Abstract
For eukaryotes, fine tuning of gene expression is necessary to coordinate complex genetic information. Recent studies have shown that noncoding RNAs (ncRNAs) play central roles in this process. For example, ncRNAs participate in multiple diverse functions such as mRNA degradation, epigenetic regulation and alternative splicing. The findings regarding this new player in gene regulation suggest that the mechanism of gene regulation is much more complicated and subtle than previously thought. In this review, new findings concerning the role of ncRNAs in gene regulation are discussed.
Keywords: chromatin, gene regulation, RNA
INTRODUCTION
Multicellular organisms must regulate the expression of genetic information to maintain the identities of different cell types. To achieve this purpose, genetic information is stored in a highly regulated structure called chromatin. Chromatin forms hierarchically higher order structure and is regulated through several mechanisms. Chromatin is assembled and disassembled by chromatin assembly complexes and chaperones according to nuclear processes, such as DNA replication, DNA repair, and transcription. The ATP-dependent chromatin-remodeling complex remodels chromatin structure to regulate the accessibility of transcription factors to DNA. The nucleosome is the fundamental packaging unit of chromatin. Within this unit, histones are subjected to several covalent modifications including acetylation, phosphorylation, ubiquitination, sumoylation and methylation. Site-specific modifications of histones lead to changes in chromatin structure or serve as beacons to recruit other chromatin modifiers. In multicellular organisms, the complexity of chromatin regulation is necessary to maintain the integrity of the genome and to regulate the expression of genetic information.
ncRNAs are functional RNA molecules that are not translated into proteins. The first ncRNA was discovered in the 1960s in studies on tRNA; however, its function was restricted to conveying genetic information, rather than regulating gene expression. In the early 1990s, several groups reported a long ncRNA, called X-inactive specific transcript (Xist), which is specifically transcribed in the inactive X chromosome and directly regulates X-chromosome inactivation (Brockdorff et al., 1991; McCarrey and Dilworth, 1992; Masui and Heard, 2006). In addition to XIST, the roX genes mediate X-chromosome hyperactivation in Drosophila and several long ncRNAs, including Air, have been implicated in genomic imprinting (Braidotti et al., 2004). Recent genome-wide analysis of transcriptome revealed that genome transcribes many long size noncoding RNAs, which are collectively called long noncodingRNAs (lncRNAs) to distinguish from small noncoding RNAs such as siRNAs and piRNAs. There are accumulating evidences that several classes of lncRNAs play critical functions in gene regulation at the chromatin level. In this review, we provide an overview of the recent findings on the functions and mechanisms of lncRNAs for gene regulation as well as the implication of small noncoding RNAs for gene regulation.
siRNA regulates heterochromatin establishment
RNA interference (RNAi) is a gene silencing mechanism mediated by small RNAs. RNAi was initially identified as a mechanism of posttranscriptional gene silencing in Caenorhabditis elegans (Fire et al., 1998); it also acts in the related processes of quelling in Neurospora and post-transcriptional gene silencing (PTGS) in plants (Hannon, 2002). RNAi involves a conserved group of proteins: Dicer and Argonaute (Hannon, 2002; Matzke et al., 2001). Double-stranded RNA (dsRNA) is processed by Dicer into small interfering RNAs (siRNAs) of 21-25 nucleotides, and siRNA then binds to Argonaute, which is a catalytic enzyme in the RNA-induced silencing complex (RISC) (Liu et al., 2004; Song et al., 2004). The siRNA-bound RISC cleaves mRNA in a sequence-specific manner to repress gene expression. However, Martienssen and Grewal’s groups showed that RNAi is also involved in the initiation of chromatin silencing and heterochromatin assembly in S. pombe (Hall et al., 2002; Volpe et al., 2002). They found that the deletion of RNAi machinery including Argonaute, Dicer and RNA-dependent RNA polymerase induces the accumulation of transcripts and transcriptional de-repression in the centromeric heterochromatin region accompanied by loss of histone H3Lys9 methylation, which is a gene repression marker. In addition, the siRNAmediated establishment of heterochromatin requires heterochromatin protein 1 (HP1/Swi6) (Iida et al., 2008). These results revealed a surprising interaction between two seemingly unrelated pathways: RNAi and chromatin regulation. RNAi-mediated chromatin regulation has been studied extensively in S. pombe, but it is not clear how the RNAi machinery works at the chromatin level in higher eukaryotes. The primary function of siRNA in heterochromatin establishment is to recruit the RNAinduced initiation of transcriptional gene silencing (RITS) complex and subsequently to recruit chromatin-modifying complexes. Although it has been speculated that siRNA recruits RITS and other complexes in a sequence-specific manner by hybridizing with nascent transcripts from heterochromatin loci, the exact mechanism is not clearly understood.
ncRNA-mediated transcriptional silencing is also observed in plants. In plants, the DCL3/HEN1 complex processes dsRNA, formed from transcripts in the sense and antisense directions, into siRNA. siRNA complexed with AGO4-PolV serves as a primer to generate more transcripts by PolV (Bies-Etheve et al., 2009). PolV transcription at the locus primed with siRNA was shown to facilitate de novo methylation by DRM2. Once the locus is methylated, PolIV transcribes aberrant RNAs from the methylated locus, which are subsequently processed by the DCL3/HEN1 enzyme complex to result in the amplification of a siRNA signal at the targeted locus. Although siRNA was initially discovered as an mRNA degradation pathway, this posttranscriptional regulation may be a minor function of RNAi; the major function may be to maintain and regulate genetic information in the nucleus.
piRNA as a tool of transposon surveillance
Several groups have recently discovered another class of small RNAs called Piwi-interacting RNA (piRNA) (Girard et al., 2006; Lau et al., 2006). Most piRNAs appear to be derived from a small number of long single-stranded RNA precursors that are often encoded by transposon-rich heterochromatic clusters (Brennecke et al., 2007; Yin and Lin, 2007). Transposons are important features of most eukaryotic genomes, and the mobilization of these elements can lead to genetic instability (McClintock, 1953). Transposon silencing is particularly critical in the germline, which maintains the genetic information that will be inherited by future generations. It has been demonstrated that the Piwi protein is genetically linked to transcriptional gene silencing in flies and that piRNA forms a complex with a protein implicated in gene silencing pathways (Lau et al., 2006). A recent study reported that piRNAs play a critical role in transposon silencing and genome maintenance during germline development (Malone et al., 2009).
piRNAs differ from siRNA in their size and in their binding partners. siRNA and miRNA are 21-22 nucleotides (nt) in size and bind to the proteins from the Ago clade of the Argonaute protein family. piRNAs are typically 24-32 nt long and associate with Piwi, Aubergine (Aub) and one PIWI clade Argonaute protein, AGO3. piRNA is generated through the “Ping-Pong cycle” by Aub and AGO3 in a slicer activity-dependent manner (Brennecke et al., 2007; Gunawardane et al., 2007; Yin and Lin, 2007). However, the mechanism of the biogenesis of Piwibound piRNA is not known. A recent study shows that there are two distinct piRNA pathways in Drosophila ovary germline and somatic cells (Malone et al., 2009). In somatic cells, only Piwi protein is expressed and is shown to regulate the gypsy family through an exclusive association with piRNAs transcribed at the flamenco cluster. In germline cells, three Piwi proteins, Piwi, Aub, and AGO3, regulate a broad range of transposon elements.
It has been reported that protein-coding genes containing transposon insertions within introns were not silenced by the piRNA pathway, suggesting that piRNA homology is removed by splicing after export from the nucleus (Brennecke et al., 2007). These data suggest that piRNA functions at the posttranscriptional level. Alternatively, several lines of evidence suggest that Piwi protein localizes to the nucleus and functions at the chromatin level. It has been shown that Piwi protein directly interacts with HP1 protein. In addition, the depletion of Piwi protein results in the loss of H3Lys9 methylation and the delocalization of HP1 protein. Furthermore, Piwi has been implicated in heterochromatin assembly in somatic cells (Brower-Toland et al., 2007; Pal-Bhadra et al., 2004). These findings suggest that piRNA and Piwi proteins interact to induce the chromatin modification of their target genes, thus imposing transcriptional silencing. Consistently, piRNA mutations reduced de novo DNA methylation of retrotransposons in fetal male germ cells (Kuramochi-Miyagawa et al., 2008). Furthermore, piRNAs have been found in polysome fractions. The mouse Piwi protein, Miwi, associates with translation initiation factors and may positively regulate translation (Grivna et al., 2006; Unhavaithaya et al., 2009). These findings raise the possibility that piRNAs also control translation. However, despite the many studies on Piwi/piRNA function, the exact mechanism of their action needs to be further investigated.
Long noncoding RNAs in X chromosome inactivation
X chromosome inactivation (XCI) is a good example for epigenetic regulation by ncRNA. It is known that there are two kinds of XCI: imprinted and random. During imprinted XCI, the paternal X chromosome is preferentially silenced in the placenta of eutherian mammals, and in all cells of earlier marsupial mammals (Martin et al., 1978; Rastan and Robertson, 1985). By contrast, random XCI takes place in the early female embryo, where both the maternal and the paternal X chromosome have the same chance of becoming inactivated (Martin et al., 1978).
XCI is regulated by a single X-inactivation center (Xic), an Xlinked locus that counts the number of X chromosomes, chooses one to remain active and silences the other (Costa, 2008). Xic is noted for its abundance of noncoding transcripts: the Xist silencer RNA (Borsani et al., 1991; Brockdorff et al., 1992; Brown et al., 1991; 1992); its antisense Tsix counterpart (Lee and Lu, 1999; Lee et al., 1999; Sado et al., 2001); and the enhancer- bearing Xite (Ogawa and Lee, 2003).
On the future active X chromosome (Xa), Xite acts on the linked Tsix allele to prolong antisense transcription, which in turn blocks Xist upregulation. On the future inactive X chromosome (Xi), Xite repression results in Tsix downregulation in cis (Ogawa and Lee, 2003), which in turn triggers induction of Xist and heterochromatinization (Sun et al., 2006). These results suggest that several ncRNAs are needed for XCI. However, research has shown that the autosomal insertion of Xist transgenes can silence genes flanking the insertion site, implicating Xist as both necessary and sufficient for X chromosome inactivation (Lee and Jaenisch, 1997; Penny et al., 1996; White et al., 1998; Wutz and Jaenisch, 2000).
During X-inactivation in ES cells, Tsix is expressed on the Xa but is downregulated on the Xi causing the expression of Xist to be upregulated (Lee and Lu, 1999). While Xist expression on the Xi is prolonged during X-inactivation maintenance, Tsix expression ceases on the Xa and is therefore not required to keep Xist expression blocked (Lee et al., 1999). The Xist RNA coating of the future Xi is followed by a series of epigenetic changes creating the characteristic chromatin signature of the transcriptionally repressed inactive X chromosome (Okamoto et al., 2004). First, active markers such as H3Lys4 methylation and H3Lys9 acetylation are gradually lost. Second, MacroH2A is incorporated in the Xi (Costanzi et al., 2000). Third, histone methyltransferase Polycomb group complex 2 (PRC2) associates with Xi and leads to the accumulation of repressive H3Lys27 trimethylation (Erhardt et al., 2003; Plath et al., 2003; 2004; Silva et al., 2003), which is later followed by H3Lys9 methylation (Okamoto et al., 2004). This observation indicates that XCI resulted from chromatin modification mediated by the long ncRNA, Xist. However, the exact mechanism of Xist recruitment of chromatin modifiers is not clear. Recently, Jeannie Lee’s group found another ncRNA involved in XCI. They found that a 1.6-kilobase ncRNA (RepA) within Xist is transcribed as a separate transcript and recruits PRC2 to its target locus, Xi. Using the RNA-chip assay, followed by biochemical approaches, they showed that RepA RNA recruits PRC2 and that the PRC2 subunit, EZH2, is responsible for direct interaction with RepA. Based on these results, they proposed a model to explain how RepA and other ncRNAs, such as Xist and Tsix, work with PRC2, which is the first ncRNA-binding protein involved in XCI. Before XCI, when only Tsix alone is transcribed, Tsix and RepA compete for PRC2 binding. Upon XCI onset, the transcription of Tsix is suppressed, and Xist is transcribed. RepA then relays PRC2 to the 5′ region of Xist, which leads to the methylation of histone H3Lys27. These data suggest that the function of ncRNA in XCI is to recruit chromatin modifiers. It would be interesting to examine if other ncRNA, such as Tsix, Xist and Xite, also recruit protein factors to modify chromatin.
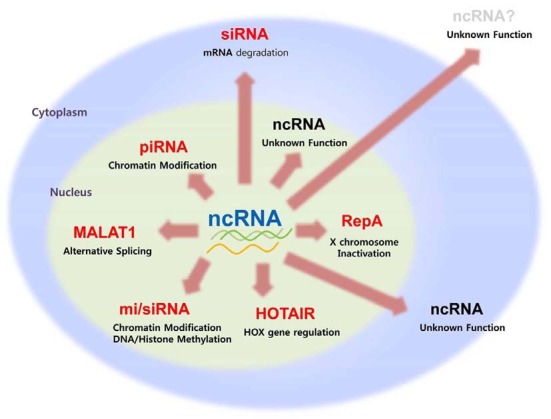
Fig. 1. The roles of noncoding RNAs.
HOTAIR regulates HOX gene transcription
Humans have 39 HOX genes grouped into HOXA, B, C, and D genes located in four separate chromosomes. HOX genes encode master regulatory transcription factors. Therefore, expression patterns of the HOX gene cluster govern the fates of cells during embryonic development. HOX genes are tightly controlled through epigenetic regulations, and ncRNAs are thought to be involved in controlling HOX genes (Rank et al., 2002). Although the presence of ncRNA in HOX gene expression has been reported previously, the function of ncRNA coded in the HOX gene cluster remained unknown until the discovery of HOTAIR by Howard Chang’s group (Rinn et al., 2007; Woo and Kingston, 2007). They characterized the transcripts from all human HOX gene loci using high resolution tiling arrays and discovered a 2.2 kilobase ncRNA transcribed in the HOXC locus, termed HOX Antisense Intergenic RNA (HOTAIR). The discovery of ncRNA in HOX gene locus was expected. However, the surprise came when they knocked down HOTAIR gene expression. The depletion of HOTAIR by RNAi has little effect on the transcription of HOXC in regions where HOTAIR is transcribed; however, the depletion of HOTAIR represses the transcription of the 40 kb region in the HOXD cluster. These data suggest that HOTAIR ncRNA transcribed in HOXC works in trans to repress HOXD located in a different chromosome. To investigate the molecular mechanism, they examined the change of the repressive marker histone, H3Lys27 and its modifying enzyme, Polycomb Repressive Complex 2. Consistent with the data that HOTAIR works in trans, the depletion of HOTAIR causes the loss of histone H3Lys27 methylation in HOXD but not in HOXC. Moreover, the level of Suz12 expression, a component of PRC2, decreases modestly but specifically in the HOXD region. These data suggest that HOTAIR ncRNA transcribed in the HOXC region recruits PRC2 to the HOXD region to methylate histone H3Lys27 leading to repression. The mechanism of HOTAIR recruitment of PRC2 may be similar to that of RepA. The paper by Howard’s group showed that HOTAIR could coordinate histone modifications by binding to multiple histone modification enzymes (Tsai et al., 2010). In this paper, HOTAIR serves as a scaffold for at least two distinct histone modification complexes. Histone H3Lys27 methylation by PRC2 is a hallmark for transcriptional repression while Histone H3Lys4 methylation by MLL complex is a hallmark for transcriptional activation. These two hallmarks function antagonistically for transcriptional regu-lation. Interestingly, HOTAIR, which induces transcriptional repression, recruits PRC2 through the 5′ domain and the LSD1/ CoREST/REST complex through the 3′ domain. The two anta-gonistic hallmarks are coordinated by the ability to recruit proteins to write a repressive mark and to erase an active mark. Many questions still remain concerning how HOTAIR specifically localizes to the HOXD cluster and whether ncRNA-mediated silencing is a common mechanism for HOX gene silencing or if this is a special case for HOTAIR.
Table 1.
The roles of noncoding RNAs
| ncRNA | Action place | Action mode |
|---|---|---|
| siRNA | Cytoplasm | mRNA degradation |
| miRNA | Cytoplasm | Translation inhibition |
| piRNA | Nucleus | Transposon surveillance |
| RepA | Nucleus | X-chromosome Inactivation |
| HOTAIR | Nucleus | Transcriptional silencing |
| MALAT1 | Nucleus/Speckles | Alternative splicing |
MALAT1 regulates alternative splicing
ncRNA-mediated gene regulation is not limited to transcriptional gene silencing. A recent paper by Prasanth’s group implicates the ncRNA MALAT1 in regulating alternative splicing. Metastasis- associated lung adenocarcinoma transcript 1 (MALAT1) (Tripathi et al., 2010), also known as NEAT2 (Hutchinson et al., 2007), was originally identified as a long ncRNA that exhibited significant expression in individuals at high risk for metastasis of many human cancers (Ji et al., 2003; Lin et al., 2007). After transcription, nascent MALAT1 is processed into a 5′-long transcript and a 3′-short tRNA-like transcript by RNase P cleavage (Sunwoo et al., 2009; Wilusz et al., 2008). The long form of MALAT-1 localizes to nuclear speckles as a structural platform while the short form is exported to the cytoplasm (Hutchinson et al., 2007).
Several lines of evidence suggest that nuclear speckles have two important functions: as compartments for assembly, modification and storage that supply alternative splicing and transcription factors to active transcription sites (Lamond and Spector, 2003); or as hubs facilitating the efficiency and integration of distinct steps in gene expression, ranging from transcription to mRNA export (Hall et al., 2006). In general, alternative splicing is regulated by trans-acting protein factors, which include the small nuclear ribonucleoproteins, the serine/arginine-rich (SR) family of nuclear phosphoproteins (SR proteins), the SR-related proteins, and the heterogeneous nuclear ribonucleoproteins (Blencowe, 2006; Long and Caceres, 2009).
Two relevant studies have described the possible functions of MALAT1. One study suggested that silencing of MALAT1 suppresses the migration of CaSki human cervical cancer cells (Tseng et al., 2009). The other study revealed that MALAT1, retained in nuclear speckles, positively regulates cell motility through the concomitant regulation of motility-related genes, such as CTHRC, CCT4, HMMR and ROD, via transcriptional and/or post transcriptional regulation (Tano et al., 2010). However, the molecular mechanism of the function of MALAT1 was not known. In Prasanth’s paper, it is revealed that MALAT1 is not required for the structural integrity of nuclear speckles, but interacts with SR splicing factors and modulates their distribution to nuclear speckles. In addition, it was suggested that MALAT1 regulates alternative splicing of pre-mRNAs by controlling the functional levels of SR splicing factors; depletion of MALTA1 increases the levels of cellular SR proteins and alters the distribution ratio of phosphorylated to dephosphorylated pools of SR proteins towards an increased fraction of the dephosphorylated forms of these proteins. These findings indicate that ncRNAs play roles in regulating the expression of genetic information not only at the transcriptional step but also at the alternative splicing step.
Future perspective
The scope of the functions of ncRNA expands from posttranscriptional silencing to transcriptional silencing and RNA splicing. It is now clear that ncRNAs play important roles at several levels of gene regulations: genomic stability maintenance, transcription, translation, and splicing. However, the mechanisms behind the actions of ncRNAs are largely unknown, and many questions remain unanswered. What is the mechanism of ncRNA biogenesis? Are RNAi machineries involved? Do ncRNAs mediate epigenetic modification of the loci to repress transcriptions as RepA does? It is likely that many other ncRNAs involved in gene regulation are yet to be discovered. It seems that RNA is situated at the center of the central dogma in regulating the genetic information flow from DNA to protein.
Acknowledgments
We apologize that we were not able to cite all related papers because of the page limit and the limited scope of this review. S.A. was supported by the Brain Korea 21 (BK21) program from the Korea Ministry of Education, Science and Technology. This work was partially supported by the World Class University (WCU) program (R31-2008-000-10071-0), grants (2010-0015253 and 2010-0007869) awarded through the National Research Foundation of Korea and funded by the Ministry of Education, Science and Technology, and a grant from the Korea Healthcare Technology R&D project of the Ministry of Health, Welfare & Family Affairs (A100525).
References
- 1.Bies-Etheve N., Pontier D., Lahmy S., Picart C., Vega D., Cooke R., Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. (2009);10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. (2006);126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Borsani G., Tonlorenzi R., Simmler M.C., Dandolo L., Arnaud D., Capra V., Grompe M., Pizzuti A., Muzny D., Lawrence C., et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. (1991);351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 4.Braidotti G., Baubec T., Pauler F., Seidl C., Smrzka O., Stricker S., Yotova I., Barlow D.P. The Air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb. Symp. Quant. Biol. (2004);69:55–66. doi: 10.1101/sqb.2004.69.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNAgenerating loci as master regulators of transposon activity in Drosophila. Cell. (2007);128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 6.Brockdorff N., Ashworth A., Kay G.F., Cooper P., Smith S., McCabe V.M., Norris D.P., Penny G.D., Patel D., Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. (1991);351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 7.Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. (1992);71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 8.Brower-Toland B., Findley S.D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S.C., Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. (2007);21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. (1991);349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 10.Brown C.J., Hendrich B.D., Rupert J.L., Lafreniere R.G., Xing Y., Lawrence J., Willard H.F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. (1992);71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 11.Costa F.F. Non-coding RNAs, epigenetics and complexity. Gene. (2008);410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Costanzi C., Stein P., Worrad D.M., Schultz R.M., Pehrson J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. (2000);127:2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- 13.Erhardt S., Su I.H., Schneider R., Barton S., Bannister A.J., Perez-Burgos L., Jenuwein T., Kouzarides T., Tarakhovsky A., Surani M.A. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. (2003);130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- 14.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. (1998);391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 15.Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. (2006);442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 16.Grivna S.T., Pyhtila B., Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA. (2006);103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. (2007);315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 18.Hall I.M., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. (2002);297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 19.Hall L.L., Smith K.P., Byron M., Lawrence J.B. Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell Evol. Biol. (2006);288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannon G.J. RNA interference. Nature. (2002);418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 21.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. (2007);8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iida T., Nakayama J., Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell. (2008);31:178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. (2003);22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 24.Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T. W., et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. (2008);22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamond A.I., Spector D.L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. (2003);4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 26.Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. (2006);313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 27.Lee J.T., Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. (1997);386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee J.T., Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. (1999);99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 29.Lee J.T., Davidow L.S., Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. (1999);21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 30.Lin R., Maeda S., Liu C., Karin M., Edgington T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. (2007);26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 31.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. (2004);305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 32.Long J.C., Caceres J.F. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. (2009);417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 33.Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., Hannon G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. (2009);137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin G.R., Epstein C.J., Travis B., Tucker G., Yatziv S., Martin D.W. Jr., Clift S., Cohen S. X-chromosome inactivation during differentiation of female teratocarcinoma stem cells in vitro. Nature. (1978);271:329–333. doi: 10.1038/271329a0. [DOI] [PubMed] [Google Scholar]
- 35.Masui O., Heard E. RNA and protein actors in Xchromosome inactivation. Cold Spring Harb. Symp. Quant. Biol. (2006);71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- 36.Matzke M., Matzke A.J., Kooter J.M. RNA: guiding gene silencing. Science. (2001);293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 37.McCarrey J.R., Dilworth D.D. Expression of Xist in mouse germ cells correlates with X-chromosome inactivation. Nat. Genet. (1992);2:200–203. doi: 10.1038/ng1192-200. [DOI] [PubMed] [Google Scholar]
- 38.McClintock B. Induction of instability at selected loci in maize. Genetics. (1953);38:579–599. doi: 10.1093/genetics/38.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogawa Y., Lee J.T. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell. (2003);11:731–743. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. (2004);303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 41.Pal-Bhadra M., Leibovitch B.A., Gandhi S.G., Rao M., Bhadra U., Birchler J.A., Elgin S.C. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. (2004);303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 42.Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. (1996);379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 43.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. (2003);300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 44.Plath K., Talbot D., Hamer K.M., Otte A.P., Yang T.P., Jaenisch R., Panning B. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J. Cell Biol. (2004);167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rank G., Prestel M., Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. (2002);22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rastan S., Robertson E.J. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J. Embryol. Exp. Morphol. (1985);90:379–388. [PubMed] [Google Scholar]
- 47.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007);129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sado T., Wang Z., Sasaki H., Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. (2001);128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 49.Silva J., Mak W., Zvetkova I., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed- Enx1 polycomb group complexes. Dev. Cell. (2003);4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 50.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. (2004);305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 51.Sun B.K., Deaton A.M., Lee J.T. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell. (2006);21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 52.Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. (2009);19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tano K., Mizuno R., Okada T., Rakwal R., Shibato J., Masuo Y., Ijiri K., Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. (2010);584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. (2010);39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. (2010);329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tseng J.J., Hsieh Y.T., Hsu S.L., Chou M.M. Metastasis associated lung adenocarcinoma transcript 1 is upregulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol. Hum. Reprod. (2009);15:725–731. doi: 10.1093/molehr/gap071. [DOI] [PubMed] [Google Scholar]
- 57.Unhavaithaya Y., Hao Y., Beyret E., Yin H., Kuramochi-Miyagawa S., Nakano T., Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. (2009);284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. (2002);297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 59.White W.M., Willard H.F., Van Dyke D.L., Wolff D.J. The spreading of X inactivation into autosomal material of an x;autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am. J. Hum. Genet. (1998);63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilusz J.E., Freier S.M., Spector D.L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. (2008);135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woo C.J., Kingston R.E. HOTAIR lifts noncoding RNAs to new levels. Cell. (2007);129:1257–1259. doi: 10.1016/j.cell.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 62.Wutz A., Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. (2000);5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 63.Yin H., Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. (2007);450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]


