), establishing a crosstalk between autophagy and the endocytic pathway. Quantitation of the number of GFP::LGG-1 puncta reveal that the average number of GFP::LGG-1 puncta was significantly increased for all three RNAi treated cases (Fig. 1B). These results indicate that autophagy can be activated upon disruption in the endocytic pathway.
Fig. 1. (A) Representative images of GFP::LGG-1 expression in the seam cells (F1 generation) fed with bacteria containing L4440 (vector) or expressing rab-5, rabx-5, rme-6 and rabx-5/rme-6 dsRNA, respectively. (B) Quantification of GFP::LGG-1 containing puncta in each case described in (A). Magnification of the images is 1,000.
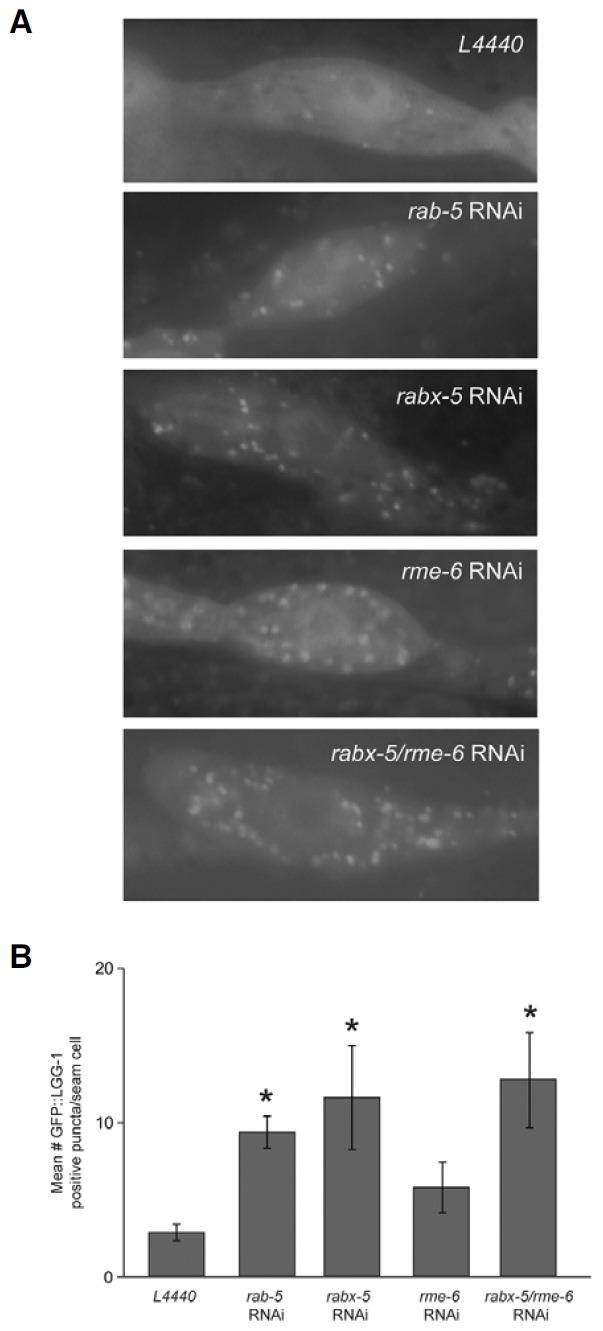
Among three genes we tested, rab-5 and rabx-5 RNAi showed more increases in the number of puncta than rme-6 RNAi (Fig. 1B), implying that crosstalk between autophagy and the endocytic pathway may occur in early endosomes predominantly because RME-6 is known to regulate RAB-5 in clathrin-coated pits. Consistent with such idea, double RNAi for rabx-5 and rme-6 did not indicate any synergistic effect on the number of GFP::LGG-1 puncta (Fig. 1B). Taken together, our data suggest that there is crosstalk between autophagy and the endocytic pathway and that the disruption of endocytic pathway at early endosomal may result in the activation of autophagic pathway.
Lifespan extension by RABX-5
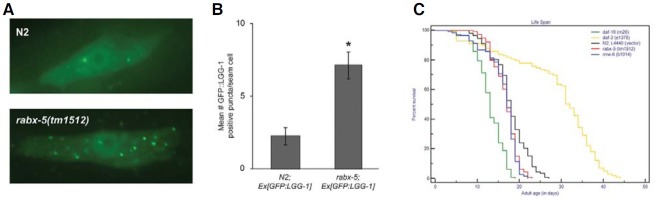
The increased autophagic puncta in worms treated with RNAi targeted against endocytic pathway genes motivated us to study the lifespan in these mutants. We first analyzed the autophagic puncta in rabx-5(tm1512) mutants and found similar increase in GFP::LGG-1 puncta as we observed after the treatment with RNAi clone when compared to wild-type worms (Figs. 2A and 2B). Interestingly the lifespans of rme-6(b1014) and rabx-5(tm1512) mutants were comparable to that of wildtype (Fig. 2C). The daf-2(e1370) and daf-16(m26) were taken as positive and negative controls for lifespan, respectively. The comparable lifespan studies of the endocytic mutants to wildtype worms suggest that all pathways leading to extended lifespan may converge to autophagy (Toth et al., 2008) but all pathways showing activated autophagy may not necessarily extend lifespan.
Fig. 2. (A) Representative images of GFP::LGG-1 expression in the seam cells in N2 strains or rabx-5(tm1512) mutant. (B) Quantification of GFP::LGG-1 containing puncta in each case described in (A). (C) Kaplan-Meier survival curve showing the life-span of N2 wild-type, daf- 16(m26), daf-2(e1370), rabx-5(tm1512) and rme-6(b1014) mutants. Magnification of the images is 1,000.
RABX-5 ubiquitin binding domain and autophagy
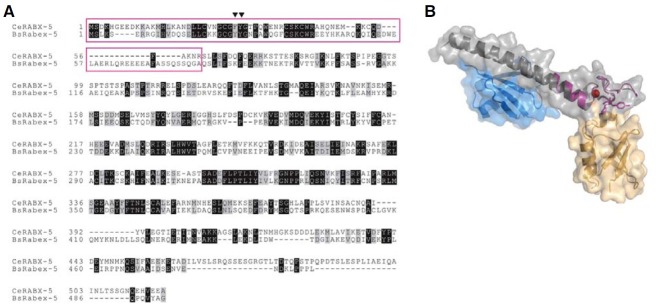
To analyze which region of RABX-5 may be responsible for the activation of autophagy, we verified the precise region in RABX- 5 disrupted in the rabx-5(tm1512) mutants. We found that the region disrupted in the rabx-5(tm1512) mutant encodes A20 zinc finger, a ubiquitin binding domain (Fig. 3A). Sequence alignment between RABX-5 and bovine Rabex-5, a mammalian ortholog, reveals that RABX-5 contains a single ubiquitin binding domain while bovine Rabex-5 revealed two such domains arranged in tandem: an A20 zinc finger domain and a motifinteracting with ubiquitin (MIU) domain. Biochemical and structural studies have established that the two tandem ubiquitin binding domains of Rabex-5 recognize ubiquitin as an intracellular trafficking signal with modest affinities (Lee et al., 2006; Penengo et al., 2006). Homology modeling of the A20 zinc finger domain of RABX-5 suggests that RABX-5 would be able to bind ubiquitin (Fig. 3B). These data suggest that the activation of autophagy by RABX-5 may be regulated by its ubiquitin recognition that in turn provides a signal for the endocytic pathway.
Fig. 3. (A) Sequence alignment between C. elegans RABX-5 and bovine Rabex-5. The region which is disrupted in the C. elegans rabx-5(tm1512) mutant is shown as a purple box. Black inverted triangles refer to the key residues in recognizing ubiquitin in bovine Rabex-5. (B) Homology modeling of C. elegans RABX-5 A20 zinc finger domain using SwissModel Server. C. elegans RABX-5 is colored in purple with zinc ion in red. Bovine Rabex-5 spanning A20 zinc finger and motif-interacting with ubiquitin (MIU) is depicted in gray with two bound ubiquitin molecules in blue and yellow.
Autophagy in dyn-1 mutants
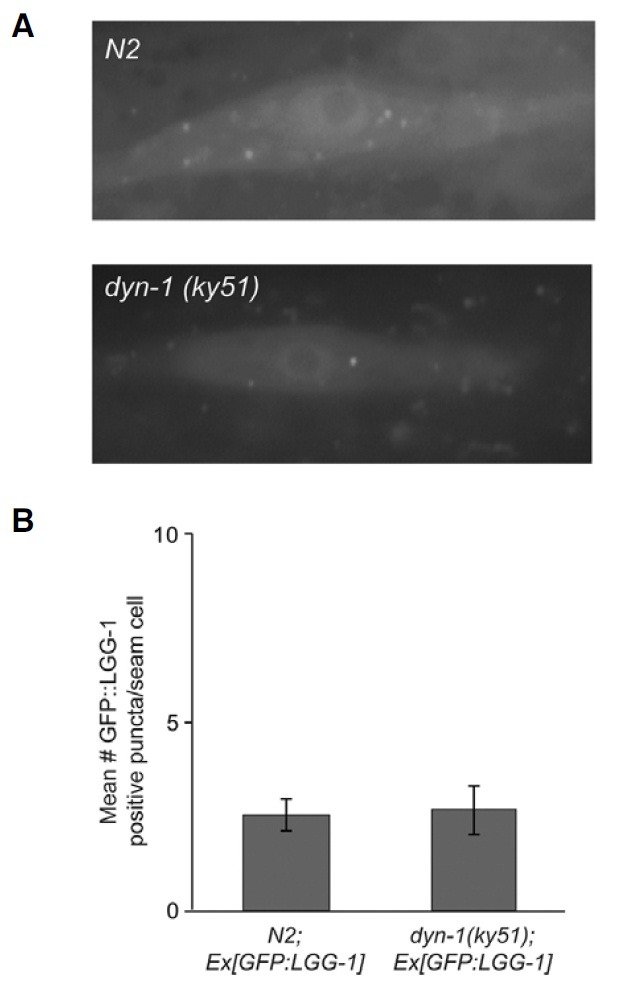
To check the sites where the levels of autophagy in rab-5, rabx- 5 and rme-6 mutants are elevated, we investigated the expression pattern of GFP::LGG-1 in dyn-1 mutants as dynamin acts at the early stages of clathrin-mediated endocytosis. We found that the number of GFP::LGG-1 puncta in dyn-1(ky51) mutants were comparable to that of wild-type worms (Figs. 4A and 4B). This result suggests that autophagy can compensate the endocytic pathway only after the formation of mature CCV. It is well established that CCVs are pinched off by dynamin, actively uncoated and transported to early endosomes. We postulate that autophagic machinery compensates the function for the degradation of cargo and clathrin molecules assembled in mature CCVs if the endocytic pathway is blocked and/or defective.
Fig. 4. (A) Representative images of GFP::LGG-1 expression in the seam cells in N2 strains or dyn-1(ky51) mutant. (B) Quantification of GFP::LGG-1 containing puncta in each case described in (A). Magnification of the images is 1,000.
Summary
Our results establish that genes involved in the endocytic pathway such as rab-5, rabx-5 and rme-6 can activate autophagy when the endocytic pathway is disrupted. Among those genes investigated, rab-5 and rabx-5 RNAi predominantly activate autophagy. Disruption of the ubiquitin binding domain in RABX- 5 is responsible for autophagy activation, suggesting that ubiquitin recognition regulates switching the involvement of RABX-5 between endocytic pathway and autophagy. However, lifespan of worms are not affected by RNAi targeted against rabx-5. Activation of autophagy by defective RABX-5 precisely appears to occur subsequent to CCV maturation. Our findings reveal for the first time the involvement of RABX-5 in autophagy.
Acknowledgments
We thank Alicia Melendez for providing the wild-type worms carrying the GFP::LGG-1 extrachromosomal array. One of the authors, M. Dwivedi, is thankful to CSIR for providing financial assistance. This work was supported by the National Research Foundation Grant funded by the Korean Government (KRF- 2008-314-C00224).
References
- 1.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. (2006);15:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S. The genetics of Caenorhabditis elegans. Genetics. (1974);77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucci C., Parton R.G., Mather I.H., Stunnenberg H., Simonks K., Hoflack B., Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. (1992);70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 4.Chavrier P., Parton R.G., Hauri H.P., Simonks K., Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. (1990);62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- 5.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. (2003);31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christoforidis S., McBride H.M., Burgoyne R.D., Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. (1999);397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 7.De Renzis S., Sonnichsen B., Zerial M. Divalent Rab effectors regulate the subcompartmental organization and sorting of early endosomes. Nat. Cell Biol. (2002);4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- 8.Dwivedi M., Ahnn J. Autophagy-is it a preferred route for lifespan extension? BMB Rep. (2009);42:65–71. [PubMed] [Google Scholar]
- 9.Dwivedi M., Song H., Ahnn J. Autophagy genes mediate the effect of calcineurin on life span in C. elegans. Autophagy. (2009);5:604–607. doi: 10.4161/auto.5.5.8157. [DOI] [PubMed] [Google Scholar]
- 10.Grant B.D., Sato M. Intracellular trafficking. In Worm- Book ed. The C. elegans Research Community. WormBook; (2006). http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horiuch H., Lippe R., McBride H.M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. (1997);90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 12.Klionsky D.J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. (2005);118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Tsai Y.C., Mattera R., Smith W.J., Kostelansky M.S., Weissman A.M., Bonifacino J.S., Hurley J.H. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat. Struct. Mol. Biol. (2006);13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., et al. Beclin1- binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. (2008);10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melendez A., Levine B. Autophagy in C. elegans. In WormBook ed. The C. elegans Research Community. WormBook; (2009). http://www.wormbook.org. [DOI] [Google Scholar]
- 16.Melendez A., Talloczy Z., Seaman M., Eskelinen E.L., Hall D.H., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. (2003);301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 17.Michelet X., Alberti A., Benkemoun L., Roudier N., Lefebvre C., Legouis R. The ESCRT-III protein CeVPS-32 is enriched in domains distinct from CeVPS-27 and CeVPS-23 at the endosomal membrane of epithelial cells. Biol. Cell. (2009);101:599–615. doi: 10.1042/BC20090025. [DOI] [PubMed] [Google Scholar]
- 18.Penengo L., Mapelli M., Murachelli A.G., Confalonieri S., Magri L., Musacchio A., Di Fiore P.P., Polo S., Schneider T.R. Crystal structure of the ubiquitin binding domains of Rabex-5 reveals two modes of interaction with ubiquitin. Cell. (2006);124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Roudier N., Lefebvre C., Legouis R. CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic. (2005);6:695–705. doi: 10.1111/j.1600-0854.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- 20.Ruck A., Attonito J., Garces K.T., Nunez L., Palmisano N.J., Rubel Z., Bai Z., Nguyen K.C., Sun L., Grant B.D., et al. The Atg6/Vps30/Beclin1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. (2011);7 doi: 10.4161/auto.7.4.14391. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato M., Sato K., Fonarev P., Huang C.J., Liou W., Grant B.D. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat. Cell Biol. (2005);7:559–569. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth M.L., Sigmond T., Borsos E., Barna J., Erdelyi P., Takacs- Vellai K., Orosz L., Kovacs A.L., Csikos G., Sass M., et al. Longevitiy pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. (2008);4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]


