Abstract
Two strains of necrotrophic Alternaria brassicicola, Ab40857 and Ab42464, are virulent on Korean cabbage and several wild types of Arabidopsis thaliana. Interaction between Ab42464 and Col-0 was compatible, whereas interaction between Ab40857 and Col-0 was incompatible. The loss of defense, no death (dnd) 1 function abrogated the compatibility between Ab42464 and Col-0, and the accelerated cell death (acd) 2 mutation attenuated the Col-0’s resistance against Ab40857. These two fungal strains induced PR1 transcription in Col-0. Ab40857 accelerated transcription of PDF1.2, THI2.1, CAT, and POX by 12 h compared to those challenged with Ab42464. More abundant cell death was observed in Col-0 infected with Ab42464, however, callose deposition was evident in the incompatible interaction. Remarkably, Ab40857-infected areas of acd2-2 underwent rampant cell death and Ab42464 triggered callose production in dnd1-1. Furthermore, the incompatibility between Ab40857 and Col-0 was nullified by the coronatine- insensitive 1 (coi1) and phytoalexin-deficient 3 (pad3) mutations but not by nonexpresser of PR genes (npr1) and pad4. Ab40857 induced abundant cell death in pad3. Taken together, cell death during the early infection stage is a key determinant that discriminates between a compatible interaction and an incompatible one, and the resistance within Col-0 against Ab40857 is dependent on a defensesignaling pathway mediated by jasmonic acid and PAD3.
Keywords: necrotroph, programmed cell death
INTRODUCTION
Programmed cell death (PCD) is ubiquitous in all organisms and is under the regulation of highly evolved intracellular signaling networks. One of the typical examples of PCD in plant is the hypersensitive response (HR) induced by pathogen invasion (Flor, 1971; Ntoukakis et al., 2009; Rusterucci et al., 2001; Staskawicz et al., 1995). After recognition of its host, a pathogen secretes virulence factors into the intercellular space (apoplast). The host perceives this pathogen-associated molecular pattern (PAMP) signal through pattern recognition receptors (PRRs). In a compatible interaction, pathogens abrogate PAMP-triggered immunity (PTI) on behalf of the effector(s)’s function and this interference results in effector-triggered susceptibility (ETS). In the presence of a complete set of resistance (R) machinery that matches specifically with the effector(s), the indirect recognition of the interaction between the effector and its host target R machinery activates intracellular defense signaling pathways and is often culminated in a host HR. Host cell death in the early infection stage is fatal for the biotrophic pathogen’s propagation in planta as the pathogen of this class is unable to obtain nutrients from dead cells. This is the core of the initial phase of plant immunity limiting the pathogen within and around the infection area. There has been a substantial amount of investigations elucidating the mode of interactions among the effectors and PAMPs from microbes and their recognitions in Arabidopsis thaliana (Jones and Dangl, 2006; Nishimura and Dangl, 2010).
A huge amount of genetic resources are available in Arabidopsis. In addition, the publicly available sequence information following the completion of the genome sequencing projects for Arabidopsis and Alternaria brassicicola (http://www.genome.wustl.edu/genome.cgi?GENOME=Alternaria%20bras-sicicola) make this combination ideal for mining potential candidates of Arabidopsis genes or signaling pathways that may be useful for the breeding of resistance against A. brassicicola infection in Brassica species (Lawrence et al., 2008). Opportunely, the updated genome sequencing results for Brassica rapa subsp. pekinensis predict a high degree of sequence similarity among Arabidopsis genes and their orthologs in Korean cabbage (Hong et al., 2006). Several works have already been undertaken to analyze the incompatible interaction between Alternaria brassicicola and Arabidopsis wild types and to explain the breakdown of this incompatibility in several mutants that lack of specific defense signaling pathways (Kagan and Hammerschmidt, 2002; Mengiste et al., 2003; Oh et al., 2005; Thomma et al., 1998; 2000).
A. brassicicola is a necrotrophic pathogen (Glazebrook, 2005; Sellam et al., 2006; Trusov et al., 2006; Zheng et al., 2006). In contrast with the biotrophic pathogens described above, necrotrophs produce and secrete PAMPs and effector(s) to kill its host’s cells during the host infection processes. They use resulting host cell leakage as a nutrient source for further propagation and subsequent disease progression in a compatible host (Cramer et al., 2006; Thomma, 2003; Van Baarlen et al., 2004; van Kan, 2006). Representative Alternaria species including A. alternata and A. brassicae produce hostspecific toxins that define their host range (Thomma, 2003). A. brassicicola also secretes cutinase/cutinolytic enzymes and lipases that are thought to be virulence factors responsible for specific interaction with its hosts (Berto et al., 1999). Up to now, several large-scale expression studies have been performed in the interaction between A. brassicicola strain MUCL20297 and its susceptible host, Brassica oleracea, and between MUCL- 20297 and resistant Arabidopsis ecotypes including Col-0 (Cramer and Lawrence, 2004; Cramer et al., 2006; Mukherjee et al., 2009). These results suggested a relationship between Col-0’s resistance and the primed state within Col-0 against MUCL20297 infection on the basis of the transcriptional patterns of MONOOXYGENASE (MO) 1.
The “defense, no death” (dnd) 1 mutant of Arabidopsis exhibits resistance against necrotrophic Botrytis cinerea (Govrin and Levine, 2002), and “accelerated cell death” (acd) 2 is resistant to biotrophic pathogens such as Pseudomonas syringae pv. maculicola due to the lack of red chlorophyll catabolite reductase activity (Greenberg et al., 1994). Altered PCD regulation by these mutations is able to change the host-pathogen interactions. Infiltration of the intercellular fluid from B. cinerea-infected Arabidopsis plants provoked more rapid and actively enlarged necrosis in acd2 (Asai et al., 2000; Govrin et al., 2006). However, the same treatment did not trigger observable alterations in dnd1. Also, acd1 is also more susceptible to Botrytis elliptica infection than its wild type parent, Col-0 (Van Baarlen et al., 2004).
Camalexin is a representative indolic phytoalexin of A. thaliana and is synthesized from indole-3-acetaldoxime (Glawischnig et al., 2004; Zhao and Last, 1996). “Phytoalexin-deficient” (pad) mutants are susceptible to biotrophic and-/or necrotrophic pathogens (Berrocal-Lobo and Molina, 2004; Glazebrook et al., 1996; 1997). A deficiency of the phytoalexin, camalexin, and insensitivity to jasmonate have resulted in the attenuation of resistance of A. thaliana against A. brassicicola (Thomma et al., 1998; 1999). Similar to these results, a comparative analysis using 24 Arabidopsis wild types indicated that there was a close relationship between the resistance against A. brassicicola and camalexin production (Kagan and Hammerschmidt, 2002).
The investigation of novel functions of active oxygen species (AOS) have revealed that, in addition to arresting pathogen proliferation in planta, AOS are involved in cell wall reinforcement (Olivain et al., 2003) and callose deposition (Huckelhoven et al., 1999). Moreover, they are able to act as signaling molecules (Alvarez et al., 1998; Bolwell et al., 1995; 1998; Chamnongpol et al., 1998; Rhee et al., 2010; Tenhaken et al., 1995; Wojtaszek, 1997).
The aims of the research presented here was to analyze Col-0 responses, including cell death, during compatible and incompatible interactions to unravel which defense signaling pathway(s) and/or factor(s) play pivotal role(s) in each interaction. To achieve this goal, we screened and selected two A. brassicicola strains that are virulent and avirulent on Col-0 and investigated the cellular/molecular responses triggered by challenges with A. brassicicola in wild type and its several mutants that lack of specific defense signaling or normal cell death regulation.
MATERIALS AND METHODS
Plants and pathogen challenge
F1 seeds of Brassica rapa subsp. pekinensis cultivar Samjin were purchased from Seminis Korea (Seoul, Korea). The 60 Arabidopsis thaliana accessions used in this study were; Aa-0, Bch-1, Br-0, C24, Chi-0, Co-1, Col-0, Cvi-0, Di-0, En-6, Gr-1, Ka-0, Kas-1, Kin-0, Lan-0, Laud-1, Li-1, Lind-1, Linnport, LIN, Lip-0, Litva, Ma-2, Mh-0, Ms-0, Na-1, Nd-0, No-0, Nok-0, Nw-0, Oy-0, Petergof, Pf-0, Pi-0, Pla-0, Pn-0, Pog-0, Pu-2-23, Pt-0, Rennes-1, Renk-1, Rome-1, Rou-0, Ri-0, Sei-0, Sf-1, Sh-0, Sha, Stw-0, Tsu-0, Tu-0, Vi-0, Vind-1, Wag1, Wc-0, Wei-1, and Ws-0. The six mutants and one transgenic plant derived from Col-0 and characterized for altered defense responses were “accelerated cell death” (acd) 2-2, “defense, no death” (dnd) 1-1, “coronatine-insensitive” (coi) 1, “nonexpresser of PR genes” (npr) 1, “phytoalexin-deficient” (pad) 3, pad4, and transgenic nahG that expresses the bacterial NahG gene. All of the genotypes were obtained from The Arabidopsis Biological Resource Center (ABRC). In particular, homozygous coi1 was selected through PCR and XcmI digestion and used for the determination of defense signaling pathways (Xie et al., 1998). Plants were grown in a growth chamber at 22℃ and 60-65% relative humidity, with 16 h of illumination daily. Arabidopsis and cabbage of 5- and 8-week-old were used for fungal inoculation, respectively.
Six strains of Alternaria brassicicola, YSH-1, Ab40034, Ab- 40036, Ab40857, Ab42464, and Ab42465, were obtained from the Korean Agricultural Culture Collection, Rural Development Administration, Korea. Fungal strains were grown on oatmeal agar (50 gram oatmeal and 25 gram Bacto agar per 1 liter distilled water) under continuous fluorescent light for 7 days at 22℃. Fungal conidia were retrieved from the medium with sterilized distilled water and 250 μg ml-1 Tween 20 and the concentration was adjusted to 1 × 105 conidia ml-1. Inoculum was sprayed until all of the leaves were covered with fine droplets. Alternatively, 5 μl-drop of spore suspension was placed on the rosette leaves. The inoculated plants were kept in a dew chamber for 16 h at 25℃ with 100% relative humidity and then transferred to a growth chamber. Fungal growth was assessed by staining the infected leaves with 2.5 mg ml-1 aniline blue resuspended in lactophenol (equal volumes of glycerol, lactic acid, and phenol) for an hour. Disease progressions were determined either by estimating water-soaked or chlorotic lesions surrounded with a yellow halo or by counting the conidia formed on each lesion at 5 days after inoculation.
Cell death, hydrogen peroxide, and callose
To investigate the hydrogen peroxide accumulation and cell death in the Arabidopsis wild type Col-0 and its mutants dnd1-1, acd2-2, pad3, and pad4, A. brassicicola-inoculated leaves were retrieved and observed microscopically. Cell death was detected by propidium iodide staining. Inoculated leaves were harvested 12 h post inoculation (hpi) and stained with 100 μg ml-1 propidium iodide (PI; Sigma, USA) resuspended in 1× phosphate buffered saline (PBS) or 2.5 mg ml-1 lactophenoltrypan blue (TB; Sigma, USA). Infected tissues were incubated in PI solution for 15 min at room temperature. The PI fluorescence at 550 nm was analyzed microscopically (Zeiss Axioplan 2). Due to the loss of normal membrane function, PI passes through the membrane and binds with the nucleic acids within dead cells. Tissues were incubated in TB solution for 1 min at 100℃ and then 16 h at room temperature. Excessive TB bound to the leaf tissues was removed using chloral hydrate as described previously (Ton et al., 2005). The histochemical detection of hydrogen peroxide and callose in the inoculated rosette leaves harvested at 12 and 24 hpi was performed according to established methods (Ahn et al., 2007). The quantitative determination of cell death, hydrogen peroxide, and callose was done using inoculated rosette leaves retrieved at 12, 24, and 24 hpi (Ahn et al., 2007).
RNA isolation and expression analyses
Total RNA was prepared using the lithium chloride precipitation method (Davis and Ausubel, 1989). Hybridization analysis was performed as described (Ahn et al., 2005b). The genes used in this study were amplified by reverse transcriptase-polymerase chain reaction (RT-PCR) using total RNA of Ab40857-inoculated Col-0 leaves harvested at 2 dpi and PR1-, PDF1.2- and THI2.1-specific primer sets (Vieira Dos Santos et al., 2003). Primer sets of POX (At1g71695) and CAT (At1g20620), encoding PEROXIDASE 12 and CATALASE 3, were designed on the basis of mRNA sequences (Schenk et al., 2003) (forward and reverse primers of POX, 5′-AGTGCGGTCAAGTCGTCTCT-3′ and 5′-AAAAACAGCTGCTGGTCGAT-3′; forward and reverse primers of CAT, 5′-TCACATGGTGCTGGATGTTT-3′ and 5′- GTTGTGGTGAGCACATTTGG-3′). First strand synthesis and amplification were done using Reverse-iT First Strand Synthesis Kit as indicated by the manufacturer’s instructions (AB gene, UK). The DNA probes were labeled with [32P]-dCTP by random primer labeling (Boehringer Mannheim, Germany).
Camalexin determination
Camalexin was extracted and measured as described by Glazebrook and Ausubel (Glazebrook and Ausubel, 1994) with some modification. Leaf samples (200 mg), pulverized using mortar and pestle and liquid nitrogen, were mixed with 700 μl of 80% methanol and kept at 80℃ for 20 min. Plant debris was removed by centrifugation at 10,000 × g for 5 min, and the methanol was evaporated under vacuum. The remaining debris was completely resuspended in 200 μl distilled water and extracted twice with the same volume of chloroform. Both chloroform extracts were combined and evaporated completely. The residue was dissolved in 15 μl of chloroform and applied to a silica thin-layer chromatography (TLC) plate (Silica gel 60, Germany) which was developed with chloroform:methanol (9:1, v/v). Camalexin was visualized by its blue fluorescence under a hand-held long-wave ultraviolet lamp (365 nm). The silica containing camalexin was retrieved from the plate and the camalexin was extracted into 1 ml of methanol. The emission of 385 nm after excitation at 315 nm was measured using a LC- 6AD HPLC (Shimadzu Corp., Japan) and RF-10A fluorescence detector (Shimadzu Corp., Japan) and the camalexin concentration was calculated by comparison with a standard curve obtained from purified camalexin.
RESULTS
Interactions between plants and Alternaria brassicicola
Among the six strains of A. brassicicola tested, Ab40034, Ab- 40036, Ab40857, and Ab42464 were highly virulent on Brassica rapa subsp. pekinensis cv. Samjin, but YSH-1 and Ab42465 were not virulent (Fig. 1A and data not shown). To investigate the compatibility and incompatibility between Cruciferaceae and A. brassicicola, interactions among 60 Arabidopsis wild types and Ab40857/Ab42464 were analyzed. Ab42464 infection progressed rapidly on C24, Col-0, Lind-1, Litva, Nd-0, No-0, Oy-0, Petergof, Sha, and Ws-0. Ab40857 also caused similar symptoms on Di-0, Kas-1, Litva, Nd-0, Oy-0, Petergof, Vind-1, and Ws-0. Conversely, Ab40857 did not induce observable alterations in Col-0. In the compatible interaction between Col-0 and Ab42464, small water-soaked infection spots appeared within 16-24 h post inoculation (hpi) and these symptoms further developed into large lesions within 36- 48 hpi (Fig. 1B). This result contrasts with the incompatible interaction between Col-0 and Ab40857 in which inoculated leaves remained unchanged. To confirm these results, conidial suspensions (2 × 104 ml-1) from the two strains were dropped onto Col- 0 rosette leaves, and the disease development was assessed (Fig. 1C). Ab42464 induced water-soaked lesions around the inoculated sites within 16 hpi and this symptom developed. The inoculated leaves were almost completely withered and dead 5 days post inoculation (dpi). In contrast, the areas around the sites inoculated with Ab40857 didn’t show evident symptom development on Col-0 leaves by 5 dpi, and only slight chlorosis was observed on the area where the inoculated area.
Fig. 1. Disease development on Korean cabbage (Brassica rapa subsp. pekinensis cv. Samjin) and Arabidopsis thaliana inoculated with Alternaria brassicicola strains Ab42464 and Ab408957. (A) Necrotic lesions on the leaves of 6-week-old Korean cabbage at seven days after sprayinoculation with conidia of A. brassicicola. (B) Disease development on the leaves of 4-week-old A. thaliana wild type Col-0 and its mutants dnd1-1 and acd2-2 24 and 72 h after spray-inoculation with A. brassicicola strain Ab42464 and Ab40857. hpi designates hours post inoculation. (C) Necrotic lesions on the leaves of Col-0 five days after drop-inoculation with conidia of A. brassicicola strain Ab42464 and Ab40857. Arrowheads indicate drop-inoculation sites. In (B, C), bars = 2 cm. All experiments were conducted more than three times with 5 replicates and almost identical tendencies were observed.
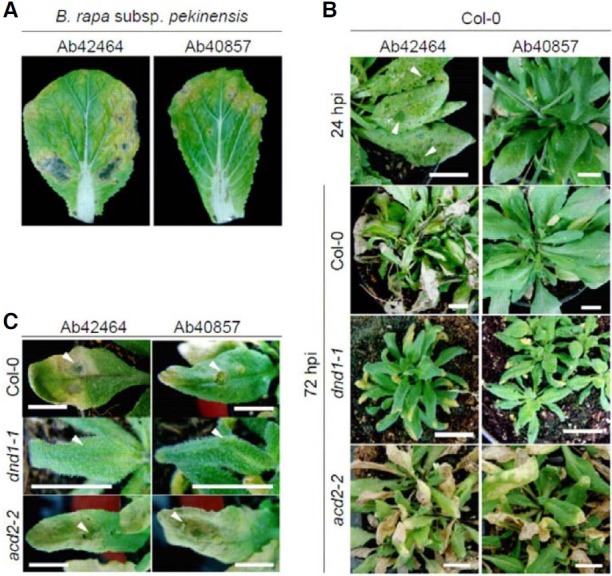
A. brassicicola is known as a necrotrophic pathogen and the above results indicated the positive relationship between cell death (water-soaked lesions) and disease progression. To evaluate the effect of cell death on compatibility and incompatibility, the interactions among Ab42464 or Ab40857 and defense, no death (dnd) 1-1 or accelerated cell death (acd) 2-2 were investigated. dnd1-1 was highly resistant to both strains, and a fungal spray challenge did not cause any alterations on the leaves in spite of the longer period of observation (Fig. 1B). acd2-2 was hyper susceptible to both strains; water-soaked lesions formed within 12 hpi and this symptom developed into spreading lesions within 24-36 hpi. Finally, the inoculated leaves turned yellow and wilted within 48 hpi. Disease development in the drop inoculation assays almost coincided with the above results (Fig. 1C).
The Defense-related gene expression in response to A. brassicicola
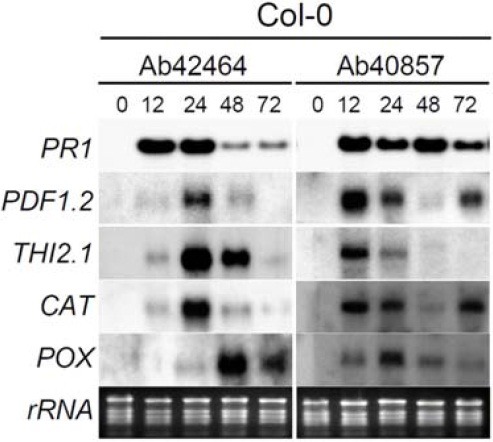
To compare the responses of Arabidopsis to Ab42464 and Ab40857 at the transcriptional level, five representative marker genes, PR1, PDF1.2, THI2.1, CAT (encoding CATALASE 3) and POX (encoding PEROXIDASE 12) were employed (Fig. 2). Previous studies showed that the transcription of these genes is induced by infection of pathogens, including A. brassicicola, or is related to oxidative stresses (Kishimoto et al., 2007; Liu et al., 2005; Manners et al., 1998; Norman-Setterbald et al., 2000; Seo et al., 2001; Vieira Dos Santos et al., 2003; Weigel et al., 2005; Yoshioka et al., 2001). PR1 transcription was induced by the two fungal strains within 12 hpi. In the compatible interaction between Col-0 and Ab42464, the transcription of PDF1.2, THI2.1, and CAT genes was also triggered within 12 hpi, reaching a maximum level at 24 hpi, and decreasing slightly thereafter. These patterns coincided with the formation of watersoaked lesions. POX transcription was also induced within 24 hpi, and peaked during 24-48 hpi. In Col-0 inoculated with Ab40857 (incompatible interaction), the transcription of PDF1.2, THI2.1, and CAT was also induced and peaked within 12 hpi. POX expression was also induced within 12 hpi and a maximum level of transcript was observed at 24 hpi.
Fig. 2. Transcript levels of defense-related genes in Arabidopsis Col-0 challenged with A. brassicicola. Five-week-old soil-grown wild-type Col-0 plants were inoculated with strains Ab42464 and Ab40857 and harvested after the indicated times. As controls, leaf samples were harvested just prior to inoculation (0 h). RNA blots were hybridized with the five probes indicated.
Cellular defense responses
Prior to penetration into the host cell, conidia of virulent Ab42464 and avirulent Ab40857 attached firmly to the surface of the plant leaf (Fig. 3A). More than 90% of the conidia germinated and formed an infection-specific structure, an appressorium, and then penetrated into host’s tissues. There were no distinctive differences between the susceptible and resistant interactions during the prepenetration development. The accumulation of hydrogen peroxide and callose deposition are common responses in incompatible interactions and are often culminated in hypersensitive responses (HRs). To investigate the cellular responses by the two strains, rosette leaves of Col-0, dnd1-1 and acd2-2 were challenged with the two fungal strains, and cell death, callose deposition, and hydrogen peroxide accumulation were assessed microscopically within and around the infection sites (Fig. 3A). The Ab42464 infection triggered robust cell death and H2O2 accumulation in Col-0; however, callose was not detectable. In contrast, avirulent Ab40857 did not induce cell death or H2O2 production, but callose deposition was evident. Inoculation with either pathogen did not trigger cell death or hydrogen peroxide accumulation in dnd1-1; however, callose deposition was constitutive and abundant. The same treatments triggered robust cell death and hydrogen peroxide production in acd2-2, yet callose deposition was barely observed and was not induced by either pathogen.
Fig. 3. Effects of A. brassicicola inoculation on the cellular defense responses in Arabidopsis wild type Col-0 , dnd1-1, and acd2-2. Arabidopsis was sprayed with a conidial suspension of A. brassicicola in 250 μg ml-1 T ween 2 0 or Tween 20 (mock) only. (A) Microscopic observation and quantification of hydrogen peroxide and callose deposition were performed at 12 h post inoculation (hpi). Cell death was analyzed on leaves harvested at 16 hpi. The brown color indicates hydrogen peroxide production. The presence of green fluorescence indicates callose deposition. Cell death was determined by the presence (dead) or absence (live) of red luminescence. Asterisks indicate cell death, callose deposition, and hydrogen peroxide accumulation. Bars = 50 μm. (B) Effects of A. brassicicola inoculation on cell death examined by staining with Evans blue. (C) Effects of A. brassicicola inoculation on callose deposition. (D) Effects of A. brassicicola inoculation on hydrogen peroxide accumulation. Data presented in (B, D) were taken in experiments performed more than three times. Bars with different letters indicate that the corresponding data are significantly different (Duncan’s multiple range test; P < 0.05).
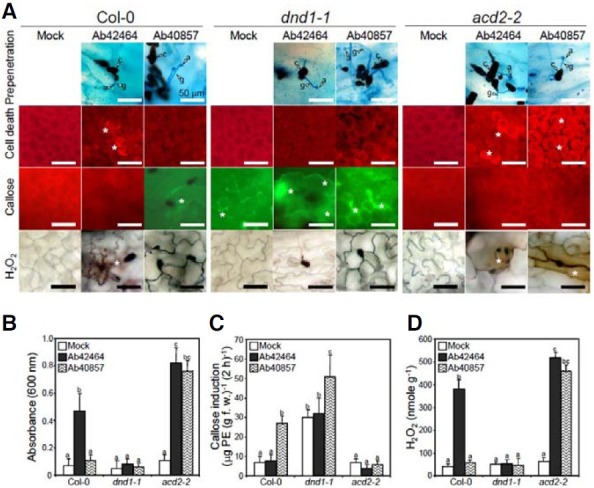
Moreover, quantitative analyses corresponded with the above observations. A spectrophotometric estimation of Evans blue bound to the dead cells indicated that cell death and H2O2 production in Col-0 infected with virulent Ab42464 were 5.8 and 7.2-fold higher, respectively, than those in the mock-treated control; however, the estimation also indicated that callose was not produced (Figs. 3B, 3C, and 3D). Callose deposition in Col- 0 challenged with incompatible Ab40857 was 3.4-fold higher. Both strains induced a high level of cell death and H2O2 in acd2-2. In addition to persistent callose deposition, avirulent strain infection increased the callose content up to 61% and 58%, as compared to those from mock-treated and virulent strain-inoculated dnd1-1, respectively, whereas neither strain induced callose deposition in acd2-2.
The effect of salicylic acid, jasmonic acid, and camalexin on the interactions
Camalexin production was assessed in wild-type Col-0 and its mutants dnd1-1 and acd2-2 (Fig. 4A). Both strains triggered camalexin production in Col-0; however, 2.7 times the amount of camalexin accumulated in the compatible interaction compared to that in the incompatible one. dnd1-1 was a higher camalexin producer than wild type and acd2-2 and the basal level was barely affected by the pathogen infection. In contrast, pathogen invasion up regulated the camalexin content in acd2-2 regardless of the strain.
Fig. 4. Camalexin production, disease development, and cell death progressions in Col-0 and defense-defective mutants. (A) The accumulation of camalexin 48 h after inoculation of Col-0, dnd1-1, and acd2-2 with A. brassicicola strains Ab42464 and Ab40857. Bars with different letters indicate that the corresponding data are significantly different (Duncan’s multiple range test; P < 0.05). The experiment was performed more than three times with similar results. (B) Disease development in Arabidopsis Col-0 wild type and mutants challenged with A. brassicicola strains Ab42464 and Ab40857. The upper panel shows the diameter of a lesion formed 5 days after A. brassicicola inoculation on 5-week-old Arabidopsis leaves. The data points represent the averages with standard errors of measurements of 50 lesions. The lower panel shows the de novo-formed conidia per lesion on the leaves inoculated with A. brassicicola. The data points represent averages with standard errors of measurements of 10 lesions. Bars with different letters indicate that the corresponding data are significantly different (Duncan’s multiple range test; P < 0.05). The experiment was performed more than three times with similar results. (C) Cell death during the early infection stage of an incompatible interaction. Leaves were harvested 12 h after challenge with Ab40857 and stained with trypan blue. Representative samples among 20 leaves from 10 plants are shown. (D) Quantitative analyses of cell death. Samples were harvested as in (C). The data represent averages with standard errors of 5 measurements from 10 plants. Bars with different letters indicate that the corresponding data are significantly different (Duncan’s multiple range test; P < 0.05).
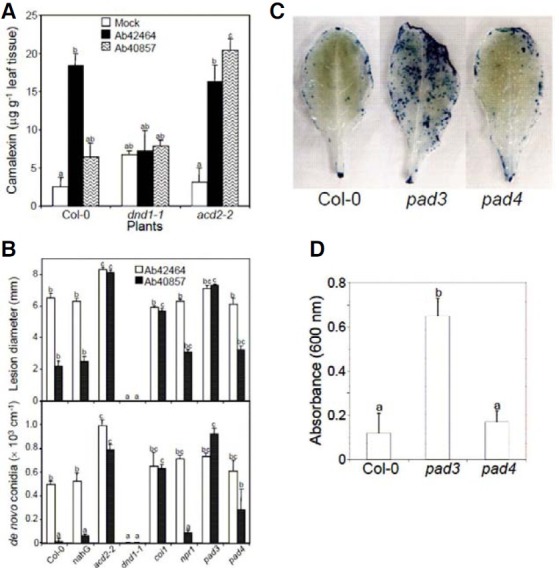
To elucidate the defense-signaling pathways for A. brassicicola, Col-0 wild type and its mutants, coi1, npr1, pad3, and pad4, and transgenic Col-0 harboring the bacterial gene, NahG, were inoculated with Ab40857 and Ab42464 (Fig. 4B). The compatibilities among the tested plants and Ab42464 were almost identical with that between Col-0 and Ab42464. The incompatibility between Col-0 and Ab40857 was also not affected by the disruptions of npr1, pad4, and NahG expression; however, the lack of COI1 or PAD3 resulted in loss of resistance against Ab40857 infection. The comparison of conidia production also coincided with the observed disease development. Additionally, the burst of cell death by avirulent Ab40857 was also abundant and evident in pad3 compared to Col-0 and pad4 (Figs. 4C and 4D).
DISCUSSION
Interactions between Arabidopsis thaliana and Alternaria brassicicola have been frequently employed to investigate the functions of plant genes in necrotrophic interactions (Cramer and Lawrence, 2004; Mengiste et al., 2003; Mukherjee et al., 2009; Oh et al., 2005; Veronese et al., 2004; Zheng et al., 2006). Based on the enigmatic innate immunity of Col-0 against MUCL20297, which is virulent on Brassica oleracea (Cramer et al., 2006), and the completion of the genome projects for both pathogen and host, a wealth of information has now accumulated on the roles of several plant genes and defense signaling pathways that are related with jasmonic acid and camalexin in the incompatible interactions. However, the factor(s) determining compatibility and incompatibility still remain unclear due to the lack of strains that are virulent on Col-0 wild type. Here, we present the defense-related responses of Col-0 and several mutants challenged with virulent and avirulent strains of A. brassicicola.
The compatible and incompatible interactions between Arabidopsis thaliana wild type Col-0 and A. brassicicola were investigated and the occurrence of host cell death during the early infection stage was analyzed because of the necrotrophic nature of the fungal pathogen. The development of symptoms on Col-0 infected with Ab42464 was evident, however, Ab40857- challenged leaves remained unchanged. The aggressive virulence of Ab40857 on Korean cabbage and several other Arabidopsis wild types indicates the specific incompatibility of this strain for Col-0. Microscopic and macroscopic cell death during the early infection stage within and around the infection site was also predominant in the compatible interaction. In contrast, incompatible Ab40857 barely induced cell death, similar to the results of MUCL20297 on Col-0 (Thomma et al., 1999; van Wees et al., 2003). Therefore, cell death is deeply related to successful disease development. When rice cells are infected with representative biotrophic pathogen that lack Avr genes, Magnaporthe oryzae, they remain alive until nearly the entire cells are filled with invasive mycelia, and they exhibit more of a delay in cell death than those challenged with pathogens expressing products of Avr genes or effectors (Dangl et al., 1996; Kankanala et al., 2007). Meanwhile, robust cell death has been shown to be indispensable for the disease progression and pathogen proliferation in Arabidopsis or rice infected with necrotrophic Pectobacterium carotovorum, B. cinerea, or Cochliobolus miyabeanus (Ahn, 2007; Ahn et al., 2005a; Govrin and Levine, 2000). Therefore, the interactions comprised with Col-0 and Ab42464/Ab40857 should also be typical examples for elucidating opposing roles of programmed cell death during the early necrotrophic infection stage.
To investigate the role of programmed cell death on the hostpathogen interaction, the disease progression and cellular and molecular responses were analyzed using dnd1 and acd2 as the positive and negative controls of cell death. dnd1-1 did not exhibit cell death and remained unchanged, regardless of the pathogen inoculation. In contrast, robust cell death was evident during the early infection stage in acd2-2, and this mutant was hyper susceptible to challenge by both pathogens. These results indicate that DND1 enhances susceptibility and ACD2 contributes to resistance. DND1 encodes the cyclic nucleotidegated ion channel 2 (AtCNGC2) protein that is involved in the influx of Ca2+, K+, and other cations into cells across the plasma membrane (Clough et al., 2000). ACD2 is a RED CHLOROPHYLL CATABOLITE REDUCTASE (RCCR) that is responsible for the breakdown of the porphyrin component of chlorophyll (Wüthrich et al., 2000), and it inhibits the PCD caused by an early mitochondrial oxidative burst (Yao and Greenberg, 2006). Phenotypically, cell death is a prerequisite in the compatible interaction between Arabidopsis and A. brassicicola. Lesion mimic mutants, such as barley mlo, show a broad-spectrum disease resistance against biotrophic pathogens but are highly susceptible to necrotrophic pathogens (Aviv et al., 2002; Huckelhoven et al., 2001; Jarosch et al., 2003; Salmeron and Vernooij, 1998; Yao et al., 2009; Yin et al., 2000). This ambivalence further supports the opposing contribution of PCD to the interaction. Taken together, these results imply that the effector(s) and/or their secretion system acclimating Col-0 to a “ready-to-death” state might be defined in Ab42464 and should be activated during its early interaction stage.
The incompatibility between Col-0 and Ab40857 was not altered by the over expression of bacterial NahG or the disruption of NPR1 and PAD4; however, coi1 and pad3 almost completely abrogated this resistance. Incompatibility between Col-0 and MUCL20297 was also shown to be dependent on intact COI1 (Thomma et al., 1998) and PAD3 genes (Narusaka et al., 2003; Thomma et al., 1999). In sum, defense signaling(s) within Col-0 against MUCL20297were almost identical with those for Ab40857 and indicate the dependency of Col-0’s resistance against Ab40857 on the jasmonic acid-mediated signaling pathway. In addition, the camalexin analysis presented here indicates that there is no relationship between phytoalexin accumulation and resistance against Ab40857. A previous investigation also described the accumulation of camalexin in the coi1 mutant, despite its loss of resistance against MUCL20297 (Thomma et al., 1999). To solve the paradox between the higher accumulation of camalexin in the compatible interaction and the attenuation of resistance against Ab40857 by pad3, the burst of cell death in pad3 and pad4 during the early infection stage was analyzed. Interestingly, only pad3 exhibited abundant cell death after challenge with Ab40857. Therefore, PAD3 might contribute to the resistance of Col-0 against Ab40857 as a component of the camalexin biosynthesis machinery and also participate in programmed cell death as a suppressor.
Callose deposition coincided with resistance in Col-0 and dnd1-1, and H2O2 accumulation correlated with susceptibility in Col-0 and acd2-2. These results suggest the role of callose as an additional plant cell wall-dependent defense barrier against pathogen infection and camalexin accumulation itself might be dispensable for the accomplishment of a defense state. Callose is a fluorescent β-1,3-glucan and a major component of papillae, which impede fungal penetration into the host cells; the induction of papillae is considered a representative defense response in the resistant state due to the reinforcement of the cell wall (Huckelhoven et al., 1999; Ton and Mauch-Mani, 2004). The abrupt induction and accumulation of active oxygen species, including H2O2, have been adopted as a reliable cellular response indicator that represents the “ready-to-defense” state of a host infected with a biotrophic pathogen; however, these responses are not helpful for enhancing resistance against A. brassicicola. In addition, our results indicate little or no relationship between callose-mediated cell wall fortification and hydrogen peroxide in this pathosystem.
The accelerated and strengthened transcription of defenserelated genes is also a type of molecular marker for whether a defense state has been accomplished, and for the signaling pathways that have been activated and/or are responsible for the observed disease resistance. The immediate activation of PDF1.2 and THI2.1 was evident in the incompatible interaction in Col-0; therefore, jasmonic acid/ethylene-dependent defense signaling might participate in this effective defense system. These results are almost identical with previous investigations that analyzed the transcriptome of Col-0 challenged with incompatible MUCL20297 (Narusaka et al., 2003; van Wees et al., 2003).
In this study, we characterized the relationship between Arabidopsis wild-type Col-0 plants and two strains of A. brassicicola. The comparative analyses of disease development and histochemical evidence confirmed the necrotrophic nature of A. brassicicola; that is, programmed cell death in the early infection processes was a key determinant overwhelming both compatibility and incompatibility. The pathosystems established in this study might be an ideal platform for the functional analysis of pathogens and Arabidopsis genes, and could also be a useful foundation for establishing control strategies in Cruciferaceae in general.
Acknowledgments
This research was supported by a grants from the National Academy of Agricultural Science (PJ907086) and BioGreen 21 (PJ00802107) to Il-Pyung Ahn. MS was supported by a grant from Brain Korea 21, Korea.
References
- 1.Ahn I.-P. Disturbance of the Ca2+/calmodulin-dependent signalling pathway is responsible for the resistance of Arabidopsis dnd1 against Pectobacterium carotovorum infection. Mol. Plant Pathol. (2007);8:747–759. doi: 10.1111/j.1364-3703.2007.00428.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahn I.-P., Kim S., Kang S., Suh S.-C., Lee Y.-H. Rice defense mechanisms against Cochliobolus miyabeanus and Magnaporthe grisea are distinct. Phytopathology. (2005a);95:1248–1255. doi: 10.1094/PHYTO-95-1248. [DOI] [PubMed] [Google Scholar]
- 3.Ahn I.-P., Kim S., Lee Y.-H. Vitamin B1 functions as an activator of plant disease resistance. Plant Physiol. (2005b);138:1505–1515. doi: 10.1104/pp.104.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn I.-P., Kim S., Lee Y.-H., Suh S.-C. Vitamin B1- induced priming is dependent on hydrogen peroxide and the NPR1 gene in Arabidopsis. Plant Physiol. (2007);143:838–848. doi: 10.1104/pp.106.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez M.E., Pennell R.I., Meijer P., Ishikawa A., Dixon R.A., Lamb C.J. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. (1998);92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 6.Asai T., Stone J.M., Heard J.E., Kovtun Y., Yorgey P., Sheen J., Ausubel F.M. Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell. (2000);12:1823–1836. doi: 10.1105/tpc.12.10.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aviv D.H., Rustérucci C., Iii B.F.H., Dietrich R.A., Parker J.E., Dangl J.L. Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J. (2002);29:381–391. doi: 10.1046/j.0960-7412.2001.01225.x. [DOI] [PubMed] [Google Scholar]
- 8.Berrocal-Lobo M., Molina A. Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant-Microbe Interact. (2004);17:763–770. doi: 10.1094/MPMI.2004.17.7.763. [DOI] [PubMed] [Google Scholar]
- 9.Berto P., Commenil P., Belingheri L., Dehorter B. Occurrence of a lipase in spores of Alternaria brassicicola with a crucial role in the infection of cauliflower leaves. FEMS Microbiol. Lett. (1999);180:183–189. doi: 10.1111/j.1574-6968.1999.tb08794.x. [DOI] [PubMed] [Google Scholar]
- 10.Bolwell G.P., Butt V.S., Davies D.R., Zimmerlin A. The origin of the oxidative burst in plants. Free Radic. Res. (1995);23:517–532. doi: 10.3109/10715769509065273. [DOI] [PubMed] [Google Scholar]
- 11.Bolwell G.P., Davies D.R., Gerrish C., Auh C.-K., Murphy T.M. Comparative biochemistry of the oxidative burst produced by rose and French bean cells reveals two distinct mechanisms. Plant Physiol. (1998);116:1379–1385. doi: 10.1104/pp.116.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamnongpol S., Willekens H., Moeder W., Langebartels C., Sandermann H. Jr., Van Montagu M., Inze D., Van Camp W. Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc. Natl. Acad. Sci. USA. (1998);95:5818–5823. doi: 10.1073/pnas.95.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clough S.J., Fengler K.A., Yu I.-C., Lippok B., Smith R.K. Jr., Bent A.F. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA. (2000);97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cramer R.A., Lawrence C.B. Identification of Alternaria brassicicola genes expressed in planta during pathogenesis of Arabidopsis thaliana. Fungal Genet. Biol. (2004);41:115–128. doi: 10.1016/j.fgb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Cramer R.A., La Rota C.M., Cho Y., Thon M., Craven K.D., Knudson D.L., Mitchell T.K., Lawrence C.B. Bioinformatic analysis of expressed sequence tags derived from a compatible Alternaria brassicicola-Brassica oleracea interaction. Mol. Plant Pathol. (2006);7:113–124. doi: 10.1111/j.1364-3703.2006.00324.x. [DOI] [PubMed] [Google Scholar]
- 16.Dangl J.L., Dietrich R.A., Richberg M.H. Death don’t have no mercy: Cell death programs in plant-microbe interactions. Plant Cell. (1996);8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis K.R., Ausubel F.M. Characterization of elicitorinduced defense responses in suspension-cultured cells of Arabidopsis. Mol. Plant-Microbe Interact. (1989);2:363–368. [Google Scholar]
- 18.Flor H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. (1971);9:275–296. [Google Scholar]
- 19.Glawischnig E., Hansen B.G., Olsen C.E., Halkier B.A. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2004);101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Ann. Rev. Phytopathol. (2005);43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 21.Glazebrook J., Ausubel F. Isolation of phytoalexindeficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA. (1994);91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glazebrook J., Rogers E.E., Ausubel F.M. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. (1996);143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glazebrook J., Zook M., Mert F., Kagan I., Rogers E.E., Crute I.R., Holub E.B., Hammerschmidt R., Ausubel F.M. Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contributes to downy mildew resistance. Genetics. (1997);146:381–392. doi: 10.1093/genetics/146.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govrin E.M., Levine A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. (2000);10:751–757. doi: 10.1016/s0960-9822(00)00560-1. [DOI] [PubMed] [Google Scholar]
- 25.Govrin E.M., Levine A. Infection of Arabidopsis with a necrotrophic pathogen, Botrytis cinerea, elicits various defense responses but does not induce systemic acquired resistance (SAR). Plant Mol. Biol. (2002);48:267–276. doi: 10.1023/a:1013323222095. [DOI] [PubMed] [Google Scholar]
- 26.Govrin E.M., Rachmilevitch S., Tiwari B.S., Solomon M., Levine A. An elicitor from Botrytis cinerea induces the hypersensitive response in Arabidopsis thaliana and other plants and promotes the gray mold disease. Phytopathology. (2006);96:299–307. doi: 10.1094/PHYTO-96-0299. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg J.T., Ailan G., Klessig D.F., Ausubel F.M. Programmed cell death in plants: a pathogen-triggered response activated coordinately with mutiple defense functions. Cell. (1994);77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 28.Hong C.P., Plaha P., Koo D.-H., Yang T.-J., Choi S.R., Lee Y.K., Uhm T., Bang J.-W., Edwards D., Bancroft I., et al. A survey of the Brassica rapa genome by BAC-end wequence analysis and comparison with Arabidopsis thaliana. Mol. Cells. (2006);22:300–307. [PubMed] [Google Scholar]
- 29.Huckelhoven R., Fodor J., Preis C., Kogel K.H. Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. (1999);119:1251–1260. doi: 10.1104/pp.119.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huckelhoven R., Dechert C., Kogel K.-H. Non-host resistance of barley is associated with a hydrogen peroxide burst at sites of attempted penetration by wheat powdery mildew fungus. Mol. Plant Pathol. (2001);2:199–205. doi: 10.1046/j.1464-6722.2001.00067.x. [DOI] [PubMed] [Google Scholar]
- 31.Jarosch B., Jansen M., Schaffrath U. Acquired resistance functions in mlo barley, which is hypersusceptible to Magnaporthe grisea. Mol. Plant-Microbe Interact. (2003);16:107–114. doi: 10.1094/MPMI.2003.16.2.107. [DOI] [PubMed] [Google Scholar]
- 32.Jones J.D.G., Dangl J.L. The plant immune system. Nature. (2006);444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 33.Kagan I.A., Hammerschmidt R. Arabidopsis ecotype variability in camalexin production and reaction to infection by Alternaria brassicicola. J. Chem. Ecol. (2002);28:2121–2140. doi: 10.1023/a:1021020512846. [DOI] [PubMed] [Google Scholar]
- 34.Kankanala P., Czymmek K., Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. (2007);19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishimoto K., Matsui K., Ozawa R., Takabayashi J. Volatile 1-octen-3-ol induces a defensive response in Arabidopsis thaliana. J. General Plant Pathol. (2007);73:35–37. [Google Scholar]
- 36.Lawrence C.B., Mitchell T.K., Craven K.D., Cho Y., Cramer R.A., Kim K.-H. At Death’s Door: Alternaria pathogenicity mechanisms. Plant Pathol. J. (2008);24:101–111. [Google Scholar]
- 37.Liu G., Holub E.B., Alonso J.M., Ecker J.R., Fobert P.R. An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. (2005);41:304–318. doi: 10.1111/j.1365-313X.2004.02296.x. [DOI] [PubMed] [Google Scholar]
- 38.Manners J.M., Penninckx I.A., Vermaere K., Kazan K., Brown R.L., Morgan A., Maclean D.J., Curtis M.D., Cammue B.P., Broekaert W.F. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. (1998);38:1071–1080. doi: 10.1023/a:1006070413843. [DOI] [PubMed] [Google Scholar]
- 39.Mengiste T., Chen X., Salmeron J.M., Dietrich R.A. The BOS1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. (2003);15:2551–2565. doi: 10.1105/tpc.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee A., Lev S., Gepstein S., Horwitz B. A compatible interaction of Alternaria brassicicola with Arabidopsis thaliana ecotype DiG: evidence for a specific transcriptional signature. BMC Plant Biol. (2009);9:31. doi: 10.1186/1471-2229-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narusaka Y., Narusaka M., Seki M., Ishida J., Nakashima M., Kamiya A., Enju A., Sakurai T., Satoh M., Kobayashi M., et al. The cDNA microarray analysis using an Arabidopsis pad3 mutant reveals the expression profiles and classification of genes induced by Alternaria brassicicola attack. Plant Cell Physiol. (2003);44:377–387. doi: 10.1093/pcp/pcg050. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura M.T., Dangl J.L. Arabidopsis and the plant immune system. Plant J. (2010);61:1053–1066. doi: 10.1111/j.1365-313X.2010.04131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norman-Setterbald C., Vidal S., Palva E.T. Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant-Microbe Interact. (2000);13:430–438. doi: 10.1094/MPMI.2000.13.4.430. [DOI] [PubMed] [Google Scholar]
- 44.Ntoukakis V., Mucyn T.S., Gimenez-Ibanez S., Chapman H.C., Gutierrez J.R., Balmuth A.L., Jones A.M.E., Rathjen J.P. Host inhibition of a bacterial virulence effector triggers immunity to infection. Science. (2009);324:784–787. doi: 10.1126/science.1169430. [DOI] [PubMed] [Google Scholar]
- 45.Oh I.S., Park A.R., Bae M.S., Kwon S.J., Kim Y.S., Lee J.E., Kang N.Y., Lee S., Cheong H., Park O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell. (2005);17:2832–2847. doi: 10.1105/tpc.105.034819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olivain C., Trouvelot S., Binet M.-N., Cordier C., Pugin A., Alabouvette C. Colonization of flax roots and early physiological responses of flax cells inoculated with pathogenic and nonpathogenic strains of Fusarium oxysporum. Appl. Environ. Microbiol. (2003);69:5453–5462. doi: 10.1128/AEM.69.9.5453-5462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhee S., Chang T.-S., Jeong W., Kang D. Methods for detection and measurement of hydrogen peroxide inside and outside of cells. Mol. Cells. (2010);29:539–549. doi: 10.1007/s10059-010-0082-3. [DOI] [PubMed] [Google Scholar]
- 48.Rusterucci C., Aviv D.H., Holt III B.F., Dangl J.F., Parker J.E. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell. (2001);13:2211–2224. doi: 10.1105/tpc.010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmeron J.M., Vernooij B. Transgenic approaches to microbial disease resistance in crop plants. Curr. Opin. Plant Biol. (1998);1:347–352. doi: 10.1016/1369-5266(88)80058-x. [DOI] [PubMed] [Google Scholar]
- 50.Schenk P.M., Kazan K., Manners J.M., Anderson J.P., Simpson R.S., Wilson I.W., Somerville S.C., Maclean D.J. Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. (2003);132:999–1010. doi: 10.1104/pp.103.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sellam A., Poupard P., Simoneau P. Molecular cloning of AbGst1 encoding a glutathione transferase differentially expressed during exposure of Alternaria brassicicola to isothiocyanates. FEMS Microbiol. Lett. (2006);258:241–249. doi: 10.1111/j.1574-6968.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 52.Seo H.S., Song J.T., Cheong J.J., Lee Y.H., Lee Y.W., Hwang I., Lee J.S., Choi Y.D. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate- regulated plant responses. Proc. Natl. Acad. Sci. USA. (2001);98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staskawicz B.J., Ausubel F.M., Baker B.J., Ellis J.G., Jones J.D.G. Molecular genetics of plant disease resistance. Science. (1995);268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 54.Tenhaken R., Levine A., Brisson L.F., Dixon R.A., Lamb C.J. Function of the oxidative burst in hypersensitive disease resistance. Proc. Natl. Acad. Sci. USA. (1995);92:4158–4163. doi: 10.1073/pnas.92.10.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomma B.P.H.J. Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. (2003);4:225–236. doi: 10.1046/j.1364-3703.2003.00173.x. [DOI] [PubMed] [Google Scholar]
- 56.Thomma B.P.H.J., Eggermont K., Penninckx I.A.M.A., Mauch- Mani B., Vogelsang R., Cammue B.P.A., Broekaert W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA. (1998);95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomma B.P., Nelissen I., Eggermont K., Broekaert W.F. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. (1999);19:163–171. doi: 10.1046/j.1365-313x.1999.00513.x. [DOI] [PubMed] [Google Scholar]
- 58.Thomma B.P.H.J., Eggermont K., Broekaert W.F., Cammue B.P.A. Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiol. Biochem. (2000);38:421–427. [Google Scholar]
- 59.Ton J., Mauch-Mani B. β-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABAdependent priming for callose. Plant J. (2004);38:119–130. doi: 10.1111/j.1365-313X.2004.02028.x. [DOI] [PubMed] [Google Scholar]
- 60.Ton J., Jakab G., Toquin V., Flors V., Iavicoli A., Maeder M.N., Metraux J.-P., Mauch-Mani B. Dissecting the β- aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell. (2005);17:987–999. doi: 10.1105/tpc.104.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trusov Y., Rookes J.E., Chakravorty D., Armour D., Schenk P.M., Botella J.R. Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol. (2006);140:210–220. doi: 10.1104/pp.105.069625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Baarlen P., Staats M., Van Kan J.A.L. Induction of programmed cell death in lily by the fungal pathogen Botrytis elliptica. Mol. Plant Pathol. (2004);5:559–574. doi: 10.1111/j.1364-3703.2004.00253.x. [DOI] [PubMed] [Google Scholar]
- 63.van Kan J.A.L. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. (2006);11:1360–1385. doi: 10.1016/j.tplants.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 64.van Wees S.C.M., Chang H.-S., Zhu T., Glazebrook J. Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. (2003);132:606–617. doi: 10.1104/pp.103.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veronese P., Chen X., Bluhm B., Salmeron J., Dietrich R.A., Mengiste T. The BOS loci of Arabidopsis are required for resistance to Botrytis cinerea infection. Plant J. (2004);40:558–574. doi: 10.1111/j.1365-313X.2004.02232.x. [DOI] [PubMed] [Google Scholar]
- 66.Vieira Dos Santos C., Letousey P., Delavault P., Thalouarn P. Defense gene expression analysis of Arabidopsis thaliana parasitized by Orobanche ramosa. Phytopathology. (2003);93:451–457. doi: 10.1094/PHYTO.2003.93.4.451. [DOI] [PubMed] [Google Scholar]
- 67.Weigel R.R., Pfitzner U.M., Gatz C. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell. (2005);17:1279–1291. doi: 10.1105/tpc.104.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wüthrich K.L., Bovet L., Hunziker P.E., Donnison I.S., Hörtensteiner S. Molecular cloning, functional expression and characterisation of RCC reductase involved in chloro-phyll catabolism. Plant J. (2000);21:189–198. doi: 10.1046/j.1365-313x.2000.00667.x. [DOI] [PubMed] [Google Scholar]
- 69.Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem. J. (1997);322:681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. COI1: An arabidopsis gene required for jasmonateregulated defense and fertility. Science. (1998);280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 71.Yao N., Greenberg J.T. Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell. (2006);18:397–411. doi: 10.1105/tpc.105.036251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao Q., Zhou R., Fu T., Wu W., Zhu Z., Li A., Jia J. Lesion mimic associates with adult plant resistance to leaf rust infection in wheat. Theor. Appl. Genet. (2009);119:1005–1012. doi: 10.1007/s00122-009-1012-7. [DOI] [PubMed] [Google Scholar]
- 73.Yin Z., Chen J., Zeng L., Goh M., Leung H., Khush G.S., Wang G.L. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant-Microbe Interact. (2000);13:869–876. doi: 10.1094/MPMI.2000.13.8.869. [DOI] [PubMed] [Google Scholar]
- 74.Yoshioka K., Nakashita H., Klessig D.F., Yamaguchi I. Probenazole induces systemic acquired resistance in Arabidopsis with a novel type of action. Plant J. (2001);25:149–157. doi: 10.1046/j.1365-313x.2001.00952.x. [DOI] [PubMed] [Google Scholar]
- 75.Zhao J., Last R.L. Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell. (1996);8:2235–2244. doi: 10.1105/tpc.8.12.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng Z., Qamar S.A., Chen Z., Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. (2006);48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]


