Abstract
Importin β1 interacts with nuclear transport factors and mediates the import of nuclear proteins. We isolated a pollen-expressed gene, rice Importin β1 (OsImpβ1), from a T-DNA insertional population that was trapped by a promoterless β-glucuronidase (GUS) gene. The GUS reporter was expressed in the anthers and ovaries from early through mature developmental stages. Its expression was also observed in all floral organs. However, these patterns changed as the spikelet developed. T-DNA was inserted into the OsImpβ1 gene at 339 bp downstream from the translation initiation site. We obtained another T-DNA insertional allele by searching the flanking sequence tag database. In both lines, the wild-type and T-DNA-carrying progeny segregated at a ratio close to 1:1. The latter genotype was heterozygous (OsImpβ1/osimpβ1). Reciprocal crosses between WT and heterozygous plants demonstrated that the mutant alleles could not be transmitted through the male gametophyte. Close examination of the heterozygous anthers revealed that the mutant pollen matured normally. However, in vitro assays showed that tube elongation was hampered in the mutant grains. These results indicate that OsImpβ1 is specifically required for pollen tube elongation.
Keywords: GUS, importin β1, pollen tube, rice, T-DNA
INTRODUCTION
Fast pollen tube elongation is essential for the fertilization of plant ovules, and pollen grains compete for success among those tubes within the styles. Several genes essential for tube growth have been studied using Arabidopsis thaliana mutants. For example, the poky pollen tube (pok) mutant exhibits very short tubes (Lobstein et al., 2004). The POK protein localizes to the Golgi apparatus, possibly being involved in the retrograde trafficking of vesicles. Furthermore, seth1 and seth2 mutations almost entirely block male transmission through reduced pollen germination and tube growth (Lalanne et al., 2004). They encode proteins that are homologous to conserved proteins involved in the first step of the glycosylphosphatidylinositol biosynthesis pathway. This finding suggests that glycosylphosphatidylinositol anchor biosynthesis is essential for pollen tube wall deposition or metabolism. The gene encoding the exocyst component ortholog, AtSEC8, is required for both pollen germination and competitive pollen tube growth (Cole et al., 2005). The significance of the calcium-signaling pathway during pollen germination and tip growth has been investigated with the no pollen germination 1 (npg1) gene, which encodes a protein related to the calmodulin-binding proteins (Golovkin and Reddy, 2003). It has been also known that a signaling regulator, PRK1 plays important roles in pollen tubes development in Petunia inflata (Park et al., 2000).
Antisense suppression of SHY, which encodes a leucine-rich repeat protein, suppresses tube growth and causes abnormal callose deposits throughout the tube and into the tips (Guyon et al., 2004). Tube growth is also affected by a T-DNA insertional mutant in AtTAF6, which encodes a TATA-binding proteinassociated factor, indicating that transcriptional regulation of only a specific subset of genes is controlled by this basal transcription factor (Lago et al., 2005). Pollen germination and tube growth also require COMATOSE, which encodes a peroximal ABC transporter required for the oxidation of storage lipids during germination (Footitt et al., 2007), as well as the nicotinate/nicotinamide mononucleotide adenyltransferase (AtNMNAT) gene, which catalyzes the synthesis of nicotinate adenine dinucleotide from nicotinate mononucleotide (Hashida et al., 2007).
In rice, no mutations defective in their pollen tube elongation have yet been reported. Instead, proteomics analysis has identified a large number of proteins that are specifically present in germinated pollen (Dai et al., 2007). Two-dimensional electrophoresis of approximately 2300 protein spots has revealed 186 that are differentially expressed in mature and germinated pollen. Differentially expressed protein spots have been found, via mass spectrometry, to match 120 diverse protein species. These proteins are involved in various cellular and metabolic processes, with an obvious functional skew toward cell wall metabolism, protein synthesis and degradation, cytoskeleton dynamics, and carbohydrate/energy metabolism.
Nuclear import and export pathways are mediated by nuclear transport receptors - importin proteins - that regulate diverse cellular functions. Most importin proteins contain nuclear localization signals (NLS) that comprise one or two short stretches of basic amino acids (Dingwall and Laskey, 1991). In the cytosol, proteins that carry NLS bind to importin α and form a nuclear pore complex. This complex interacts with the nuclear membrane protein importin β via an amino-terminal IBB domain. Then, this trimeric import complex docks to the cytoplasmic face of the nuclear pore complex via importin β, and subsequently is translocated to the nucleoplasmic side. In the nucleus, Ran-GTP binds to importin β and induces the release of import cargoes. This nuclear transport receptor family shares an amino-terminal Ran-GTP-binding domain, which shows limited sequence conservation. Members bind very specifically to the different classes of macromolecules that must be transported across the nuclear envelope. The specificity of these transport processes is ensured by the variety of receptors that recognize different substrates (for a review, see Görlich and Kutay, 1999).
The first plant Importin β genes were identified from rice by cloning two (OsImpβ1 and OsImpβ2) cDNA clones (Matsuki et al., 1998). Transcripts of both genes were detected in roots, shoots, and calli. In vitro binding assays have indicated that rice importin β1 assembles a complex with importin α1 and an NLS protein, and also binds to the nuclear envelope (Jiang et al., 1998). Moreover, Ran-GTP, but not Ran-GDP, interacts with rice importin β1 and dissociates the heterodimer formed between rice importins α1 and β1. In HeLa cells, the rice importin β1 mediates nuclear envelope-docking of the NLS proteins and their subsequent translocation into the nucleus (Jiang et al., 1998).
Although many importin β-like proteins exist in the Arabidopsis genome, only a few have been characterized. The nuclear export receptor XPO1, which specifically binds to leucine-rich nuclear export signal (NES), has been functionally analyzed (Haasen et al., 1999). The Arabidopsis protein interacts with Arabidopsis Ran and with NESs of plant and human proteins. Export activity within a plant cell has been demonstrated in vivo using an assay system that employs green fluorescent protein (GFP) fusion proteins. This finding demonstrates the high conservation of that nuclear export pathway among animals, yeast, and plants (Haasen et al., 1999). Among the 17 members of the importin β family, HASTY was the first shown to have an essential role in plants. A loss-of-function mutation of HASTY affects many different processes in Arabidopsis growth and accelerates its developmental program (Bollman et al., 2003; Telfer and Poethig, 1998). In contrast, mutation of another Arabidopsis exportin, PAUSED, causes delayed leaf initiation and transition to flowering (Hunter et al., 2003). Furthermore, a hasty/paused double mutant has a more severe phenotype than either single mutant (Hunter et al., 2003), suggesting that these two exportins act on different pathways. Sensitive to ABA and Drought 2 (SAD2) mutations in another member of the importin β family cause ABA hypersensitivity in seed germination and seedling growth (Verslues et al., 2006). This suggests a role for nuclear transport of importin β proteins in ABA signal transduction.
In this study, we examined the role of Importin β1 in rice development, using loss-of-function mutants to investigate their defects in pollen tube elongation and fertilization rates.
MATERIALS AND METHODS
Plant growth
Seeds from wild-type japonica rice (Oryza sativa cv. Dongjin) and heterozygous (OsImpβ1/osimpβ1) plants were germinated on a Murashige and Skoog (MS) medium containing 0.44% MS basal salts, 3% sucrose, 0.2% phytagel, and 0.55 mM myoinositol (Sigma, USA). The seedlings were grown for 1 week at 27℃ under continuous light, then transplanted to soil in the greenhouse and raised to maturity.
Histochemical GUS assay and microscopic analyses
Histochemical GUS staining was performed as described by Kim et al. (2009). For light-microscopic analysis, the tissues were fixed in a solution containing 50% ethanol, 5% acetic acid, and 3.7% formaldehyde, and then embedded in Technovit 8100 Resin (Kulzer and Co., Germany). These samples were sectioned to 3- to 10-μm thickness with a microtome (Leica, Germany), and observed under a microscope (Nikon, Japan) using bright- and dark-field illumination.
Isolation of T-DNA flanking sequence by TAIL-PCR and identification of a fusion transcript between the OsImpβ1 and GUS genes
PCRs were performed in 50 μl of a mixture containing 2 ng of cDNA, 10× Ex Taq buffer, 0.2 mM dNTP, 0.5 unit of Ex Taq polymerase (Takara, Japan), and 1 μM of the primers. Thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) was conducted as described previously (Jung et al., 2005; Liu et al., 1995). The template was cDNA prepared from the panicles of OsImpβ1/osimpβ1-1 plants collected at the mature stage. The specific primers were gus1 (5′-GCCGTAATGAGTGAC CGCATCG-3′), gus2 (5′-ATCTGCATCGGCGAACTGATCG-3′), and gus3 (5′-CACGGGTTGGGGTTTCTACAGG-3′). Arbitrary primers included ad1 [5′-NTCGA(G/C)T(G/C)G(A/T)GTT-3′], ad2 [5′-NGTCGA(G/C)(A/T)GANA(A/T)GAA-3′], and ad3 [5′- (A/T)GTGNAG(A/T)ANCANAGA-3′]. The third PCR product was subjected directly to sequencing using the gus3 primer. The fusion transcript between OsImpβ1 and GUS was amplified by the p1 primer (5′-TCCTGCTATCAGCTCAATCT-3′, 17 bp downstream from the translation initiation site of the OsImpβ1 gene; Fig. 1) and by the p2 primer (5′-CATCACTTCCTGATTATTG ACC-3′, 316 bp downstream from the translation initiation site of the GUS gene; Fig. 1).
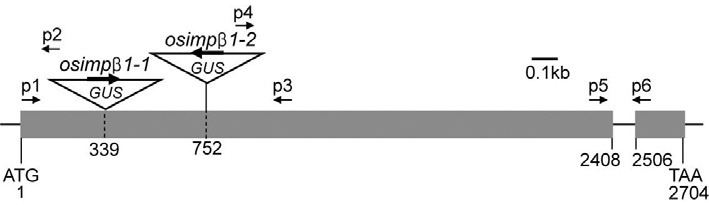
Fig. 1. Schematic diagram of OsImpβ1 gene and T-DNA insertion positions in osimpβ1 mutants. Black boxes represent exons; intervening lines, introns. Positions of T-DNA insertion are indicated with triangles. Numbers indicate nucleotide positions from the start ATG codon. Primer sets of p1 and p3, and of p1 and p2 were used for genotyping progenies of OsImpβ1/osimpβ1-1 selfed, OsImpβ1/ osimβ1-1 (female) × WT (male), and WT (female) × OsImpβ1/osimpβ1-1 (male) lines. Primer sets of p1 and p3, and of p3 and p4 were used for genotyping progenies of OsImpβ1/osimpβ1-2 selfed line.
Genotyping the mutant plants
PCRs for genotyping were performed in 50 μl of a mixture containing 20 ng of plant DNA, 10× Ex Taq buffer, 0.2 mM dNTP, 0.5 unit of Ex Taq polymerase (Takara), and 1 μM of the primers. The protocol included 35 cycles of 94℃ for 60 s, 60℃ for 90 s, and 72℃ for 120 s. Primers for genotyping were 5′- TCCTGCTATCAGCTCAATCT-3′ (17 bp downstream of the translation initiation site of OsImpβ; p1 in Fig. 1), 5′-TAGCAAT GAGCCCAAGGCAT-3′ (1050 bp downstream from the translation initiation site of OsImpβ1; p3 in Fig. 1), 5′-CATCACTT CCTGATTATTGACC-3′ (316 bp downstream from the translation initiation site of GUS; p2 in Fig. 1), and 5′-ATGGCAGTGA ATTAACATAGC-3′ (2924 bp downstream from the translation initiation site of the GUS gene; p4 in Fig. 1).
Quantitative real-time RT-PCR
Total RNAs were isolated using Tri Reagent (MRCI Inc., USA). For the first-strand cDNA synthesis, 1 μg of total RNA was reverse- transcribed in a total volume of 20 μl that contained 10 ng of oligo(dT)12-18 primer, 2.5 mM deoxynucleotide triphosphate (dNTP), and 200 units of Moloney Murine Leukemia Virus reverse transcriptase (Promega, USA) in a 5× reaction buffer. Quantitative real-time PCR was performed with a Roche Lightcyler, using SYBR premix ExTaq (Takara). PCR reactions were carried out in a final volume of 20 μl of reaction solution containing 10 μl PCR mixture, 1 μl cDNA, and 0.5 μM of each primer, under the following conditions: 1 min at 95℃, then 45 cycles at 94℃ for 15 s, 60℃ for 13 s, and 72℃ for 12 s. At the end of the reaction, the specificity of the primer set was confirmed by a melting curve. The Ubiquitin 1 (Os03g0221500, LOC_Os03g12140) mRNA level was used to normalize the expression ratio for each gene. Primers for the Ubiquitin 1 gene were 5′-CACGGTTCAA CAACATCCAG-3′ and 5′-TGAAGACCCTGACTGGGAAG-3′. The primers for OsImpβ1 were 5′-GGCATACTCCAGGGTAT AAA-3′ (2311 bp downstream from the translation initiation site of the gene; p5 in Fig. 1) and 5′-ACTCCCTCAGGAACTCAA CA-3′ (2608 bp downstream from the translation initiation site of the gene; p6 in Fig. 1). Changes in gene expression were calculated according to the delta-delta Ct method.
In vitro pollen germination
Pollen grains from dehisced anthers (both wild-type and mutant) were placed on a slide glass at 35℃ for 2 h in a pollen germination medium consisting of 1 mM CaCl2, 1 mM KCl, 0.8 mM MgSO4, 1.6 mM H3BO3, 30 μM CaSO4, 0.03% casein, 0.3% 2-(N-morpholino) ethanesulfonic acid, 10% sucrose, and 12.5% polyethylene glycol. The humidity was maintained above 90%. Germinated pollen grains were then stained in a GUS assay solution at 37℃ for 3 h. The grains were observed with a microscope (Nikon), using bright-field illumination.
4′, 6-daimidino-2-phenylindole (DAPI) staining
Pollen grains were fixed in an ethanol:acetic acid (3:1) solution for 1 h at room temperature, then dehydrated through an ethanol series (75, 55, and 35%). They were transferred to Tris-HCl buffer (pH 7.5) with 200 ng ml-1 DAPI, and incubated for 1 h at 60℃. The stained pollen was monitored under UV light with a Zeiss Airplane 2 microscope.
Ultrastructural analyses by transmission electron microscopy
Anthers sampled from OsImpβ1-1/osimpβ1-1 plants at the mature stages were fixed for 4 h in cacodylate buffer (pH 7.2) that contained 2% paraformaldehyde (Sigma) and 2% glutaraldehyde (Sigma). They were then rinsed with the same buffer and post-fixed for 1 h in cacodylate buffer containing 1% osmium tetroxide (Pelco International, USA). After dehydration, the specimens were embedded in London Resin White (London Resin Co., UK). Ultra-thin sections (40 to 60 nm thick) collected on uncoated nickel grids (300 mesh) were stained with 4% uranyl acetate and examined at 60 to 80 kV with a JEOL 1200 transmission electron microscope (JEOL Ltd., Japan).
Transient expression of GFP and RFP fusion constructs in protoplasts
Protoplasts were prepared from rice mesophyll cells. They were harvested and incubated in an enzyme solution (1.5% cellulase RS, 0.5% macerozyme, 0.1% pectolyase, 0.6 M mannitol, 10 mM MES, 1 mM CaCl2, and 0.1% bovine serum albumin) for 4 to 5 h at 26℃ with gentle agitation (50 to 75 rpm). The protoplasts were isolated from the undigested material with a nylon mesh (20 μm) and centrifuged at 40 to 60 × g for 2 min. They were then re-suspended in 5 ml of KMC solution (117 mM KCl, 82 mM MgCl2, and 85 mM CaCl2) and pelleted again by centrifugation for 90 s at 40 to 50 × g. These pelleted protoplasts were re-suspended in an EP3 solution (70 mM KCl, 5 mM MgCl2, 0.4 M mannitol, and 0.1% MES; pH 5.6). For our transfection experiment, 1 × 106 protoplasts were electroporated with 20 μg each of the fusion constructs, using the Gene Pulser Xcell (BioRAD, USA) at 300 V and 450 μF. Following transformation, expression of the fusion constructs was monitored at various times with a Zeiss LSM510 confocal laser scanning microscope. The primers for constructing the OsImpβ1-GFP fusion vectors were 5′-CCCGGGGGTACAGCCATGAATATC AC-3′ (the ATG start codon of OsImpβ1 is underlined) and 5′- TCGAGCTTAAGAAACCAGTGCTTGGTT-3′ (the TAA stop codon is underlined).
RESULTS
Isolation of a promoter trap line using GUS activity in a pollen-expressed gene
We have previously generated insertional mutant populations by T-DNA (Jeon et al., 2000; 2002; Jung et al., 2003; Ryu et al., 2004). Within that T-DNA, a promoterless GUS gene is placed immediately next to the T-DNA border so that insertion of the TDNA within a gene can generate a fusion between an endogenous gene and the GUS reporter. GUS analysis of the developing spikelets has allowed us to identify several lines with GUS activity in their pollen (Han et al., 2006).
Here, we report the characterization of Line 0-19302. In the spikelets, GUS expression was observed in the anthers and ovary from the early through mature developmental stages (Fig. 2A). The activity was also observed in the palea/lemma and lodicules during earlier stages, but disappeared later (Fig. 2A). The assay indicated that the gene was also expressed in the embryo of mature seeds (Fig. 2B) and shoots of 4 day old seedling (Fig. 2C). Finally, GUS was strongly detected in the root tips of the seedling (Fig. 2C).
Fig. 2. Expression profiles of the OsImpβ1 gene using GUS assay. (A) Temporal and spatial expression patterns of the OsImpβ1-GUS gene fusion product in OsImpβ1/osimpβ1-1 spikelets at various developmental stages: Samples 1, pollen mother cell stage; 2, meiosis stage; 3, tetrad stage; 4, late vacuolated stage; 5, mature pollen stage. (B) Expression patterns in a mature seed. (C) Expression patterns in a seeding. Bar = 2 mm. Cross sections of anthers at pollen mother cell stage (D), meiosis stage (E), tetrad stage (F), late vacuolated stage (G), and mature pollen stage (H). Photographs were taken with dark-field microscope (D-H), where GUS activity appeared as red coloring. An, anther; Br, bract; Ca, callose; EC, endothecial cell layer; Em, embryo; En, endosperm; Ep, epidermal cell layer; Lo, lodicules; M, middle layer; MMC, microspore mother cells; MSp, microspores; O, ovary; Pa, palea; PG, pollen grain; Ro, root; Sh, shoot; T, tapetal cell layer; Tds, tetrads; V, vascular region. Bars = 40 μm.
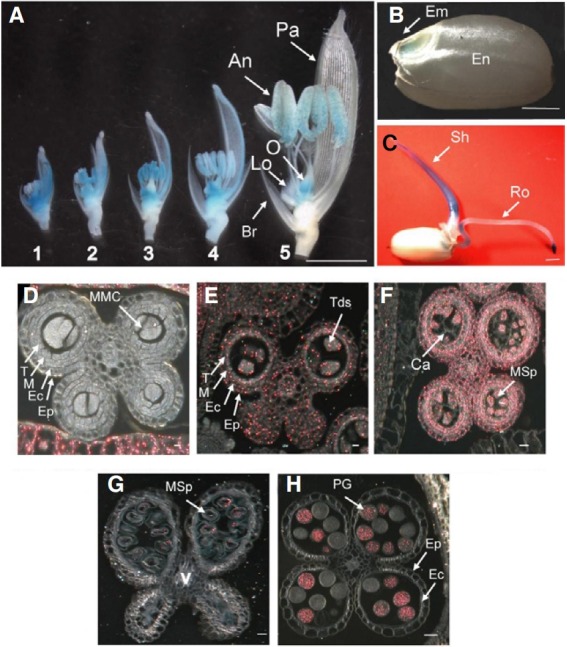
To observe these expression patterns at the cellular level, we cross-sectioned the anthers at five developmental stages. At the microspore mother cell stage, GUS expression was not detected in the anthers (Fig. 2D). GUS expression was found in all cell types of anthers at the meiotic (Fig. 2E) and tetrad stages (Fig. 2F), but was restricted to only the pollen at the latevacuolated (Fig. 2G) and mature-pollen stages (Fig. 2H). About half the pollen grains were stained by GUS assay, indicating segregation of the GUS reporter gene in male gametophyte (Figs. 2G and 2H). The cross-section analysis showed that GUS expression in the palea/lemma was the highest at the microspore mother cell stage, and the expression disappeared at mature-pollen stage, indicating that GUS patterns in developing pollens were antiparallel with those in developing palea/ lemma (Figs. 2D-2H).
T-DNA insertion occurs in a gene encoding Importin β1
We determined the T-DNA flanking sequence for Line 0-19302 via thermal asymmetric interlaced PCR (Jung et al., 2005). TDNA was inserted into the rice Importin β1 (OsImpβ1) gene at 339 bp downstream from the translation initiation site (Fig. 1A). The OsImpβ1 protein was previously reported to mediate nuclear envelope docking of nuclear localization signal (NLS) containing proteins and their subsequent translocation into the nucleus (Jiang et al., 1998; Matsuki et al., 1998). In order to obtain another allele, we searched our flanking sequence tag (FST) database (An et al., 2003; Jeong et al., 2006). This resulted in the identification of Line 2C-10311, which carries the T-DNA insertion at 752 bp downstream from the OsImpβ1 translation initiation site (Fig. 1).
Analyses of OsImpβ1 expression
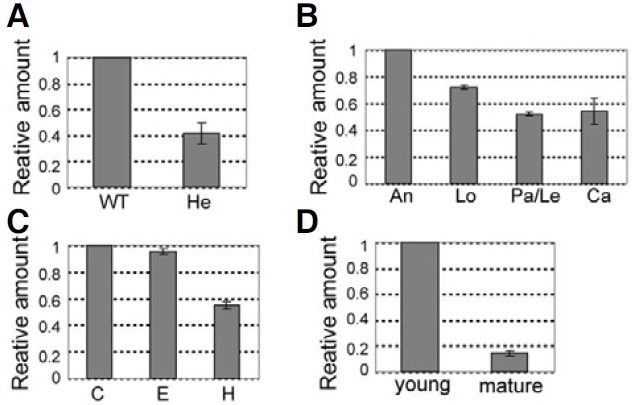
We measured transcripts of OsImpβ1 by using quantitative real-time RT-PCR with gene-specific primers with total RNAs prepared from various organs (P5 and P6 in Fig. 1). As expected, the transcript level in heterozygous anthers (OsImpβ1/osimpβ1-1) was about half of the amount measured from the wild-type anthers (Fig. 3A). Quantitative real-time RT-PCR analyses in mature spikelets showed that transcription was the highest in the anthers but only moderate in the other floral organs (Fig. 3B). Furthermore, amount of transcript was higher in the root tip and root elongation zone than in the root hair zone (Fig. 3C). In the palea/lemma, transcription was greater at the young stage compared with the mature stage (Fig. 3D). Overall, these results are in good agreement with our GUS data.
Fig. 3. Quantitative real-time RT-PCR analyses of OsImpβ1 expression. (A) Quantitative real-time RT-PCR analysis of OsImpβ1 in wild-type and hetero (OsImpβ1/ osimpβ1-1) anthers at mature pollen stage. (B) Spatial expression levels of OsImpβ1 in mature floral organs (C) Temporal expression patterns of OsImpβ1 in wild-type roots at 10 days after germination. (D)Temporal expression patterns of OsImpβ1 in wild-type palea/lemma at young and mature stages. An, anther; C, root cap; Ca, carpel; E, elongation zone; H, root hair zone; Le, lemma; Lo, lodicule; Pa, palea. The OsImpβ1 expression levels were normalized with respect to the internal control Ubiquitin 1(Os03g0221500, LOC_Os03g12140) gene. The gene expression levels were calculated by the delta Ct Method.
T-DNA insertions in OsImpβ1 cause aberrant transmission through male gametophytes
All of the progeny from the T-DNA tagging lines were normal. Therefore, we determined the T-DNA segregation in T2 progeny of Line 0-19302 by analyzing GUS expression and hygromycin resistance (Table 1). As a result, the segregation ratio of wild-type and T-DNA-carrying progeny was 143:150, that is, close to 1:1. We observed a similar pattern (94:90) from Line 2C-10311. These results indicate that transmission of T-DNA was hampered in the gametophyte. We genotyped the progeny from the selfed population of the primary transgenic plants and observed that all the T-DNA-carrying progeny were heterozygous (Table 1). Therefore, we could predict that either the male or female gametophytes would be defective in the insertional lines.
Table 1.
Genetic analyses of male or female gene transfer caused by mutations of Osimpβ1
| Cross (female x male) | No. plants WT | No. plants OsImpβ1/osim pβ1 | Observed ratio | Expected ratio | χ2 |
|---|---|---|---|---|---|
| OsImpβ1/osimpβ1-1 selfed Line 0-19302 | 143 | 150 | 1 : 1.05 | 1 : 1 | 0.17 |
| OsImpβ1/osimpβ1-2 selfed Line 2C-10311 | 94 | 90 | 1 : 0.96 | 1 : 1 | 0.09 |
| OsImpβ1/osimβ1-1 x WT | 26 | 29 | 1 : 1.12 | 1 : 1 | 0.16 |
| WT x OsImpβ1/osimpβ1-1 | 52 | 0 | N/A | N/A | N/A |
To determine whether this defect was due to a problem in the development and production of male or female gametophytes, we performed reciprocal crosses between wild-type and OsImpβ1/osimpβ1-1 heterozygous plants (Line 0-19302). When the latter were used as the female, both WT and heterozygous progeny were obtained (Table 1). However, when OsImpβ1/osimpβ1-1 plants were used as the pollen donor, only wild-type progeny were produced (Table 1). These experiments indicate that the osimpβ1 mutation caused a defect in the male gametophyte.
OsImpβ1 is not essential for pollen maturation
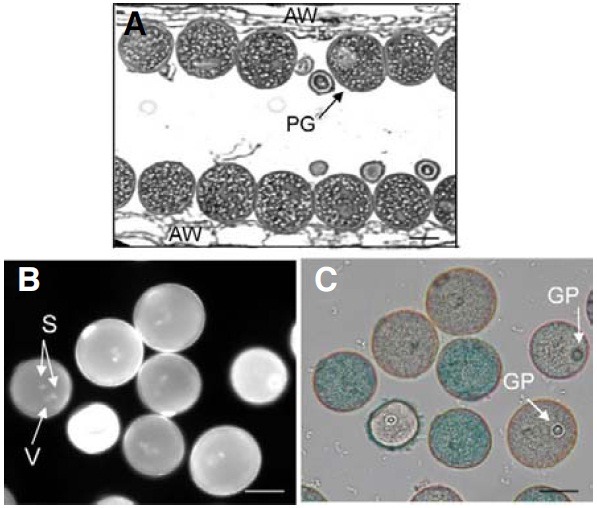
Because mutations in OsImpβ1 prevented the transmission of genetic materials through the male gametophyte, we examined ultramicrostructure of OsImpβ1/osimpβ1-1 anthers at the mature pollen stage. Longitudinal sections, analyzed by transmission electron microscopy (TEM), revealed that all the pollen grains were morphologically normal (Fig. 4A), thereby indicating that the osimpβ1-1 pollen reached the mature stage without any developmental defect. Using DAPI staining to determine whether the grains successfully achieved the trinuclear stage, we noted that all contained three nuclei, with the two more brightly stained small organelles being the sperm nuclei, and the larger and diffusely stained one being the vegetative cell nucleus (Fig. 4B). GUS staining enabled us to distinguish osimpβ1-1 pollen grains from the wild-type (Fig. 4C). We also tested pollen viability via fluorescein diacetate staining and observed that both types of pollen were equally viable (data not shown).
Fig. 4. Transmission electron microscopy analysis of OsImpβ1/osimpβ1-1 anther. Longitudinal sections of OsImpβ1/osimpβ1-1 anther at mature stage (A). Pollen grains from the OsImpβ1/osimpβ1-1 anther at mature stage were stained with DAPI (B) and GUS solutions (C). Stained pollen grains were monitored with a fluorescence microscope (B) and a bright-field microscope (C). AW, anther wall; GP, germination pore; PG, pollen grain; S, sperm nuclei; V, vegetative nucleus. Bars = 50 μm.
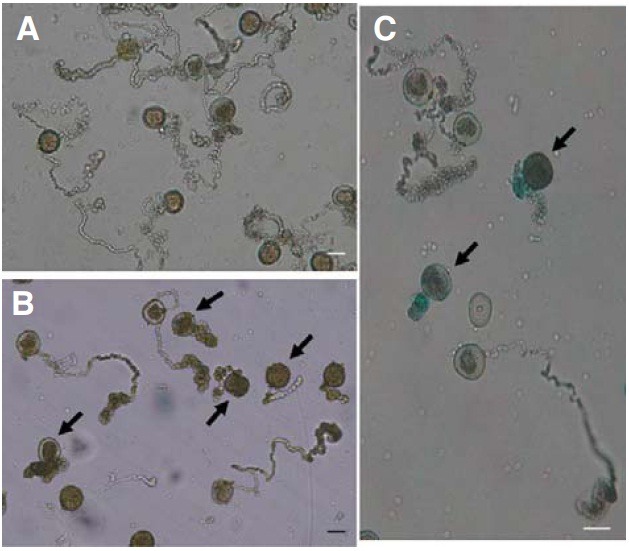
Pollen tube elongation is delayed in osimpβ1-1
Because the osimpβ1-1 pollen grains matured normally, we analyzed pollen germination and elongation under in vitro conditions. Here, 98% (501/509) of the wild-type grains germinated and elongated normally (Fig. 5A), compared with a rate of 46% (347/755) for the OsImpβ1/osimpβ1-1 grains (Fig. 5B). GUS staining of these germinated pollens showed that those unelongated grains were mostly GUS-positive while those that lengthened normally were GUS-negative (Fig. 5C). Therefore, we may conclude that mutations of the OsImpβ1 gene affect pollen tube elongation.
Fig. 5. In vitro pollen germination assays. Mature pollen grains from wild-type (A) and OsImpβ1/osimpβ1-1 (B) anthers were germinated for 2 h in the pollen germination medium. Germinated pollens from the OsImpβ1/osimpβ1-1 anther were stained in GUS solution for 3 h before microscope observations (C). Thick arrows indicate the osimpβ1-1 pollen. Bars = 50 μm.
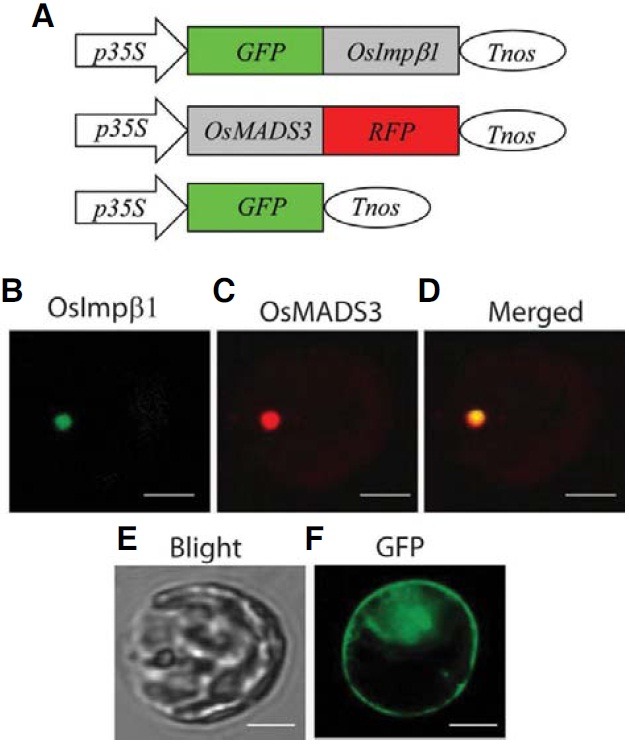
OsImportin β1 is located at the nucleus
Jiang et al. (1998) have previously reported that the Cy3- labeled rice Importin β1 is accumulated predominantly in the nuclear envelope of tobacco BY-2 cells. To confirm that observation, we made a fusion molecule by linking the carboxyterminal end of the full-length OsImpβ1 cDNA to the green fluorescent protein (GFP) gene. This construct was placed under the control of the 35S promoter and the nos terminator (Fig. 6A). As a positive control, the cDNA of a previously characterized nuclear protein, OsMADS3, was fused to the red fluorescent protein (RFP) gene and located between the same regulatory elements (Kang et al., 1998). When the chimeric molecules were co-introduced into protoplasts prepared from rice mesophyll cells, both green and red fluorescence signals were detected in the nucleus (Figs. 6B-6D).
Fig. 6. Subcellular localization of OsImpβ1. (A) Schematic diagrams of fusion constructs of GFP and OsImpβ1 cDNA (upper) and of OsMADS3 cDNA and RFP (middle). Fusion constructs were placed between the CaMV 35S promoter (p35S) and nos terminator (Tnos). Protoplasts isolated from mesophyll cells were co-transfected with p35S-GFP-OsImpβ1-Tnos and p35S-OsMADS3-RFP-Tnos vectors, and green fluorescent signal (B) or red fluorescent signal (C) was examined 30 h after transfection. (D) Merged photo of (B) and (C). (E) Bright field microscopy of the protoplast observed in (B)-(D). (F) Control GFP signal in a mesophyll protoplast obtained by transiently expressing p35S-GFP-Tnos. Bars = 10 μm.
DISCUSSION
T-DNA tagging using the GUS reporter gene is an efficient way to identify gametophyte-defective mutants
Screening of gametophytic mutants is difficult because it generally requires a large number of plants to be assayed under a microscope (Howden et al., 1998; Lalanne et al., 2004; Procissi et al., 2001). Therefore, we have previously utilized a method of enriching such mutants by screening the lines that express the GUS reporter gene in male gametophytes. In doing so, we have identified mutants defective in that reproductive organ. For example, this strategy has led to the discoveries of mutations in the Undeveloped Tapetum 1(Udt1) gene, a major regulator of early tapetum development (Jung et al., 2005), and in the Waxdeficient anther (Wda1) gene that is involved in very-long-chain fatty acid biosynthesis (Jung et al., 2006). This protocol has also enabled us to identify another mutant in the RICE IMMATURE POLLEN 1 (RIP1) gene, a regulator of late pollen development (Han et al., 2006). Here, we identified osimpβ1 mutants defective in their pollen tube elongation using the GUS trap system.
The OsImpβ1 gene is ubiquitously expressed in proliferating cells
Our T-DNA vector used for the gene trap has promoterless GUS gene near the right border of T-DNA. Therefore, it could generate a translational fusion when it is inserted within a gene in a correct orientation. The resulting GUS expression pattern should represent endogenous activity of the tagged gene. It is different from transgenic expression from exogenously introduced promoter-GUS constructs, which may show position effect. GUS analysis of our T-DNA tagged line showed that the OsImpβ1 gene was expressed in all floral organs, from early developmental stages to maturity. Expression was higher in young floral buds, with transcript levels decreasing thereafter. Upon maturation, expression could no longer be detected in any organs except pollen grains and ovaries. In vegetative organs, the gene was expressed in seedling roots and shoots, but declined in its induction in mature tissues. Therefore, it was apparent that OsImpβ1 is expressed and functional in actively proliferating cells.
The osimpβ1 is a male-gametophytic mutant
Our osimpβ1 mutant lines exhibited segregation distortion. The osimpβ1-1 homozygous progeny of Line 0-19302 were not present during four consequent generations produced by selfing OsImpβ1/osimpβ1-1 plants. Homozygous progeny also were lacking in another allele, Line 2C-10311. Reciprocal crosses between heterozygous plants and the wild type indicated a problem in transmitting the OsImpβ1 gene through the male gametophyte. Such a male-specific defect can result from damage during microsporogenesis or gametogenesis. However, our microscopic examinations revealed no characteristics that would distinguish the mutant pollen from the wild-type, nor did any affect sperm cell development and division.
Even if there are two more genes closely linked to OsImpβ1 in the phylogenic tree (Supplementary Fig. S1), expression pattern of the genes suggest that OsImpβ1 plays predominant roles in reproductive stages. Although OsImpβ1 (LOC_Os05g 28510) and LOC_ Os03g18350 showed similar expression patterns, OsImpβ1 expression was higher than LOC_Os03g18350 especially during floral development. The third gene LOC_ Os12g38110 was highly expressed in germinating and developing seeds, but weakly during floral development.
OsImpβ1 is necessary for pollen tube growth
Pollen tube growth requires changes in gene expression, and the proteins needed for this must be transported into the nucleus. Within 1 h of pollination, a large number of proteins are synthesized in the germinating pollen (Taylor and Hepler, 1997). Many of these contain a nuclear localization signal and are probably transported by importin protein family members. It is likely that the failure to transfer these specific proteins affects tube elongation.
Under in vitro conditions, the osimpβ1 pollen germinated normally but its tube elongation was hampered. Therefore, the mutant grains were unable to compete with wild-type gametophytes. This resulted in a male-specific transmission defect. Because this mutant phenotype was not observed during pollen maturation and germination, and not at all in the female gametophytes, it is evident that OsImpβ1 is absolutely required for pollen tube elongation. It is possible that the rice Importin β1 protein is necessary for nuclear-targeting of a group of proteins that are needed specifically for tube elongation. For example, the pollen tube interacts intimately with the nutrient-rich extracellular matrix of the stylar tract, and adhesion factors implicated in this tube growth have been identified (Lord and Russell, 2002). Studies of nucleocytoplasmic receptors in yeast and mammals have indicated that some receptors regulate the transport of a large number of functionally diverse proteins, whereas others have a more limited range of cargo molecules (for reviews, see Görlich, 1997; Nigg, 1997). Possible targets are the MIKC* MADS-proteins, which bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters (Verelst et al., 2007). A considerable number of putative direct-target genes for the AtMIKC* transcription factor complexes are either known or have proposed functions in pollen tube growth. We investigated the nuclear localization of OsImpβ1 by transiently expressing GFP-OsImpβ1 in mesophyll cells. OsImpβ1-GFP was observed mostly in and around the nucleus, similar to subcellular localization of the GFP-tagged SAD2 encoding Arabidopsis Importin β. However, our result is different from those from tobacco BY-2 cells, where Impβ1 was localized to the nuclear envelope (Jiang et al., 1998). Therefore, we do not rule out the possibility that the N-terminal fusion interfered with a proper localization of OsImpβ1.
Alternatively, OsImportin β1 may not have target specificity but is required when a cell is in large demand for protein import to the nucleus. Pollen tubes grow at extremely high rates in vivo and, consequently, have a very great need for energy (Lord and Russell, 2002). It will be interesting to determine whether OsImpβ1 controls general translocation of nuclear proteins by testing various GFP-nuclear proteins in the osimpβ1 mutants. Because we did not obtain any homozygous progeny, however, we were unable to examine the effect of this osimpβ1 mutation on seedling growth. In particular, it will be interesting to determine whether the OsImpβ1 gene is required for root tip growth.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
We express our thanks to Priscilla Licht for critical reading of the manuscript. This work was supported, in part, by grants from the Basic Research Promotion Fund (KRF-2007-341-C00028) and Kyung Hee University (20100791).
References
- 1.An S., Park S., Jeong D.H., Lee D.Y., Kang H.G., Yu J.H., Hur J., Kim S.R., Kim Y.H., Lee M., et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. (2003);133:2040–2047. doi: 10.1104/pp.103.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollman K.M., Aukerman M.J., Park M.Y., Hunter C., Berardini T.Z., Poethig R.S. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development (Cambridge, England) (2003);130:1493–1504. doi: 10.1242/dev.00362. [DOI] [PubMed] [Google Scholar]
- 3.Cole R.A., Synek L., Zarsky V., Fowler J.E. SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol. (2005);138:2005–2018. doi: 10.1104/pp.105.062273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai S., Chen T., Chong K., Xue Y., Liu S., Wang T. Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol. Cell. Proteom. (2007);6:207–230. doi: 10.1074/mcp.M600146-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Dingwall C., Laskey R. Nuclear targeting sequence-a consensus? Trends Biochem. Sci. (1991);16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 6.Footitt S., Dietrich D., Fait A., Fernie A.R., Holdsworth M.J., Baker A., Theodoulou F.L. The COMATOSE ATPbinding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. (2007);144:1467–1480. doi: 10.1104/pp.107.099903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golovkin M., Reddy A.S. A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc. Natl. Acad. Sci. USA. (2003);100:10558–10563. doi: 10.1073/pnas.1734110100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Görlich D. Nuclear protein import. Curr. Opin. Cell Biol. (1997);9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 9.Görlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Biol. (1999);15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 10.Guyon V., Tang W.H., Monti M.M., Raiola A., Lorenzo G.D., McCormick S., Taylor L.P. Antisense phenotypes reveal a role for SHY, a pollen-specific leucine-rich repeat protein, in pollen tube growth. Plant J. (2004);39:643–654. doi: 10.1111/j.1365-313X.2004.02162.x. [DOI] [PubMed] [Google Scholar]
- 11.Haasen D., Köhler C., Neuhaus G., Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. (1999);20:695–705. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- 12.Han M.J., Jung K.H., Yi G., Lee D.Y., An G. Rice immature pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol. (2006);47:1457–1472. doi: 10.1093/pcp/pcl013. [DOI] [PubMed] [Google Scholar]
- 13.Hashida S.N., Takahashi H., Kawai-Tamada M., Uchimiya H. Arabidopsis thaliana nicotinate/nicotinamide mononucleotide adenyltransferase (AtNMNAT) is required for pollen tube growth. Plant J. (2007);49:694–703. doi: 10.1111/j.1365-313X.2006.02989.x. [DOI] [PubMed] [Google Scholar]
- 14.Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics. (1998);149:621–631. doi: 10.1093/genetics/149.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter C.A., Aukeman M.J., Sun H., Fokina M., Poethig R.S. PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol. (2003);132:2135–2143. doi: 10.1104/pp.103.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. (2000);22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- 17.Jeong D.H., An S., Kang H.G., Moon S., Han J.J., Park S., Lee H.S., An K., An G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. (2002);130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong D.H., An S., Park S., Kang H,G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. (2006);45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 19.Jiang C.J., Imamoto N., Matsuki R., Yoneda Y., Yamamoto N. In vitro characterization of rice importin beta1: Molecular interaction with nuclear transport factors and mediation of nuclear protein import. FEBS Lett. (1998);437:127–130. doi: 10.1016/s0014-5793(98)01207-1. [DOI] [PubMed] [Google Scholar]
- 20.Jung K.H., Hur J., Ryu C.H., Choi Y., Chung Y.Y., Miyao A., Hirochika H., An G. Characterization of a rice chlorophyll- deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. (2003);44:463–472. doi: 10.1093/pcp/pcg064. [DOI] [PubMed] [Google Scholar]
- 21.Jung K.H., Han M.J., Lee Y.S., Kim Y.W., Hwang I., Kim M.J., Kim Y.K., Nahm B.H., An G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. (2005);17:2705–2722. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung K.H., Han M.J., Lee D.Y., Lee Y.S., Schreiber L., Franke R., Faust A., Yephremov A., Saedler H., Kim Y.W., et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell. (2006);18:3015–3032. doi: 10.1105/tpc.106.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H.G., Jeon J.S., Lee S., An G. Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. (1998);38:1021–1029. doi: 10.1023/a:1006051911291. [DOI] [PubMed] [Google Scholar]
- 24.Kim S.R., Lee D.Y., Yang J.I., Moon S., An G. Cloning vectors for rice. J. Plant Biol. (2009);52:73–78. [Google Scholar]
- 25.Lago C., Clerici E., Dreni L., Horlow C., Caporali E., Colombo L., Kater M.M. The Arabidopsis TFIID factor AtTAF6 controls pollen tube growth. Dev. Biol. (2005);285:91–100. doi: 10.1016/j.ydbio.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Lalanne E., Honys D., Johnson A., Borner G.H., Lilley K.S., Dupree P., Grossniklaus U., Twell D. SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell. (2004);16:229–240. doi: 10.1105/tpc.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. (1995);8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- 28.Lobstein E., Guyon A., Ferault M., Twell D., Pelletier G., Bonhomme S. The putative Arabidopsis homolog of yeast vps52p is required for pollen tube elongation, localizes to Golgi, and might be involved in vesicle trafficking. Plant Physiol. (2004);135:1480–1490. doi: 10.1104/pp.103.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lord E.M., Russell S.D. The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. (2002);18:81–105. doi: 10.1146/annurev.cellbio.18.012502.083438. [DOI] [PubMed] [Google Scholar]
- 30.Matsuki R., Iwasaki T., Shoji K., Jiang C.J., Yamamoto N. Isolation and characterization of two importin-β genes from rice. Plant Cell Physiol. (1998);39:879–884. doi: 10.1093/oxfordjournals.pcp.a029448. [DOI] [PubMed] [Google Scholar]
- 31.Nigg E.A. Nucleocytoplasmic transport: Signals, mechanism and regulation. Nature. (1997);386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 32.Park S.W., Yu S.H., Kim M.I., Oh S.C., Kao T., Pai H. Interaction of PRK1 Receptor-like Kinase with a Putative elF2B β-Subunit in Tobacco. Mol. Cells. (2000);10:626–632. doi: 10.1007/s10059-000-0626-z. [DOI] [PubMed] [Google Scholar]
- 33.Procissi A., de Laissardiere S., Ferault M., Vezon D., Pelletier G., Bonhomme S. Five gametophytic mutations affecting pollen development and pollen tube growth in Arabidopsis thaliana. Genetics. (2001);158:1773–1783. doi: 10.1093/genetics/158.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu C.H., You J.H., Kang H.G., Hur J., Kim Y.H., Han M.J., An K., Chung B.C., Lee C.H., An G. Generation of TDNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol. Biol. (2004);54:489–502. doi: 10.1023/B:PLAN.0000038257.93381.05. [DOI] [PubMed] [Google Scholar]
- 35.Taylor L.P., Hepler P.K. Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. (1997);48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- 36.Telfer A., Poethig R.S. HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development (Cambridge, England) (1998);125:1889–1898. doi: 10.1242/dev.125.10.1889. [DOI] [PubMed] [Google Scholar]
- 37.Verelst W., Saedler H., Munster T. MIKC* MADSprotein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. (2007);143:447–460. doi: 10.1104/pp.106.089805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verslues P.E., Guo Y., Dong C.H., Ma W., Zhu J.K. Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J. (2006);47:776–787. doi: 10.1111/j.1365-313X.2006.02833.x. [DOI] [PubMed] [Google Scholar]


