Abstract
MCRS2 is an oncoprotein that is sequestered in the nucleolus. When in the nucleolus, it promotes the transcription of the rRNA gene. MCRS2 also brings proteins into the nucleolus to change their function. This study analyzes the sequence of MCRS2 and determines that the nuclear localization signal, which has the sequence KRKK, is situated between amino acids 66 and 69. Meanwhile, MCRS2 contains a bipartite nucleolar localization signal, which comprises a KKSK motif, located between amino acids 133 and 136, and a downstream 152-amino acid region, from amino acid 314 to 465. The results of this study are important to understand the function of MCRS2.
Keywords: MCRS2, nucleolus, nucleolar localization
INTRODUCTION
The nucleolus, which is the site of ribosome biogenesis and assembly, is one of the most prominent structures in the nucleus that can be observed under a light microscope (Boisvert et al., 2007; Emmott and Hiscox, 2009). The nucleolus not only regulates cell cycle and cell growth but also is involved in the response to stress (Boisvert et al., 2007). Earlier studies have established that the transport of a protein across the nuclear membrane into the nucleus involves a nuclear localization signal (NLS), which is usually abundant in basic amino acids, and dedicated transport systems that interact with NLS and carry the protein into the nucleus (Boulikas, 1993). Unlike the nucleus, the nucleolus does not have a membrane, and the nucleolar localization of proteins appears to involve mechanisms that differ from that of nuclear transport (Emmott and Hiscox, 2009; Pederson and Tsai, 2009). The sequences on which nucleolar localization in many nucleolar proteins depends, nucleolus localization signals (NoLS), have been determined. Although NoLSs often contain clusters of basic amino acids, they commonly vary in context and in length (Emmott and Hiscox, 2009). Scott et al. (2010) analyzed NoLSs from 46 proteins and demonstrated that the length of the signals varies from 6 to 45 amino acids. The percentage of basic amino acids in the signals also varies. For example, the NoLS in promyelocytic leukemia protein (PML), which has 13 amino acids, contains only two basic amino acids (Condemine et al., 2007), although many NoLSs contain more than 50% basic amino acids (Scott et al., 2010). In some cases, a protein may contain a bipartite or tripartite NoLS sequence. For instance, the NoLSs of herpesvirus saimiri (HVS) ORF57 and parafibromin are bipartite and tripartite, respectively (Boyne and Whitehouse, 2006; Hahn and Marsh, 2007). Additionally, nucleolar trafficking of a protein is likely caused by the retention by rDNA, rRNA, or other nucleolar proteins (Carmo-Fonseca et al., 2000).
Microspherule protein 58 kDa (MSP58), reportedly an oncoprotein, and its two isoform proteins, MCRS1 and MCRS2, all c olocalize with microspherules in the nucleolus and seem to have similar functions (Bader et al., 2001; Okumura et al., 2005; Ren et al., 1998). An earlier investigation found that MSP58 interacts with Daxx and brings it to the nucleolus, relieving its repression, and thereby activating the transcription of the glucocorticoid receptor gene (Lin and Shih, 2002). Another protein that is carried by MSP58 to the nucleolus is FMRP Iso6, and the interaction between the protein and MSP58 can directly participate in the translocation of FMRP-containing mRNP to the nucleolus (Davidovic et al., 2006). Furthermore, when presents in the nucleolus, MCRS2 interacts with UBF, Mi-2β, and RET Finger Protein (RFP) to activate the promoter of the rRNA gene (Shimono et al., 2005). This investigation determines the sequences in MCRS2 that are required for nuclear translocation and nucleolar targeting.
MATERIALS AND METHODS
Cell lines
293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) that was supplemented with 10% fetal calf serum.
Plasmids
A DNA fragment that encodes MCRS2 was amplified by PCR using primers 5′-CCGGAATTCATGACACGTGGCACCG-3′ and 5′-CCGCTCGAGTCACTGTGGTGTGATCTTG-3′; pCMV6-XL (OriGene) was used as a template. Plasmid pEGFP-MCRS2 was constructed by inserting a PCR fragment that contains the entire MCRS2 gene sequence into the EcoRI-SalI sites in pEGFP-C2 (BD Clontech). Plasmids that expressed deleted GFP-MCRS2, including GFP-N108, -N257, -N369, -N465, -N455, -C375/475, -C255/475 and -C109/475, which contain the MCRS2 regions from amino acids 1 to 108, 257, 369, 465, 455, and 375 to 475, 255 to 475 and 109 to 475, respectively, were constructed by inserting a DNA fragment that was amplified by PCR into pEGFP-C2. Internal deletions in the MCRS2 genes were generated by inverse PCR using pEGFP-MCRS2 as a template. Plasmids that encoded MCRS2 with alanine substitutions were also formed by inverse PCR using primers that contain mutated sequences.
Indirect immunofluorescence analysis
The 293T cells were transfected with a plasmid that expresses GFP-MCRS2 or its deletion derivatives using Lipofectamine 2000 (Invitrogen). The cells were grown on glass coverslips in 6-well tissue culture plates. At 18 h after transfection, the cells were fixed with 4% paraformaldehyde in PBS at 4℃ for 30 min. They were then incubated with anti-fibrillarin monoclonal antibody for 1 h, followed by Alexa Fluor 594-conjugated goat antimouse IgG polyclonal antibody (Molecular Probes). Then the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 15 min and finally washed in PBS, mounted in Citifluor (Agar Scientific) and observed under a confocal laser-scanning microscope (model LSM 510 META; Zeiss) (Lee et al., 2008).
RESULTS
Deletion analysis of MCRS2
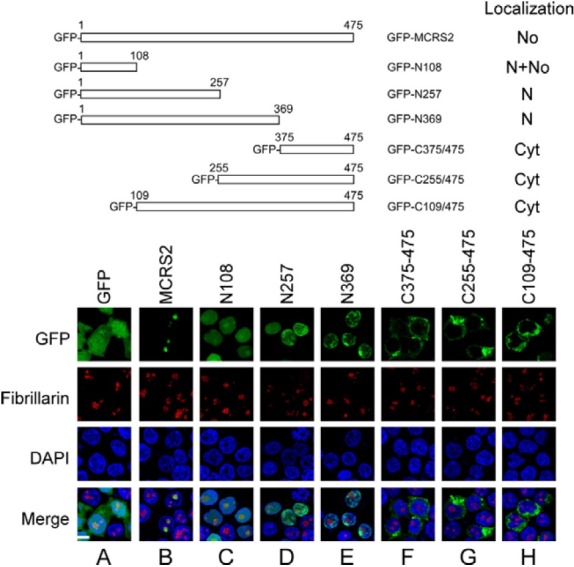
The purpose of this investigation was to determine the sequence in MCRS2 on which nuclear translocation depends. To achieve this goal, MCRS2 was deleted from the N-terminal or C-terminal end. The 293T cells were then transfected with plasmids that expressed these mutant proteins. GFP-MCRS2 and GFP were expressed in 293T cells, which were then used as positive and negative controls, respectively. Analyzing these proteins under a confocal microscope revealed that although GFP that was expressed from pEGFP-C2 was present in both the nucleus and the cytoplasm (Fig. 1A), GFP-MCRS2 was present in the nucleolus (Fig. 1B), verifying that MCRS2 is a nucleolar protein. This study also found that deleting the Cterminal region in MCRS2 from amino acids 109 (GFP-N108), 258 (GFP-N257), and 370 (GFP-N369) did not prevent the protein from entering the nucleus (Figs. 1C-1E), indicating that the N-terminal 108-amino acid region contains an NLS. Deletions were also created in the N-terminal region in MCRS2. These mutants, i.e. GFP-C375/475, GFP-C255/475, and GFPC109/ 475, were present in the cytoplasm but absent from the nucleus (Figs. 1F-1H). These results suggest that nuclear translocation depends on the N-terminal 108-amino acid region. Notably, GFP-N369, although present in the nucleus, was absent from the nucleolus (Fig. 1E), revealing that the region from amino acid 370 to 475 is required for the nucleolar localization of MCRS2.
Fig. 1. Map and distribution of MCRS2 mutants. 293T cells were transfected with plasmids that express GFP, GFP-MCRS2, GFPN108, GFP-N257, GFP-N369, GFP-C375/ 475, GFP-C255/475, and GFP-C109/475. At 18 h after transfection, proteins in the cell were stained with anti-fibrillarin antibody and Alexa 594-conjugated goat anti-mouse IgG antibody to reveal the nucleolus (No). The nucleus (N) was stained using DAPI. Cyt, cytoplasm; bar, 10 micrometer.
NLS in MCRS2
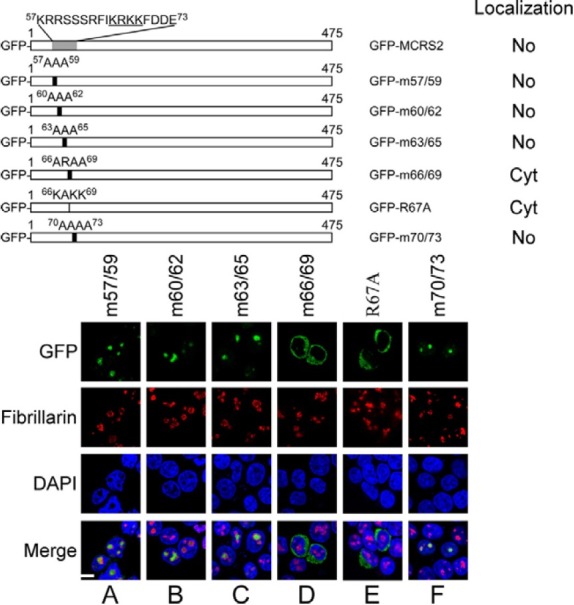
An earlier investigation predicted that the NLS in MSP58 is located between amino acids 113 and 123 (Davidovic et al., 2006). This region is present in MCRS2 between amino acids 126 and 136. However, this prediction is inconsistent with the results of this study, which reveals that the NLS is present in the N-terminal 108-amino acid region in MCRS2 (Fig. 1C). Therefore, this study analyzed the sequence of MCRS2 and found that the region between amino acids 66 and 69 in MCRS2 contained a KRKK sequence, which resembles an NLS (Hsu et al., 2005; Kalderon et al., 1984). Accordingly, the KRKK sequence was mutated and the way in which the mutation affected the localization of MCRS2 was studied. Confocal microscopic work revealed that GFP-m57/59, GFP- m60/62, and and GFP-m63/65, in which the amino acids between amino acids 57 and 65 were substituted with alanines, were present in the nucleolus (Figs. 2A-2C), showing that replacing the amino acids in this region did not influence protein localization. However, MCRS2 did not enter the nucleus when the three lysines at amino acid positions 66, 68, and 69 were substituted for alanines (GFP-m66/69) (Fig. 2D). Replacing Arg-67 with alanine (GFP-R67A) had a similar effect (Fig. 2E), indicating the importance of the KRKK sequence to nuclear translocation. Mutating the sequence between amino acids 70 and 73 (GFPm70/ 73) from FDDE to AAAA did not alter the ability of the protein to target the nucleolus (Fig. 2F). These results reveal that the KRKK sequence is an NLS of MCRS2.
Fig. 2. Identifying NLS in MCRS2. Sequence between amino acids 57 and 73 was mutated by substituting these amino acids for alanines. At 18 h after transfection, proteins in the cells were stained with antifibrillarin antibody and Alexa 594-conjugated goat antimouse IgG antibody to reveal the nucleolus (No). The nucleus was stained using DAPI. Cyt, cytoplasm; bar, 10 micrometer.
Analyzing the regions in MCRS2 that influence nucleolar localization
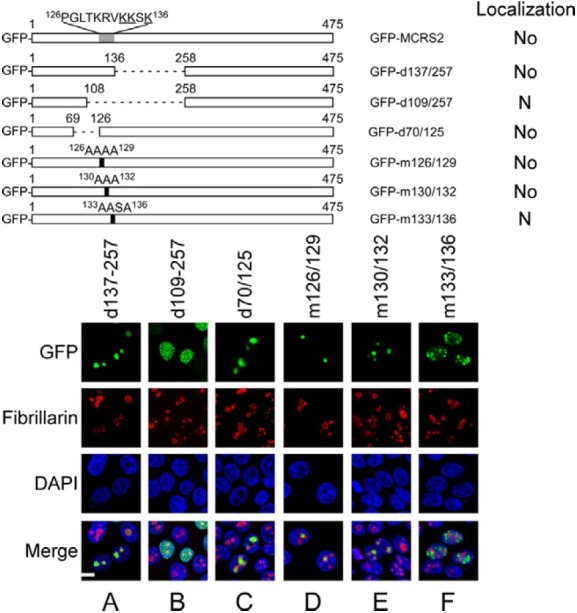
An earlier study predicted that the sequence between amino acids 126 and 136 of MCRS2 contains an NLS (Davidovic et al., 2006). Therefore, in this study, the region that contains this sequence was deleted to examine how the deletion affects the distribution of MCRS2 in the cell. This study found that GFPMCRS2 entered the nucleus but was not sequestered in the nucleolus after the region between amino acids 109 and 257 had been deleted (GFP-d109/257) (Fig. 3B), revealing that the region contains an NoLS rather than an NLS. Furthermore, deleting the regions from amino acid 137 to 257 (GFP-d137/ 257) and 70 to 125 (GFP-d70/125) influenced neither nuclear translocation nor nucleolar sequestration (Figs. 3A and 3C), indicating that the sequence that is important to nucleolar targeting is located between amino acid 126 and 136. Changing the sequence from amino acid 126 to 129 (GFP-m126/129) and 130 to 132 (GFP-m130/132) did not have an impact on nucleolar localization (Figs. 3D and 3E), showing that an NoLS is present in the region between amino acids 133 and 136. This study further mutated Lys-133, Lys-134, and Lys-136 to alanines (GFP-m133/136). This mutant protein was found in the nucleus rather than the nucleolus (Fig. 3F), demonstrating that Lys-133, Lys-134, and Lys-136 are important to the nucleolar localization of MCRS2.
Fig. 3. Analysis of region between amino acids 126 and 136. The regions from amino acids 137 to 257, 109 to 257 and 70 to 125 in GFP-MCRS2 were deleted (GFP-d137/257, GFP-d109/257 and GFP-d70/125). The sequence between 126 and 136 was substituted for alanines (GFP-m126/129, GFP-m130/ 132 and GFPm133/ 136). At 18 h after transfection, proteins in the cell were stained with anti-fibrillarin antibody and Alexa 594-conjugated goat anti-mouse IgG antibody to reveal the nucleolus (No). The nucleus (N) was stained using DAPI. Bar, 10 micrometer.
C-terminal deletions and nucleolar localization of MCRS2
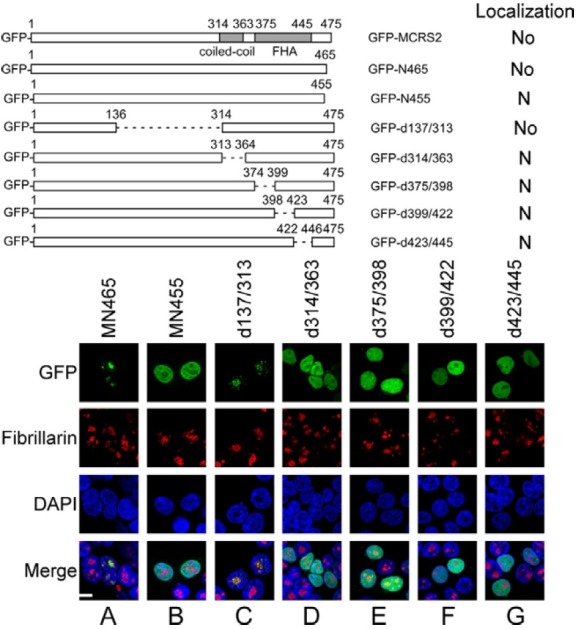
This investigation finds that GFP-N369, although present in the nucleus, is not present in the nucleolus (Fig. 1E), revealing that the C-terminal region in MCRS2 is also important to nucleolar targeting. Therefore, this region was analyzed to identify the sequence that governs nucleolar localization. Deleting the region from amino acid 466 to 475 (GFP-N465) did not alter the distribution of the protein in the cell (Fig. 4A). However, a mutant protein with a deletion from amino acid 456 to 475 (GFPN455) was present in the nucleus but not in the nucleolus (Fig. 4B). Moreover, although a deletion from amino acid 137 to 313 (GFP-d137/313) did not affect the localization of the protein (Fig. 4C), deleting the region from amino acids 314 to 363 (GFPd314/ 363), which is the coiled-coil domain in MCRS2, from the protein prevented it from being able to target the nucleolus (Fig. 4D). In this study, three mutants, in which the FHA domain was deleted, i.e. GFP-d375/398, GFP-d399/422 and GFP-d423/445, were also generated. These three proteins were found in the nucleus but not in the nucleolus (Figs. 4E-4G), indicating the importance of the FHA domain. These results indicate that the sequence in MCRS2 from amino acid 314 to 465 is important to nucleolar localization.
Fig. 4. Delineating C-terminal NoLS sequence in MCRS2. Deletions were generated in the C-terminal and middle regions in MCRS2. At 18 h after transfection, proteins in the cell were stained with anti-fibrillarin antibody and Alexa 594-conjugated goat antimouse IgG antibody to reveal the nucleolus (No). Nucleus (N) was stained using DAPI. Coiled-coil, coiled-coil domain; FHA, fork head-associated domain. Bar, 10 micrometer.
DISCUSSION
MCRS2 is a nucleolar protein, which often interacts with other proteins to influence their localization in a cell, as well as their biological functions. Our unpublished work finds that the Rta of Epstein-Barr virus (EBV), which is a transcription factor that activates the EBV lytic genes, interacts with MCRS2, and that this interaction causes nucleolar sequestration of Rta (results not shown). Accordingly, mutations were generated according to the NLS and NoLS in MSP58 that were reported by Davidovic et al. (2006) to investigate how MCRS2 affects the function of Rta. However, mutating these sequences did not yield the expected results, prompting an investigation of how MCRS2 enters the nucleus and localizes in the nucleolus. The present study finds that MCRS2 contains a typical NLS sequence, KRKK, from amino acid 66 to 69 and shows that mutating this sequence eliminates the ability of the protein to enter the nucleus (Figs. 2D and 2E). This study also finds that MCRS2 contains a bipartite NoLS from amino acids 133 to 136 and 314 to 465; mutating either sequence destroys the capacity of MCRS2 to localize in the nucleolus (Figs. 3 and 4). A bipartite NoLS is also present in the HVS ORF57 (Boyne and Whitehouse, 2006), in which the upstream bipartite sequence is KRPR; the sequence between amino acids 133 and 136 in MCRS2 has a similar length and is also rich in the basic amino acid, KKSK (Fig. 3). However, the downstream NoLS sequence in ORF57 has a length of only ten amino acids, whereas the sequence in MCRS2 is 152 amino acids long, from amino acid 314 to 465 (Fig. 4). In fact, MCRS2 contains a coiled-coil domain and a fork head-associated (FHA) domain in this region (Fig. 4) (Davidovic et al., 2006). This investigation finds that deleting the coiled-coil domain from GFP-MCRS2 yields a mutant protein, GFP-d314/363, which is present only in the nucleus but not in the nucleolus (Fig. 4D), indicating the importance of the domain to nucleolar localization. Furthermore, the amino acids in the FHA domain, which covers the region from amino acid 375 to 445, were mutated in triplets with alanines. With the exceptions of RATK, PAW and EGR sequences from amino acids 380 to 383, 396 to 398, and 420 to 422, respectively, other mutations prevents the GFP-MCRS2 from being able to localize in the nucleolus, indicating that the FHA domain is also important to the tethering of MCRS2 to the nucleolus (data not shown). Furthermore, the sequence between the coiled-coil and FHA domain, from amino acid 364 to 374, is important because the protein does not target the nucleolus after the sequence has mutated (data not shown). Accordingly, the region that contains these domains in MCRS2 and the KKSK sequence from amino acid 133 to 136 are required for the sequestration of MCRS2 to the nucleolus. Notably, GFPN108 is present in both the nucleus and the nucleolus (Fig. 1C), probably owing to the nonspecific binding to the nucleolus, since GPF-257 and GFP-369 are present only in the nucleus rather than the nucleolus. This investigation identifies the sequences in MCRS2 that are important to nuclear and nucleolar translocation. This information will be important to understanding the functions of MCRS2.
Acknowledgments
This study was supported by the National Science Council of the Republic of China (NSC99-3112-B-185-005), National Health Research Institute of the Republic of China (NHRI-EX96-9417BI) and Chang-Gung Memorial Hospital Grant (CMRPD-160113).
References
- 1.Bader A.G., Schneider M.L., Bister K., Hartl M. TOJ3, a target of the v-Jun transcription factor, encodes a protein with transforming activity related to human microspherule protein 1 (MCRS1). Oncogene. (2001);20:7524–7535. doi: 10.1038/sj.onc.1204938. [DOI] [PubMed] [Google Scholar]
- 2.Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. (2007);8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 3.Boulikas T. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. (1993);3:193–227. [PubMed] [Google Scholar]
- 4.Boyne J.R., Whitehouse A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA. (2006);103:15190–15195. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmo-Fonseca M., Mendes-Soares L., Campos I. To be or not to be in the nucleolus. Nat. Cell Biol. (2000);2:E107–112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 6.Condemine W., Takahashi Y., Le Bras M., de The H. A nucleolar targeting signal in PML-I addresses PML to nucleolar caps in stressed or senescent cells. J. Cell Sci. (2007);120:3219–3227. doi: 10.1242/jcs.007492. [DOI] [PubMed] [Google Scholar]
- 7.Davidovic L., Bechara E., Gravel M., Jaglin X.H., Tremblay S., Sik A., Bardoni B., Khandjian E.W. The nuclear microspherule protein 58 is a novel RNA-binding protein that interacts with fragile X mental retardation protein in polyribosomal mRNPs from neurons. Hum. Mol. Genet. (2006);15:1525–1538. doi: 10.1093/hmg/ddl074. [DOI] [PubMed] [Google Scholar]
- 8.Emmott E., Hiscox J.A. Nucleolar targeting: the hub of the matter. EMBO Rep. (2009);10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn M.A., Marsh D.J. Nucleolar localization of parafibromin is mediated by three nucleolar localization signals. FEBS Lett. (2007);581:5070–5074. doi: 10.1016/j.febslet.2007.09.050. [DOI] [PubMed] [Google Scholar]
- 10.Hsu T.Y., Chang Y., Wang P.W., Liu M.Y., Chen M.R., Chen J.Y., Tsai C.H. Reactivation of Epstein-Barr virus can be triggered by an Rta protein mutated at the nuclear localization signal. J. Gen. Virol. (2005);86:317–322. doi: 10.1099/vir.0.80556-0. [DOI] [PubMed] [Google Scholar]
- 11.Kalderon D., Roberts B.L., Richardson W.D., Smith A.E. A short amino acid sequence able to specify nuclear location. Cell. (1984);39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y.H., Chiu Y.F., Wang W.H., Chang L.K., Liu S.T. Activation of the ERK signal transduction pathway by Epstein-Barr virus immediate-early protein Rta. J. Gen. Virol. (2008);89:2437–2446. doi: 10.1099/vir.0.2008/003897-0. [DOI] [PubMed] [Google Scholar]
- 13.Lin D.Y., Shih H.M. Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J. Biol. Chem. (2002);277:25446–25456. doi: 10.1074/jbc.M200633200. [DOI] [PubMed] [Google Scholar]
- 14.Okumura K., Zhao M., Depinho R.A., Furnari F.B., Cavenee W.K. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proc. Natl. Acad. Sci. USA. (2005);102:2703–2706. doi: 10.1073/pnas.0409370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pederson T., Tsai R.Y. In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. (2009);184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren Y., Busch R.K., Perlaky L., Busch H. The 58-kDa microspherule protein (MSP58), a nucleolar protein, interacts with nucleolar protein p120. Eur. J. Biochem. (1998);253:734–742. doi: 10.1046/j.1432-1327.1998.2530734.x. [DOI] [PubMed] [Google Scholar]
- 17.Scott M.S., Boisvert F.M., McDowall M.D., Lamond A.I., Barton G.J. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. (2010);38:7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimono K., Shimono Y., Shimokata K., Ishiguro N., Takahashi M. Microspherule protein 1, Mi-2beta, and RET finger protein associate in the nucleolus and up-regulate ribosomal gene transcription. J. Biol. Chem. (2005);280:39436–39447. doi: 10.1074/jbc.M507356200. [DOI] [PubMed] [Google Scholar]
- 19.Song H., Li Y., Chen G., Xing Z., Zhao J., Yokoyama K.K., Li T., Zhao M. Human MCRS2, a cell-cycle-dependent protein, associates with LPTS/PinX1 and reduces the telomere length. Biochem. Biophys. Res. Commun. (2004);316:1116–1123. doi: 10.1016/j.bbrc.2004.02.166. [DOI] [PubMed] [Google Scholar]


