Abstract
Some genetic studies indicate that plant homologues of proteins involved in chromatin modification and remodeling in other organisms may regulate plant development. Previously, we described an Arabidopsis mutant with altered cold-responsive gene expression (acg1) displaying a late flowering phenotype, a null allele of fve. FVE is a homologue of the mammalian retinoblastoma-associated protein (RbAp), one component of a histone deacetylase (HDAC) complex involved in transcriptional repression, and has been shown to be involved in the deacetylation of the FLOWERING LOCUS C (FLC) chromatin encoding for a repressor of flowering. In an effort to gain insight into the biochemical functions of FVE, we overexpressed FVE tagged with the hemagglutinin (HA) and FLAG epitope at the N-terminus in acg1 mutants. The results of physiological and molecular analyses demonstrated that FVE overexpression in acg1 rescued the mutant phenotypes, including late flowering and alterations in floral pathway gene expression such as FLC, SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), and FLOWERING LOCUS T (FT), and also super-induced cold-responsive reporter gene expression. The chromatin immunoprecipitation experiments revealed the amplification of specific DNA regions of FLC and COLD-REGULATED 15A (COR15A), indicating that FVE may bind to the FLC and COR15A chromatin. Gel-filtration chromatography and the immunoprecipitation of putative FVE complexes showed that FVE forms a protein complex of approximately 1.0 MDa. These results demonstrate that FVE may exist as a multiprotein complex, similar to the mammalian HDAC complex harboring RbAp, to regulate flowering time and cold response by associating with the FLC and COR chromatin.
Keywords: chromatin, flowering, FVE, histone deacetylase, retinoblastoma-associated protein
INTRODUCTION
The transition to flowering is genetically controlled by several pathways, and involves the integration of environmental cues and the endogenous developmental state of the plant. Five major pathways regulate floral transition (Ausin et al., 2005; Boss et al., 2004; Putterill et al., 2004). These include photoperiod, light quality, ambient temperature cues, gibberellin hormone signaling, and autonomous pathways. Signals from these five flowering regulatory pathways converge into the floral pathway integrator genes, FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), and LEAFY (LFY), which promote floral transition.
The autonomous pathway promotes flowering by repressing FLC, which encodes for a MADS box transcriptional regulator which acts as a floral repressor through the repression of the floral pathway integrators FT, SOC1, and LFY (Boss et al., 2004). Seven autonomous pathway genes, FCA, FLOWERING LOCUS D (FLD), FLOWERING LOCUS K (FLK), FPA, FVE, FY, and LUMINIDEPENDENS (LD), have been identified in Arabidopsis thaliana (Simpson, 2004). FCA, FPA, and FLK harbor RNA-binding domains and may interact with RNA (Lim et al., 2004; Macknight et al., 1997; Schomburg et al., 2001). FY is a protein partner of FCA and is involved in the 3’ processing of mRNA (Simpson et al., 2003). LD is a homeodomain protein (Lee et al., 1994). LD binds to SUPPRESSOR OF FRIGIDA4 (SUF4) in the absence of FRI, thereby preventing SUF4 from acting on FLC (Kim et al., 2006). Two components of the autonomous pathway, FLD and FVE, are similar to the components of the mammalian histone deacetylase (HDAC) 1, 2 co-repressor complexes involved in the repression of gene expression via the deacetylation of histone residues (Ausin et al., 2004; He et al., 2003; Kim et al., 2004). FLD, a homologue of human KIAA0601, is similar to the polyamine oxidases and harbors the SWI3p, Rsc8p, and Moira (SWIRM) domains, which are found in the enzymes associated with chromatin remodeling (He et al., 2003).
FVE is a homologue of the yeast protein multicopy suppressor of IRA1 (MSI1) and of the mammalian retinoblastomaassociated proteins RbAp46 and RbAp48 (Qian and Lee, 1995) and is also referred to as AtMSI4 or the nucleosome/chromatin assembly factor group C4 (NFC4) (Ausin et al., 2004; Hennig et al., 2005; Kim et al., 2004). In the fve mutants, the FLC chromatin was enriched in acetylated histones H3 and H4, thereby indicating that FVE is required for histone deacetylation at the FLC chromatin (Ausin et al., 2004). A similar result was obtained with the fld mutants, which indicates that both FVE and FLD negatively regulate FLC expression via the histone deacetylation of FLC chromatin as a component of the HDAC complex (He et al., 2003). It is, however, not currently known whether FVE and FLD are recruited to FLC chromatin as a protein complex. We previously isolated an Arabidopsis mutant with altered cold-responsive gene expression harboring four copies of a cold responsive C-repeat/dehydration-responsive element (CRT/DRE) fused to the GUS (β-glucuronidase) reporter construct, and determined that acg1 is a null allele of fve, and that FVE negatively regulates the CRT/DRE-binding factor/ DRE-binding protein (CBF/DREB) cold response pathway as well as FLC (Kim et al., 2004).
In this study, we demonstrate, using chromatin immunoprecipitation and gel-filtration chromatography, that FVE is associated with the promoter and the intron region of the FLC chromatin and the promoter, the intron and the exon regions of the COR15A chromatin, a representative CBF/DREB target, and forms a protein complex of approximately 1.0 MDa in size. This result indicates that FVE can bind to the FLC chromatin as well as the COR15A chromatin as a large multiprotein complex, thereby regulating both flowering time and cold response.
MATERIALS AND METHODS
Plant growth conditions
4CRT/DRE-GUS and acg1 have been described previously (Kim et al., 2002; 2004) and fve-3 and fca-9 were obtained from the Arabidopsis Biological Resource Center. We employed the Columbia-0 ecotype in all experiments. Arabidopsis was grown on 0.5× Murashige-Skoog (MS) agar media under long-day conditions (16 h light-8 h dark) at 23℃ as previously described (Kim et al., 2002), and employed for various measurements. For flowering time analysis, the plants were grown and measured as described previously (Kim et al., 2004). Time to flowering was measured as the number of rosette leaves formed on the main shoot when the main stem had bolted 1 cm.
Plasmid construction and Arabidopsis transformation
To generate FVE-overexpressing acg1 plants, the DNA fragment encoding for the full-length FVE protein was PCR-amplified from the plasmid harboring FVE cDNA (RINKEN RAFL09-46-N10) using the primers FVE F1 (5′-ATGAAAGA AAGTGGGAAGAA-3′) and FVE R2 (5′-TTAAGGCTTGGAGG CACAA-3′). The amplified DNA products were subsequently digested with BamHI and SacI, then cloned into the pBluescript II KS (pBSIIKS) (-) vector (Stratagene), yielding pBSIIKS(-)- FVE. The DNA fragment encoding for the HA (amino acids: YPYDVPDYA) and FLAG epitope (amino acids: DYKDDD DKG) were ligated in tandem into pBSIIKS(-)-FVE at the 5′-end of the FVE DNA. The HA:FLAG:FVE DNA fragment was excised and ligated into the pBI121 (Clontech) vector, yielding Pro35S:HA:FLAG:FVE. Pro35S:HA:FLAG:FVE was introduced into acg1 by Agrobacterium-mediated transformation. The acg1 overexpressing HA:FLAG:FVE was isolated on the basis of complementation of the late flowering phenotype of the parental-type acg1. The homozygous lines were isolated via PCR analysis of the transgene FVE and amplified.
RNA-gel blot analysis and immunoblot analysis
RNA-gel blot analysis was conducted with 20 μg of total RNA extracted with TRI reagent (Molecular Research Center) as described previously (Kim et al., 2004). For immunoblot analysis, 50 μg of total proteins extracted by standard procedures (Gusmaroli et al., 2004) were separated on 10% SDS-PAGE and transferred to Immuno-blot PVDF membranes (Bio-Rad, USA), and then detected with ECL™ in conjunction with the Western blotting detection system (GE Healthcare, UK).
Real time RT-PCR analysis for the quantification of FLC, SOC1, and FT expression
Real-time RT-PCR was carried out using a QuantiTect SYBR Green RT-PCR kit (Qiagen) in a Rotor-Gene 2000 real-time thermal cycling system (Corbett Research) as previously described (Jeon et al., 2010). Real-time RT-PCR conditions and primer sequences are shown in Supplementary Table 1.
Analysis of GUS expression
We conducted quantitative GUS assays in tissue extracts by fluorometric measurements of the 4-methylumbelliferone (MU) generated from the β-D-glucuronide precursor, as previously described (Jefferson and Wilson, 1991).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were conducted essentially as previously described (Gendrel et al., 2005). Twoweek old plants grown on 0.5× MS agar medium were harvested. Two grams of seedlings were cross-linked in 37 ml of 1% formaldehyde in vacuo, washed with distilled water, and ground in liquid N2. The ground seedlings were added to 30 ml of extraction buffer 1 [0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM β-mercaptoethanol, 0.1 mM phenylmethylsulphonylfluoride (PMSF)]. The slurry was then filtered through two layers of Miracloth (Calbiochem) and centrifuged for 20 min at 3,000 × g at 4℃. The pellet was resuspended in 1 ml of extraction buffer 2 (0.25 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF) and centrifuged for 10 min at 12,000 × g at 4℃. The pellet was subsequently resuspended in 300 μl of extraction buffer 3 (1.7 M sucrose, 10 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF), layered on top of a 300 μl cushion of extraction buffer 3, and centrifuged for 1 h at 12,000 × g at 4℃. The chromatin pellet was then resuspended in 500 μl of ice-cold nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, 1 mM PMSF), sonicated on ice, and centrifuged for 10 min at 12,000 × g at 4℃. The supernatant (300 μl) was subsequently added to 2700 μl of ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl) and precleared with equilibrated Protein A-agarose beads (Upstate Biotechnology). After centrifugation, 1 ml of the supernatant was immunoprecipitated with anti-HA antibody (Santa Cruz Biotechnology). The immunoprecipitated DNA was recovered by ethanol precipitation and resuspended in 30 μl of distilled water. PCR analysis was conducted with the immunoprecipitated DNA using Takara Ex-taq polymerase under the following conditions: the mixtures were melted at 94℃ for 5 min followed by 30 cycles of 94℃ of 30 s, 56℃ of 45 s, 72℃ of 30 s, and a final 7-minute extension at 72℃ for FLC and 35 cycles at 94℃ for 30 s, 50℃ for 45 s, 72℃ for 30 s, and a final 7-minute extension at 72℃ for COR15A. The products were visualized with ethidium bromide in 2% agarose gel. Quantitative real-time PCR analysis was conducted using SsoFast EvaGreen Supermix (Bio-Rad, USA) on a CFX96 real-time system machine (Bio-Rad, USA) under the following conditions: the mixtures were melted for 5 min at 65℃ to 94℃ followed by 30 s at 95℃, 2 s at 95℃, 5 s at 56℃ for FLC and 95℃ for 30 s, 95℃ for 2 s, and 50℃ for 2 s for COR15A. The primer sequences used for all ChIP analyses are described in Supplementary Table 2.
Gel-filtration chromatography and immunoprecipitation
Two-week-old Pro35S:HA:FLAG:FVE plants were harvested after growing on 0.5× MS agar medium. Ground tissues were homogenized in EB1 extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2.5 mM EDTA, 1mM DTT, 10% glycerol, 0.1% Nonidet P-40) with freshly added protease inhibitors, 1 mM PMSF, protease inhibitor cocktail (3 μg ml-1 pepstatin, 3 μg ml-1 leupeptin, 5 μg ml-1 aprotinin), and the metalloprotease inhibitor, 2 mM o-phenanthrolin hydrochloride monohydrate. The homogenates were microcentrifuged twice for 15 min and the supernatants were filtered through 0.2 μm filters (PALL, USA). For gel fractionation analyses, 100 mg of total proteins were loaded onto a Sephacryl S-300 (HiPrep16/60) gel-filtration column (GE Healthcare, Sweden). The column was then equilibrated with 240 ml of EB1 with final concentrations of 1 mM PMSF and 25 mM β-glycerophosphate. The proteins were eluted in the same buffer at a flow rate of 0.2 ml min-1. All the procedures were carried out at 4℃. One-ml fractions were collected, finishing with 120 fractions. The fractions were concentrated using 20% trichloroacetic acid and washed in acetone at least three times. Equal volumes of each fraction were boiled for 5 min in SDS sample buffer, separated by 10% SDS-PAGE, and transferred onto PVDF Immobilon membranes (Bio-Rad, USA). The elute was incubated overnight with 50 μl of anti-FLAG M2 affinity resin (Sigma) at 4℃ on a rotary shaker. The resins were washed three times in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0% CA-630, 0.5% sodium deoxycholate, 0.1% SDS) and twice in TBS buffer. The immunoprecipitated proteins were eluted with 3× FLAG peptide (Sigma) and loaded on 12% SDS-PAGE and silver-stained.
MALDI-TOF analysis and database search
The FVE complexes immunoprecipitated with anti-FLAG antibody (Sigma) were loaded on 12% SDS-PAGE and silverstained. The protein band corresponding to FVE at 60 kDa was enzymatically digested in-gel using modified porcine trypsin as described previously (Shevchenko et al., 1996). Gel pieces were washed in 50% acetonitrile, dried, rehydrated with trypsin (8-10 ng μl-1), and incubated overnight at 37℃. The proteolytic reaction was terminated by the addition of 5 μl of 0.5% trifluoroacetic acid. Tryptic peptides were recovered by combining the aqueous phase from several extractions of gel pieces with 50% aqueous acetonitrile. The peptide mixture was concentrated using a SpeedVac concentrator and desalted using C18ZipTips (Millipore), and then the peptides were eluted in 1 to 5 μl of acetonitrile. An aliquot of this solution was mixed with an equal volume of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% aqueous acetonitrile, after which 1 μl of mixture was spotted onto a target plate. Protein analysis was carried out using a Voyager DE-STR MALDI-TOF mass spectrometer (PerSeptive Biosystems, USA). Mass spectra were obtained in the positive-ion reflector mode with delayed extraction. Spectra were calibrated with angiotensin I (1296.79 Da), adrenocorticotropic hormone (ACTH clip 1-17, 2093.01 Da), and ACTH clip 18-39 (2465.20 Da) as internal standards. The Swiss-Prot database using the MS-Fit search program (http://prospector.ucsf.edu/prospector/mshome.htm) was used to search the MALDIpeptide- mass fingerprinting data.
RESULTS
Overexpression of FVE in acg1 mutants
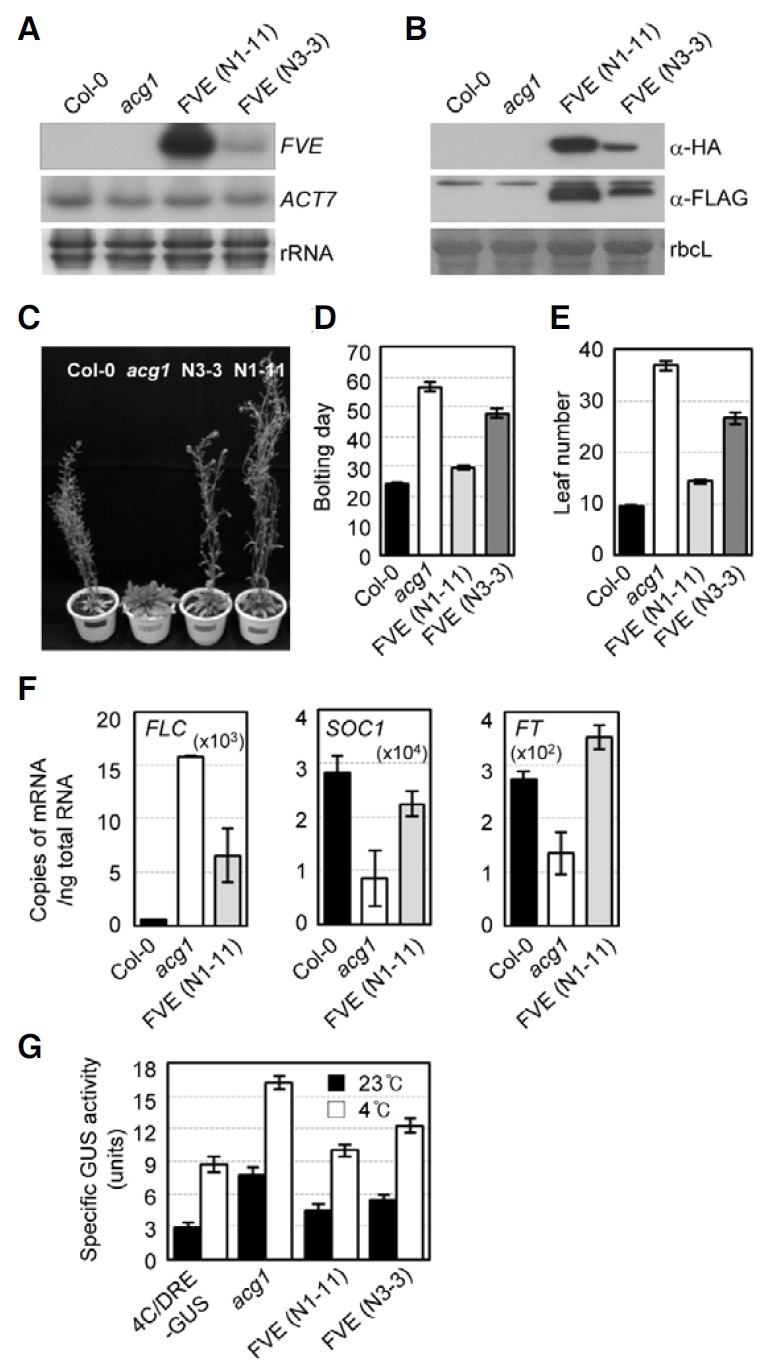
In order to determine whether FVE binds to the FLC chromatin by the formation of a protein complex, we generated transgenic Arabidopsis plants that overexpressed the FVE proteins tagged with the HA and FLAG epitope in tandem at the N-terminus under the control of the CaMV 35S promoter in acg1 mutants, Pro35S:HA:FLAG:FVE/acg1, and then isolated two independent transgenic lines (N1-11 and N3-3). RNA-gel blot analysis using a FVE DNA probe showed that both lines, N1-11 and N3-3, overexpressed the FVE mRNA, and higher FVE mRNA levels were expressed in N1-11 as compared with N3-3 (Fig. 1A). Immunoblot analysis with antibodies against the HA or FLAG epitope demonstrated that the HA and FLAG-tagged FVE proteins are produced at high levels in Pro35S:HA:FLAG:FVE/acg1 Arabidopsis plants lacking endogenous FVE proteins (Fig. 1B). The late flowering phenotype of acg1 was complemented to the wild-type in the N1-11 line and to a significant degree in N3-3, which correlates with FVE expression levels (Figs. 1C-1E). We previously noted markedly elevated FLC expression levels in acg1 with consequent significant reductions in the expression of SOC1 and FT via negative regulation by FLC (Kim et al., 2004). Therefore, we analyzed the expression of FLC as well as SOC1 and FT in the N1-11 line by real-time RT-PCR, and demonstrated that FLC expression in acg1 was significantly reduced and that SOC1 and FT expression were increased to almost wild-type levels by FVE overexpression, evidencing a complementation of mutant phenotypes (Fig. 1F). The fact that FLC expression was not fully rescued to the wild-type via ectopic expression of FVE under CaMV 35S promoter might be attributable to the different levels of tissue expression of FVE under the control of the CaMV 35S promoter and that of FLC, as, for example, FLC is expressed most abundantly in the vegetative apex (Michaels and Amasino, 1999) as opposed to the expression of the GUS reporter gene fused to the CaMV 35S promoter in the larger proportion of phloem-associated cells in roots and stems as compared to leaves (Jefferson et al., 1987). However, the reduced FLC expression induced by ectopic FVE expression in Pro35S:HA:FLAG:FVE/acg1 plants proved sufficient to release the repression of SOC1 and FT by FLC in acg1 mutants to wild-type levels. In addition, we observed no phenotypes other than flowering time. Finally, we determined that FVE rescued the increased expression of the cold-responsive GUS reporter gene in acg1 with and without the application of cold treatment (Fig. 1G). Collectively, these results demonstrated that FVE can negatively regulate both FLC expression and cold-responsive gene expression via CRT/DRE.
Fig. 1. Overexpression of HA:FLAG:FVE in acg1. (A) RNA-gel blot analysis of acg1 overexpressing HA: FLAG:FVE. Total RNAs isolated from 10-day-old plants, Col-0, acg1, and two lines [FVE (N1-11) and FVE (N3- 3)] of Pro35S:HA:FLAG:FVE/acg1, were subjected to RNA-gel blot analysis with FVE or ACTIN7 DNA probe. (B) Immunoblot analysis of acg1 overexpressing HA: FLAG:FVE. Total proteins extracted from 10-day-old plants, Col-0, acg1, and two l ines of Pro35S: HA:FLAG: FVE/acg1, were subjected to immunoblot analysis with anti-HA or anti-FLAG antibody. The rbcL protein encoding for the large subunit of rubisco is shown as a loading control. (C) Eight-week-old Col-0, acg1, and acg1 overexpressing HA:FLAG:FVE. Plants were grown for eight weeks under long-day conditions and photographed. (DE) Bolting day (D) and the rosette leaf numbers (E) of Col-0, acg1, and acg1 overexpressing HA:FLAG:FVE. n ≥ 16. (F) Real-time RT-PCR analysis of FLC, SOC1, and FT from Col-0, acg1, and acg1 overexpressing HA:FLAG:FVE. Total RNAs isolated from 10-day-old plants, Col-0, acg1, and Pro35S:HA:FLAG:FVE/acg1 (N1-11), were quantified by real-time RT-PCR. Copies of the transcripts were plotted per ng of total RNA after the normalization of ACTIN7 RNA. Multiplication of the number in the brackets inside the graph by the number in the y-axis generates copy numbers of the corresponding transcripts. The mean values and standard errors from biological triplicate experiments were plotted. (G) Fluorometric quantification of the GUS activities of 4CRT/ DRE-GUS, acg1, and acg1 overexpressing HA:FLAG: FVE. Ten-day-old plants were used for fluorometric quantification of GUS activities. Specific activities (units) are expressed as the pmol reaction product (4-methylumbelliferone) generated per min per mg of total protein. The data are expressed as the means ± standard errors of three independent treatments. The open column and closed column indicate the sample treated at 4℃ for 2 days and the sample treated at 23℃, respectively.
Analysis of FVE binding to FLC and COR15A chromatin by ChIP assays
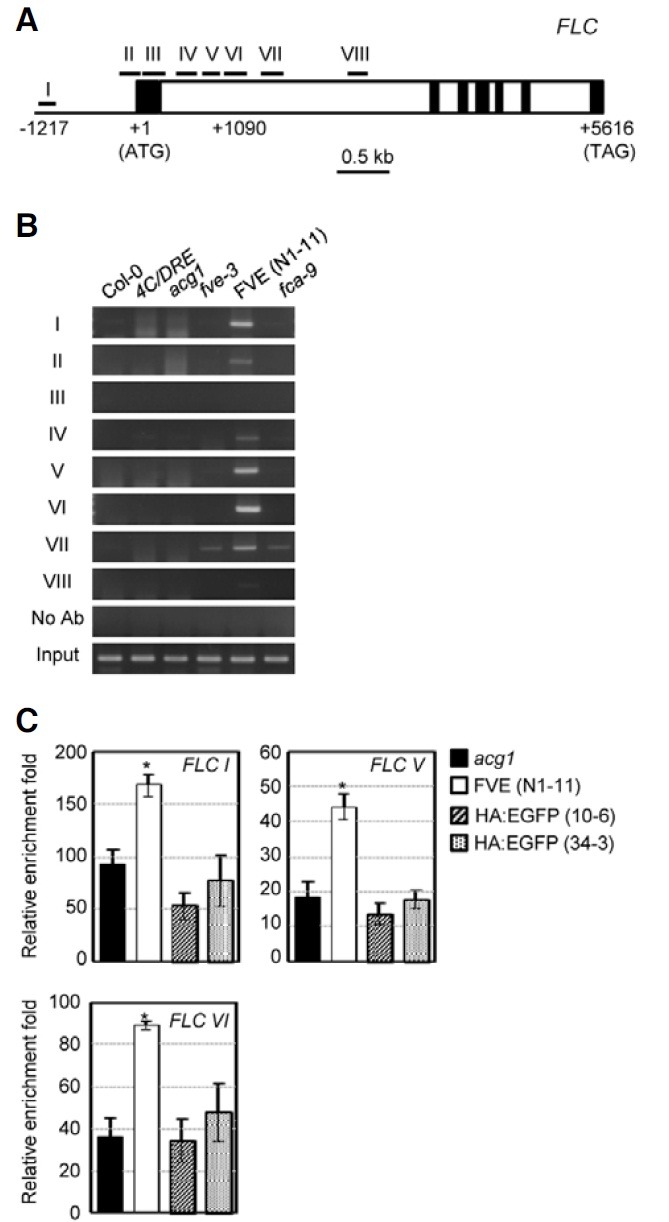
We then employed ChIP assays to determine whether FVE can be recruited to the FLC chromatin. The chromatins of the Pro35S:HA:FLAG:FVE/acg1 N1-11 line along with various controls, Col-0, 4CRT/DRE-GUS, acg1, fve-3, and fca-9 plants, were immunoprecipitated with anti-HA antibody. We amplified eight DNA fragments spanning the promoter, the first exon, and the first intron of FLC from the precipitated chromatin (Figs. 2A and 2B). Specific DNA fragments were amplified in the promoter region (I), the region close to the translation initiation point (II), and the first intron (IV-VI) of FLC. By contrast, we noted no amplification of the DNA fragments in these regions from Col-0, acg1, and other mutant plants. In order to eliminate the possibility that the precipitation of the chromatin might be attributable to the overexpression of nonspecific proteins, we employed additional control Arabidopsis plants that overexpressed modified green fluorescent protein (EGFP) tagged with the HA epitope in the N-terminus under the control of the CaMV 35S promoter in these ChIP assays. We selected two lines of transgenic Pro35S:HA:EGFP plants that exhibited the highest expression of the HA:EGFP transcripts among 13 lines generated (data not shown). We carried out quantitative PCR analysis for three genomic DNA regions, in which the most significant amplification of the DNA fragments was assessed without nonspecific PCR products in the ChIP assays (Fig. 2C). We noted significant enrichments in these three DNA regions from the Pro35S:HA:FLAG:FVE/acg1 plants but not from the two Pro35S: HA:EGFP lines. (Fig. 2C). These results indicate that FVE is recruited to promoter region I and intron regions V and VI of the FLC chromatin.
Fig. 2. ChIP analyses of the FLC chromatin in acg1 overexpressing HA:FLAG:FVE. (A) Schematic structure of the FLC region. I to VIII represent the regions in which FVE binding was evaluated by a ChIP. The translation initiation point is +1. The filled boxes represent exons and the open boxes represent introns. (B) ChIP analyses of FVE binding to the FLC chromatin. No Ab indicates ChIP carried out without antibody. The input is the chromatin prior to immunoprecipitation. ChIP analyses were carried out with three biological replicates; representative PCR data are shown. (C) Quantitative PCR analysis of FVE binding to the FLC chromatin compared with EGFP. Genomic DNA obtained by ChIP using HA antibody in acg1 plants overexpressing HA:FLAG:FVE or acg1, or HA:EGFP-overexpressing plants were analyzed by quantitative PCR. Values are expressed as the means of triplicate biological replications with error bars indicating standard errors. *denotes statistically significant changes with p < 0.05 compared with the acg1 control.
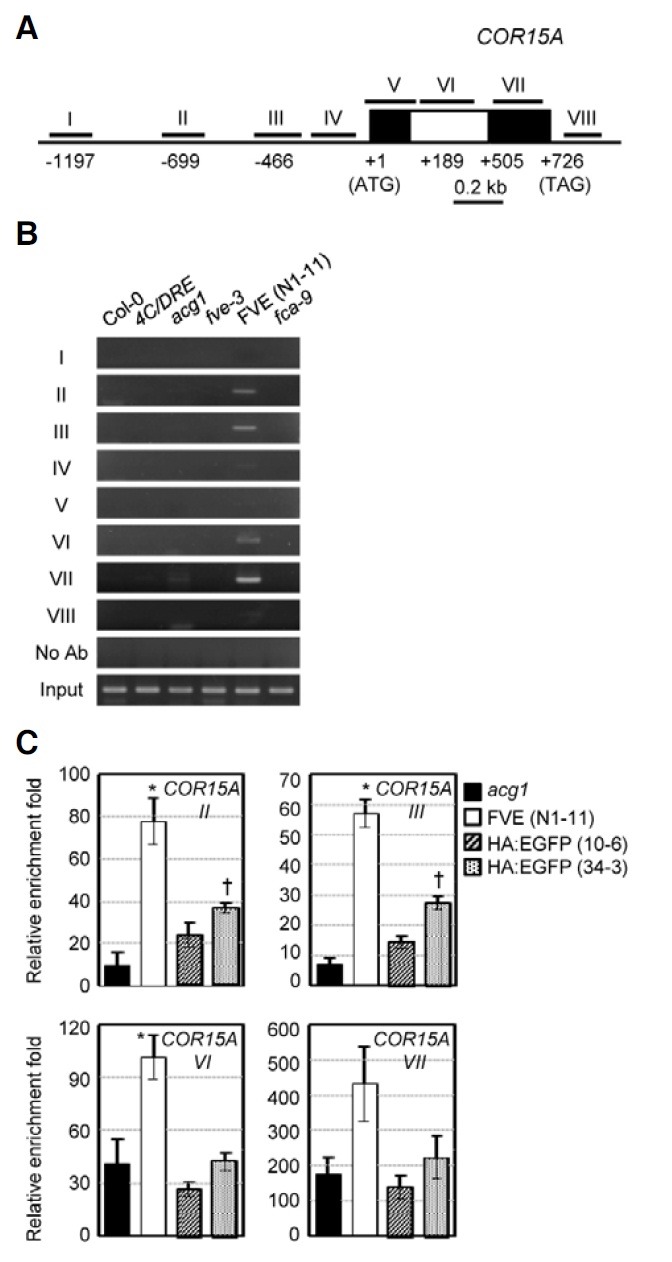
In the acg1 mutants, COR15A is upregulated and superinduced in response to cold (Kim et al., 2004). Ectopic FVE expression in acg1 partially complemented the elevated expression of GUS with and without cold treatment through CRT/DRE (Fig. 1G), thereby indicating that FVE may be associated with COR15A chromatin, like FLC, to regulate cold response. To assess this possibility, ChIP was also conducted with eight DNA fragments spanning the promoter to the coding region of COR15A (Fig. 3A). The amplification of specific DNA fragments was noted in the promoter regions (II and III) and the intron (VI) and the second exon (VII) regions (Fig. 3B). No DNA fragments in these regions were detected in any other control plants. The transgenic Pro35S:HA:EGFP lines were also employed as additional controls to further verify the binding of FVE to the COR15A chromatin. As shown in Fig. 3C, we observed greater enrichment of the DNA fragments in regions II, III, VI, and VII from Pro35S:HA:FLAG:FVE/acg1 plants relative to those of the two Pro35S:HA:EGFP lines upon quantitative PCR analysis of the ChIP assays. These data indicate that FVE is recruited to the COR15A chromatin.
Fig. 3. ChIP analyses of the COR15A chromatin in acg1-overexpressing HA:FLAG:FVE. (A) The schematic structure of the COR15A region. (B) ChIP analyses of FVE binding to the COR15A chromatin. (C) Quantitative PCR analysis of FVE binding to the COR15A chromatin compared with EGFP. Experiments were conducted as described in Fig. 2C.
Analysis of FVE complex by gel-filtration chromatography
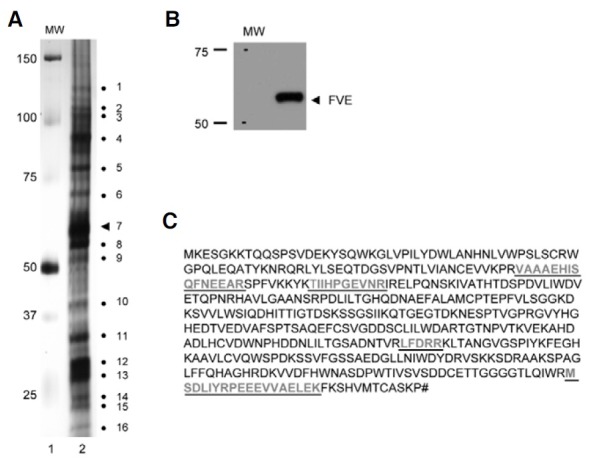
We next attempted to determine, by gel-filtration chromatography, whether or not FVE forms a multiprotein complex. We extracted total proteins from Pro35S:HA:FLAG:FVE/acg1 plants that lacked endogenous FVE proteins, but generated high levels of the HA and FLAG-tagged FVE proteins, and conducted gel-filtration chromatography of the extracted proteins followed by immunoblot analyses of each fraction with HA antibody (Fig. 4). The results showed that FVE is eluted at a size of approximately 1.0 MDa in addition to a monomer. The combined fractions corresponding to 1.0 MDa of FVE were pooled from five rounds of gel-filtration chromatography and subjected to immunoprecipitation with FLAG antibody to further verify the presence of FVE in the complex and to detect the polypeptides associated with FVE. After running the immunoprecipitates on SDS-PAGE and silver-staining, 15 polypeptides ranging from 25 to 150 kDa in size (Fig. 5A) were revealed, in addition to an FVE band (indicated by dot number 7, Fig. 5A) by immunoblot analysis (Fig. 5B). The FVE band was corroborated further by peptide sequencing (bold letters underlined in Fig. 5C). The protein fractions corresponding to 1.0 MDa acquired from the acg1 plants exhibited no significant bands on SDS-PAGE and silver staining after immunoprecipitation with FLAG antibody (data not shown). These results show that FVE exists as a multiprotein complex in Arabidopsis.
Fig. 4. Gel-filtration chromatography of the total protein extracts from acg1 overexpressing HA:FLAG:FVE. (A) Chromatogram of the total protein extracts from acg1 overexpressing HA:FLAG:FVE. The total proteins from two-week-old Pro35S:HA:FLAG:FVE/acg1 (N1-11) plants were separated on a Sephacryl S-300 gel filtration column and fractionated at 1 ml. The eluted proteins were monitored at 280 nm. (B) Immunoblot analyses of each fraction received from gel-filtration chromatography. Equal volumes of each fraction were separated by 10% SDS-PAGE and subjected to immunoblot analysis with HA antibody (A). Each fraction was stained by Coomassie blue dye (B).
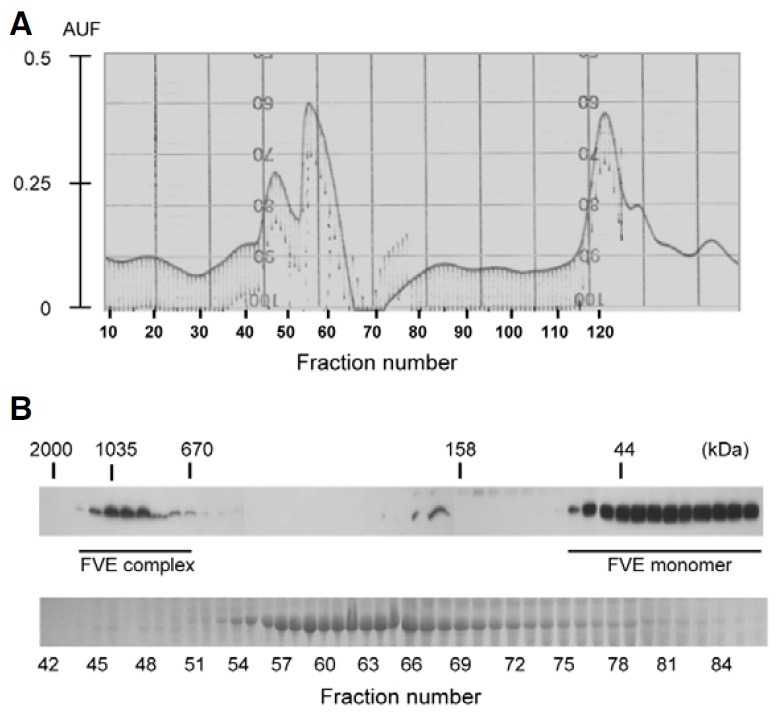
Fig. 5. SDS-PAGE analysis of proteins associated in the FVE complex. (A) SDS-PAGE analysis of the fractions corresponding to the FVE complexes after immunoprecipitation with FLAG antibody. The FVE protein complexes collected by gel-filtration chromatography from 44 to 52 fractions were incubated with 50 μl of anti-FLAG M2 affinity resin, precipitated, and eluted with the 3× FLAG peptide. The elutes (lane 2) were then loaded on 12% SDS-PAGE and silver-stained. The numbers to the left are the sizes of the molecular weight marker proteins (lane 1). Numbers to the right show the apparent molecular weights of the polypeptides detected by silver staining in decreasing order (lane 2). The triangle numbered 7 indicates FVE. (B) Immunoblot analysis of the FVE complexes immunoprecipitated with FLAG antibody. The immunoprecipitated FVE protein complexes were loaded onto 12% SDS-PAGE and subjected to immunoblot analysis with HA antibody. (C) Peptide sequence of FVE detected by MALDI-MS. The FVE complexes immunoprecipitated with FLAG were used for 1-D gel electrophoresis. The number 7 band (A) was identified as FVE. Underlined bold letters indicate the peptides detected by MALDI-MS. The matched peptides represent 11% amino acid sequence coverage.
DISCUSSION
Hyperacetylation was noted on histones H3 and H4 of the FLC chromatin in fve mutants, suggesting that FVE might be a component of an HDAC transcriptional co-repressor complex, and negatively regulate the FLC chromatin (Ausin et al., 2004). In order to function as a component of the HDAC complex modifying the FLC chromatin, FVE should be associated with FLC chromatin. To evaluate this model, we overexpressed FVE tagged with the HA and FLAG epitopes in acg1 mutants lacking endogenous FVE and conducted ChIP assays. Our results showed that FVE can be recruited to the promoter and the first intron of the FLC chromatin. The locations of FVE in these regions defined by our ChIP assays are consistent with the hyperacetylation sites of the FLC chromatin in fve mutants (He et al., 2003). We also demonstrated that FVE is recruited to the COR15A chromatin in the promoter regions, the first intron, and the second exon, thus suggesting that FVE regulates flowering time and cold response by modulating the chromatin state. Considering that COR15A promoter regions III and IV (Fig. 2) harbor CRT/DRE cis-acting elements to which CBF/DREB transcription factors bind (Stockinger et al., 1997), the HDAC complex containing FVE might deacetylate the histone proteins around the regions to which CBF/DREB binds for the regulation of COR15A expression.
In mammalian systems, the Rb protein represses the E2F family of transcription factor-mediated transcription by recruiting a histone deacetylase (Luo et al., 1998; Magnaghi-Jaulin et al., 1998). A maize HDAC was also shown to interact physically with Rb-related proteins to repress transcription (Rossi et al., 2003). Mammalian RbAp48 is a component of the HDAC1 recruited by RB (Nicolas et al., 2000). In yeast, the Sin3/RPD3- HDAC complex exists as a large protein complex (larger than 2 MDa in size) (Alland et al., 1997; Hassig et al., 1997; Heinzel et al., 1997; Kasten et al., 1997; Laherty et al., 1997; Nagy et al., 1997; Taunton et al., 1996; Wade et al., 1998; Zhang et al., 1997). The mammalian Sin3 complex consists of at least seven subunits, including HDAC1, HDAC2, RbAp48, RbAP46, and other polypeptides (Zhang et al., 1998). FVE, which is homologous to RbAp46 and RbAp48, interacted with the maize Rb protein in the immunoprecipitation assays, thereby indicating that FVE might be a component of the retinoblastoma-containing complexes in Arabidopsis (Ausin et al., 2004). These results, coupled with the fact that FVE is not a specific DNAbinding protein, predicted that FVE should form a multiprotein complex to be recruited to the chromatin. In order to determine whether or not FVE forms a large protein complex, we employed gel-filtration chromatography with total protein extracts harboring FVE tagged with the HA and FLAG epitopes that were overexpressed in acg1 mutants. Consistent with that prediction, we showed that FVE is a component of a large (1.0 MDa) multiprotein complex. The immunoprecipitation of these complexes with FLAG antibody and SDS-PAGE analysis demonstrated that the complex consists of 16 polypeptides, including FVE. A very recent study showed that FVE functionally interacts with the cullin-RING ubiquitin ligase (CUL4-DDB1) and a CLF-Polycomb Repressive Complex 2 (PRC2) at the FLC chromatin, thereby regulating flowering time in Arabidopsis (Pazhouhandeh et al., 2011). It will be interesting to determine the molecular identities of the components that form a complex with FVE isolated herein, and to elucidate their biochemical functions, thereby shedding light on the mechanism by which the chromatin state is regulated by histone deacetylation in response to environmental signals and developmental cues.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
This work was supported by a grant from the World Class University project funded by the Ministry of Education, Science, and Technology of Korea (grant no. R31-2009-000-20025-0) to J. Kim. We thank Su-Jin Ku and Min-Jung Kim for the plasmid construct, Pro35S:HA:FLAG:FVE and Pro35S:HA:EGFP transgenic lines, respectively.
References
- 1.Alland L., Muhle R., Hou H., Jr., Potes J., Chin L., Schreiber- Agus N., DePinho R.A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. (1997);387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Ausin I., Alonso-Blanco C., Jarillo J.A., Ruiz-Garcia L., Martinez- Zapater J.M. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. (2004);36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 3.Ausin I., Alonso-Blanco C., Martinez-Zapater J.M. Environmental regulation of flowering. Int. J. Dev. Biol. (2005);49:689–705. doi: 10.1387/ijdb.052022ia. [DOI] [PubMed] [Google Scholar]
- 4.Boss P.K., Bastow R.M., Mylne J.S., Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell. (2004);16:S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gendrel A.V., Lippman Z., Martienssen R., Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. (2005);2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- 6.Gusmaroli G., Feng S., Deng X.W. The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell. (2004);16:2984–3001. doi: 10.1105/tpc.104.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassig C.A., Fleischer T.C., Billin A.N., Schreiber S.L., Ayer D.E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. (1997);89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 8.He Y., Michaels S.D., Amasino R.M. Regulation of flowering time by histone acetylation in Arabidopsis. Science. (2003);302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 9.Heinzel T., Lavinsky R.M., Mullen T.M., Soderstrom M., Laherty C.D., Torchia J., Yang W.M., Brard G., Ngo S.D., Davie J.R., et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. (1997);387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 10.Hennig L., Bouveret R., Gruissem W. MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. (2005);15:295–302. doi: 10.1016/j.tcb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Jefferson R.A., Wilson K.J. The GUS gene fusion system. Plant Mol. Biol. Manual. Kluwer Academic Publishers; Dordrecht: (1991). pp. 1–33.B14 [Google Scholar]
- 12.Jefferson R.A., Kavanagh T.A., Bevan M.W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. (1987);6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon J., Kim N.Y., Kim S., Kang N.Y., Novák O., Ku S.J., Cho C., Lee D.J., Lee E.J., Strnad M., et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. (2010);285:23371–23386. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasten M.M., Dorland S., Stillman D.J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol. Cell. Biol. (1997);17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H.J., Kim Y.K., Park J.Y., Kim J. Light signalling mediated by phytochrome plays an important role in coldinduced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. (2002);29:693–704. doi: 10.1046/j.1365-313x.2002.01249.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim H.J., Hyun Y., Park J.Y., Park M.J., Park M.K., Kim M.D., Lee M.H., Moon J., Lee I., Kim J. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. (2004);36:167–171. doi: 10.1038/ng1298. [DOI] [PubMed] [Google Scholar]
- 17.Kim S., Choi K., Park C., Hwang H.J., Lee I. SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell. (2006);18:2985–2998. doi: 10.1105/tpc.106.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laherty C.D., Yang W.M., Sun J.M., Davie J.R., Seto E., Eisenman R.N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. (1997);89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 19.Lee I., Aukerman M.J., Gore S.L., Lohman K.N., Michaels S.D., Weaver L.M., John M.C., Feldmann K.A., Amasino R.M. Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell. (1994);6:75–83. doi: 10.1105/tpc.6.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim M.H., Kim J., Kim Y.S., Chung K.S., Seo Y.H., Lee I., Kim J., Hong C.B., Kim H.J., Park C.M. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell. (2004);16:731–740. doi: 10.1105/tpc.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo R.X., Postigo A.A., Dean D.C. Rb interacts with histone deacetylase to repress transcription. Cell. (1998);92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 22.Macknight R., Bancroft I., Page T., Lister C., Schmidt R., Love K., Westphal L., Murphy G., Sherson S., Cobbett C., et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. (1997);89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 23.Magnaghi-Jaulin L., Groisman R., Naguibneva I., Robin P., Lorain S., Le Villain J.P., Troalen F., Trouche D., Harel- Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. (1998);391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 24.Michels S.D., Amasino R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. (1999);11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy L., Kao H.Y., Chakravarti D., Lin R.J., Hassig C.A., Ayer D.E., Schreiber S.L., Evans R.M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. (1997);89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 26.Nicolas E., Morales V., Magnaghi-Jaulin L., Harel-Bellan A., Richard-Foy H., Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J. Biol. Chem. (2000);275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- 27.Pazhouhandeh M., Molinier J., Berr A., Genschik P. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2011);108:3430–3435. doi: 10.1073/pnas.1018242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putterill J., Laurie R., Macknight R. It’s time to flower: the genetic control of flowering time. Bioessays. (2004);26:363–373. doi: 10.1002/bies.20021. [DOI] [PubMed] [Google Scholar]
- 29.Qian Y.W., Lee E.Y. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J. Biol. Chem. (1995);270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 30.Rossi V., Locatelli S., Lanzanova C., Boniotti M.B., Varotto S., Pipal A., Goralik-Schramel M., Lusser A., Gatz C., Gutierrez C., et al. A maize histone deacetylase and retinoblastoma- related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. (2003);51:401–413. doi: 10.1023/a:1022090916446. [DOI] [PubMed] [Google Scholar]
- 31.Schomburg F.M., Patton D.A., Meinke D.W., Amasino R.M. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. (2001);13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. (1996);68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 33.Simpson G.G. The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. (2004);7:570–574. doi: 10.1016/j.pbi.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Simpson G.G., Dijkwel P.P., Quesada V., Henderson I., Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. (2003);113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 35.Stockinger E.J., Gilmour S.J., Thomashow M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cisacting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA. (1997);94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taunton J., Hassig C.A., Schreiber S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. (1996);272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 37.Wade P.A., Jones P.L., Vermaak D., Wolffe A.P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. (1998);8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. (1997);89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Sun Z.W., Iratni R., Erdjument-Bromage H., Tempst P., Hampsey M., Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. (1998);1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]


