Abstract
Hoxc8 is a homeobox gene family member, which is essential for growth and differentiation. Mgl1, a mouse homologue of the Drosophila tumor suppressor gene lgl, was previously identified as a possible target of Hoxc8. However, the biological effects and underlying molecular mechanism of Hoxc8 regulation on Mgl1 has not been fully established. The endogenous expression patterns of Hoxc8 were inversely correlated with those of Mgl1 in different types of cells and tissues. Here we showed that Hoxc8 overexpression downregulated the Mgl1 mRNA expression. Characterization of the ~2 kb Mgl1 promoter region revealed that the upstream sequence contains several putative Hox core binding sites and chromatin immunoprecipitation assay confirmed that Hoxc8 directly binds to the 5′ upstream region of Mgl1. The promoter activity of this region was diminished by Hoxc8 expression but resumed by knockdown of Hoxc8 using siRNA against Hoxc8. Functional study of Mgl1 in C3H10T1/2 cells revealed a significant reduction in cell adhesion upon expression of Hoxc8. Taken together, our data suggest that Hoxc8 downregulates Mgl1 expression via direct binding to the promoter region, which in turn reduces cell adhesion and concomitant cell migration.
Keywords: cell adhesion, direct downstream target gene, Hoxc8, Mgl1, tumor suppressor
INTRODUCTION
Hox transcription factors are not only master regulators for the embryonic body pattern formation during animal development, but are also known to have regulatory functions in adults (Lee et al., 2008; Morgan, 2006; Pearson et al., 2005). Since it becomes increasingly evident that Hox plays crucial roles in numerous cellular processes such as cell division, adhesion, proliferation and apoptosis, it becomes more important to understand how Hox modulates the downstream transcriptional events under specific conditions. During mouse development, Hoxc8, a member of the Hox family of transcription factors, is expressed in both ectoderm-derived neural tube and mesoderm- derived somites, which are further restricted to the sclerotome (Shashikant and Ruddle, 1996). Although there has been no Hoxc8 natural mutant identified, deregulation of Hoxc8 expression has been reported in several different types of human cancers, such as prostate, cervical, and esophageal cancers, emphasizing the importance of Hoxc8 in cancer development (Alami et al., 1999; Chen et al., 2005; Miller et al., 2003).
Until recently, numerous attempts have been applied to determine the Hox downstream target genes by either low resolution analysis using traditional proteomics, ChIP, ChIP-PCR or high resolution analysis combined with microarray profiling (Chung et al., 2010; Kang et al., 2010; Kwon et al., 2003; Lei et al., 2005; 2006; 2007; Min et al., 2010). However, only a few have been identified thus far. Interestingly, long before these attempts, Tomotsune et al. (1993) isolated in vivo Hoxc8 target sequences from mouse spinal cords by screening immunopurified lambda gt10 library and identifying a clone containing a significantly homologous sequences to the Drosophila lethal (2) giant larvae (lgl) gene, after which “Mgl1” was named in mice. Drosophila lgl was originally discovered as a tumor suppressor gene and was found to be necessary for the maintenance of cell polarity and asymmetric cell division. Loss of lgl caused massive tumor-like overgrowth, tissue disorganization, and lethal phenotypes in both Drosophila and mice (Klezovitch et al., 2004; Vasiokhin, 2006). In addition, a human homologue Hugl1 was often lost or downregulated in a variety of human solid tumors, supporting its role as a tumor suppressor in humans as well (Grifoni et al., 2004; Kuphal et al., 2006). However, the significance of this incident in carcinogenesis, and the correlation between Hoxc8 and Mgl1 remains unknown (Grifoni et al., 2004; Kupha et al., 2006; Schimanski et al., 2005).
Since the in situ expression patterns of Hoxc8 and Mgl1 have been implied to be mutually exclusive in a previous report (Tomotsune et al., 1993) that indicated negative regulation by Hoxc8, we wanted to prove whether Mgl1 is a real downstream direct target gene of Hoxc8. First, we compared the in vivo expression pattern of Mgl1/Hugl1 and Hoxc8 in developing mouse embryos, tissues, and then further in human cancer cell lines. Direct binding of Hoxc8 to the promoter region of Mgl1 was confirmed by ChIP analysis and the functional activity of this binding sequence was demonstrated by the luciferase reporter assay. We further investigated the implications of Mgl1, a cytoskeletal protein, in cancer progression and the effects of Hoxc8 on this process.
MATERIALS AND METHODS
Animal preparation, tissue source and storage
ICR male mice about two months old were dissected to obtain the brain, heart, kidney, spleen and liver tissues under aseptic conditions. These tissues were collected in 1.5 ml eppendorf tubes and immediately frozen in liquid nitrogen and stored at -80℃ until we isolated the total RNA. To obtain E11.5 embryos, male and female ICR mice were caged together for mating around 6 pm. The next morning, when the vaginal plugs were present, we defined it as 0.5 day postcoitum (dpc) or E0.5 embryo. After 11 days, the pregnant female mice were sacrificed, and the E11.5 embryos were extracted. The maternal and extra- embryonic tissues were removed. The embryos were divided into five parts along the anterior-posterior axis and then used for total RNA preparation.
Cell cultures, transfection and Western analysis
All the cells we used in our studies were cultured in the recommended cell culture medium (Dulbecco’s Modified Eagles Medium; WelGENE Inc., Korea) supplemented with 10% FBS (Fetal bovine serum, WelGENE Inc., Korea) and 1× of penicillin- streptomycin from 100X stock (WelGENE Inc., Korea), at 37℃, 5% CO2.
The plasmid pcDNA3.1-Hoxc8 harboring mouse Hoxc8 gene has been previously described (Kwon et al., 2003). pcDNA3.1- Hoxc8 or pcDNA3.1 empty vector was transfected into C3H10T1/2 murine mesenchymal progenitor cells using Lipofectamine 2000™ reagent (Invitrogen, USA) as indicated by the manufacturer. To establish stable cell lines, pcDNA3.1-myc-His-Hoxc8 or control empty vector was transfected into NIH3T3 cells. Stable cells were selected in the presence of G418 antibiotic (GIBCO, Invitrogen, USA). The medium was changed every three days. Cells were subcultured when the cells reached 90% confluency.
For Western blot analysis, cells were lysed in lysis buffer (17081, iNtRON Biotechnology Inc.) and then protease inhibitor cocktail (P8340, Sigma-Aldrich) was added. Samples were quantified and equal amount of proteins were boiled for 5 min at 95℃. Proteins were resolved by SDS-PAGE, transferred to a membrane (Amersham Hybond™-P, GE Healthcare Limited, Amearshamplace, Little Chalfont, Buckinghamshire HP7 9NA) and subjected to immunoblotting with the Hoxc8 monoclonal antibody MMS-286R (Covance), or mouse monoclonal beta Actin antibody AC-15 (Abcam ab6276). Immunoreactive proteins were detected using SuperSignal West Pico Chemiluminescent Substrate (PIERCE).
Total RNA isolation and semi-quantitative RT-PCR
Total RNA was isolated using Trizol reagent (Invitrozen, USA) and reverse transcription (RT) was performed with 2 ug of the RNA using ImProm-llTM Reverse Transcriptase (Promega, USA). PCR amplification was performed in triplicate using GTaq polymerase (Cosmo Genetech Co., LTD, Korea) under the following conditions: initial denaturation for 5 min at 95℃, then 30 cycles of 94℃ for 30 s (denaturation), 58℃ (for Hoxc8 and β-Actin) or 62℃ (for Mgl1) for 30 s (annealing) and 72℃ for 30 s (polymerization). The primer sequences were as follows: for the mouse Hoxc8, 5′-CAC GTC CAA GAC TTC TTC CAC CAC GGC-3′ (forward) and 5′-CAC TTC ATC CTT CGA TTC TGG AAC C-3′ (reverse); Mgl1, 5′-AGG AGG ACA TCA GTG GCA TC-3′ (forward) and 5′-TGT CTC TGG TCC CAT TAG CC-3′ (reverse); control β-Actin, 5′- CTT CTT GGG TAT GGA ATC CTG -3′ (forward) and 5′- TCA GGA GGA GCA ATG ATC TTG-3′ (reverse); for human HOXC8, 5′-CCT ATT ACG ACT GCC GGT TC-3′ (forward) and 5′-TTG GCG GAG GAT TTA CAG TC-3′ (reverse); Hugl1, 5′-GCC TGC CTC ACC AAC CTG-3′ (forward) and 5′-TGG CTC AGC TTG GGT GAC TC-3′ (reverse); β-ACTIN, 5′-CAT GTT TGA GAC CTT CAA CAC CCC-3′ (forward) and 5′-GCC ATC TCT TGC TCG AAG TC-3′ (reverse).
Sequence analysis
The upstream region of Mgl1 (about 2 kb) was analyzed using the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgNear) gene sorter program. Hox consensus binding sites (TAAT/ATTA/TTAT/ATAA/TTAC) were searched manually in the 2 kb upstream region of Mgl1.
Chromatin immunoprecipitation (ChIP)-PCR
NIH3T3 stable cells overexpressing Hoxc8 and mouse embryonic tissues (brain and trunk) were used for ChIP analysis. The ChIP protocol was slightly modified from the user manual of the ChIP assay kit (Upstate Biotechnology, USA). Crosslinking was performed using 1% formaldehyde in DMEM. Sonicated product (~500 bp) was used as input after reverse cross linking. Immunoprecipitation was performed with anti-Hoxc8 antibody (Covance MMS-286R, Covance Research Product Inc., USA) or normal mouse IgG antibody (SC-2026; Santa Cruz Biotechnology Inc., USA) as a control. PCR was performed with the Mgl1 primers: 5′-GGT GCA AAC CGA AGT TCT TTG T-3′ (forward) and 5′-TGT TTT CCA CTC ACG ACT GAG-3′ (reverse).
Mgl 1 reporter plasmid construction and effector-reporter analysis
The ~2 kb promoter for Mgl1 was amplified by PCR using mouse genomic DNA as template and specific primers, 5′-ACA CTG TGT CTT CAG GCA CAC CA-3′ (forward) and 5′-CGG AAC CGA AAC TTC ATC AT-3′ (reverse), and then cloned into the TA vector (RBC lifesciences, Real Biotech Corporation, Taiwan). It was then subcloned into the pGL3-basic vector (Promega, USA) using the HindIII restriction endonuclease site to generate a luciferase reporter construct called pGL3-Mgl1-luc.
C3H10T1/2 (4 × 104 cells in 24-well plates) were transiently transfected using Lipofectamine 2000 reagent (Invitrogen, USA) with a total of 400 ng of luciferase reporter plasmid and the indicated combination of effector expression plasmids (i.e., pcDNA3.1-Hoxc8) plus 10 ng of pRL-TK vecctor. After 4 h of transfection, the media were changed and Hoxc8-siRNA (Samchully Pharm Co. Ltd., Korea) was transfected using Hi-perfect transfection reagent (Qiagen, USA). Luciferase activity was measured 48 h after transfection using the Dual Luciferase assay kit (Promega, USA) on a GLOMAX 20/20 luminometer (Promega, USA), according to the manufacturer’s instructions.
Cell adhesion assay
The C3H10T1/2 cells were seeded in a 24 well plate (4 × 104 cells/well) on the day before transfection. Cells were transfected in triplicate with 400 ng of pcDNA3.1 or pcDNA3-Hoxc8 vectors and 100 nM of Mgl1-siRNA (Genolution Pharmaceuticals, Korea) or negative control-siRNA using G-fectin (Genolution Pharmaceuticals, Korea) according to the manufacturer’s instructions. After 60 h of transfection, the adherent cells were trypsinized and counted using a hemocytometer (C-chip; iNCYTO, Korea). Untransfected cells were used as controls.
Statistical analysis
Statistical analysis was performed using Sigma plot 10 software. T-tests were performed and the results were expressed as mean ± standard error (SE). For all t-tests, a P-value of < 0.05 was considered to be statistically significant.
RESULTS
Mgl1 mRNA expression is negatively correlated with that of Hoxc8
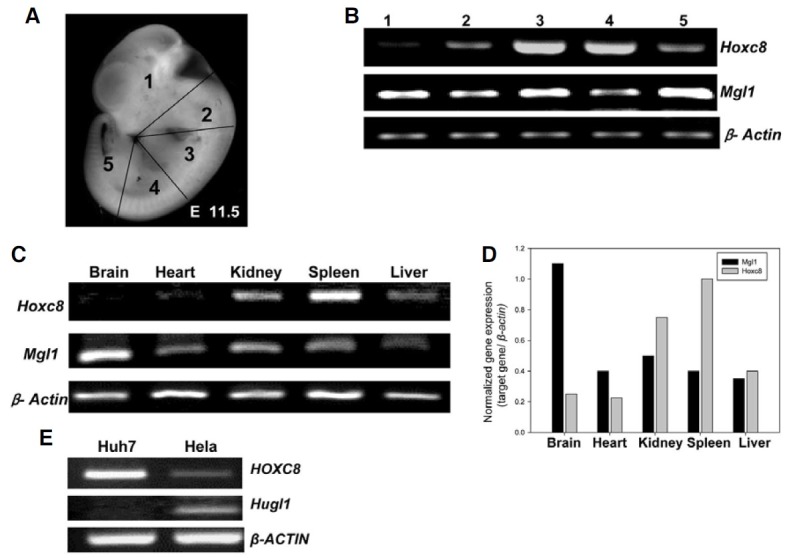
Hox genes are not only involved in pattern formation during embryogenesis but also have regulatory roles in adult tissues. Since we aimed to study the relationship between Hoxc8 and Mgl1, we first examined the endogenous expression pattern of these genes in mouse embryos as well as in adult tissues. Using E11.5 embryos, the expression levels of Hoxc8 and Mgl1 were analyzed using semi-quantitative RT-PCR along the anterior to posterior axis (slices 1 through 5 in Fig. 1A). As shown in Fig. 1B, position specific expression pattern of Hoxc8, which were already shown in previous reports (Chung et al., 2010; Kwon et al., 2005; Min et al., 2010) was detected: strong expression in the thoracic region (slices 3 and 4) and low expression in the area containing the brain and tail (slices 1 and 5). In the case of Mgl1, the opposite expression patterns were detected; high levels of expression in slices 1 and 5 (Figs. 1A and 1B). When adult tissues were analyzed in mice, the trend of negative correlation between these two genes was also detected, i.e. strong expression of Mgl1 but low expression of Hoxc8 in brain tissue and the opposite results in the spleen, etc. (Figs. 1C and 1D). When we looked at the expression patterns of HOXC8 and Hugl1, a human homologue of Mgl1, in two different human cancer cells, Huh7 (a human hepatoma cell line) and Hela (a human epithelial cervical cancer cell line), the opposite correlation of HOXC8 and Hugl1 expression level was still noticed in both cell lines, although HOXC8 was intensively expressed in Huh7 but not in the other depending on cell context (Fig. 1E). Together, these data strongly suggest that Hoxc8/ HOXC8 may negatively regulate Mgl1/Hugl1 gene expression.
Fig. 1. In vivo endogenous expression patterns of Hoxc8 and Mgl1 in different mouse tissues and human cells. (A) E11.5 mouse embryos indicating five regions (1-5) sliced along the anterior-posterior (AP) axis as indicated. (B) Endogenous gene expression levels of Hoxc8 and Mgl1 in each region along the anterior-posterior axis through RT-PCR. (C) Gene expression patterns in different adult mouse tissues and their quantification normalized with those of β-Actin using a densitogram. (D) Endogenous expression levels of human HOXC8, Hugl, and β-ACTIN were anayzed by RT-PCR in two different human cancer cells, Huh7 (hepatoma) and Hela (cervical cancer).
Overexpression of Hoxc8 downregulates Mgl1 expression in vitro
Since the expression patterns of Mgl1 and Hoxc8 showed a negative correlation, we examined the effect of Hoxc8 on Mgl1 expression in vitro using Hoxc8 expression vector, pcDNA3.1- Hoxc8. When Hoxc8 expression vector (pcDNA3.1-Hoxc8) was transiently transfected into the C3H10T1/2 cells along with the empty vector (pcDNA3.1), Mgl1 expression decreased depending on Hoxc8 expression: the amount of Mgl1 transcript was initially high, because C3H10T1/2 mouse embryonic cells do not express Hoxc8, but then decreased with overexpression of Hoxc8 (Fig. 2). This in vitro result further confirmed the in vivo results (Fig. 1) in which the endogenous expression level of Hoxc8 and Mgl1 was maintained in an opposing manner.
Fig. 2. Overexpression of Hoxc8 down-regulates Mgl1 expression in vitro. C3H10T1/2 cells were transfected with either empty (pcDNA3.1) or Hoxc8-expression vector (pcDNA3.1-Hoxc8) with different concentrations and were harvested after 48 h. Total RNA was isolated and RT-PCR was performed. PCR amplification was carried out using specific primers described in the materials and methods section. The protein level of Hoxc8 was also confirmed by Western blot analysis (see bottom).

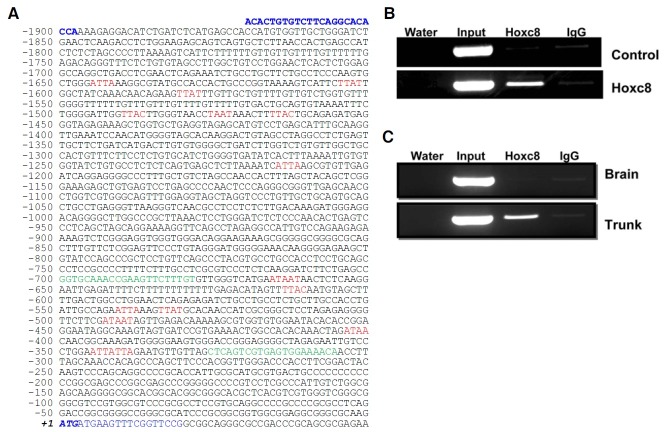
Hoxc8 directly binds to the promoter region of the Mgl1
To test whether Hoxc8 could function as a direct transcriptional regulator for Mgl1 gene expression, putative Hoxc8 core binding sites (TAAT/ATTA/TTAT/ATAA/TTAC) were analyzed in the promoter regions of Mgl1 spanning from ~1920 bp upstream (-1920) to ~50 bp downstream (+50) of the Mgl1 translation start site. Total 18 putative binding sites were detected, and interestingly enough, 11 sites were enriched in a ~330 bp region (Fig. 3A). So we further confirmed whether Hoxc8 really binds to this region using ChIP-PCR analysis. Hoxc8-bound chromatin was isolated from in vitro cells stably overexpressing Hoxc8 or in vivo embryonic tissue, and then immunoprecipitated with anti-Hoxc8 antibody. The Hoxc8-bound fragment was amplified with specific primer set. As shown in Figs. 3B and 3C, 395 bp PCR product (-700~-306 from the translation initiation site of Mgl1) was detected in Hoxc8 expressing cells (trunk and Hoxc8 transfectant), but not in the controls (brain and control empty vector transfectant), indicating that this region was directly bound to Hoxc8 protein.
Fig. 3. Sequence analysis and direct binding of Hoxc8 in the 5′ upstream promoter region of Mgl1 through ChIP-PCR. (A) Sequence analysis of the ~2 kb upstream regulatory region of Mgl1: NT_096135.5 (26,016,321..26,018,290). Putative Hox core consensus binding elements (TAAT/ ATTA/ TTAC/ TTAT/ ATAA) are written in red. The translation start codon (ATG) is written in italic bold. Primers used for ChIP-PCR and upstream promoter amplification for reporter (pGL3-Mgl1-Luc) construction are indicated in green and blue letters, respectively. ChIP-PCR analysis of in vitro (B) and in vivo cells (C). Chromatins were precipitated in the presence of anti-Hoxc8 monoclonal antibody (lane 3) or antimouse IgG (lane 4), and then the precipitated chromatin was subjected to PCR using ChIP-specific primers. Input (prior to immunoprecipitation) was used as an internal control (lane 2). Water indicates a no-template control (lane 1).
Mgl1 promoter activity is regulated by Hoxc8
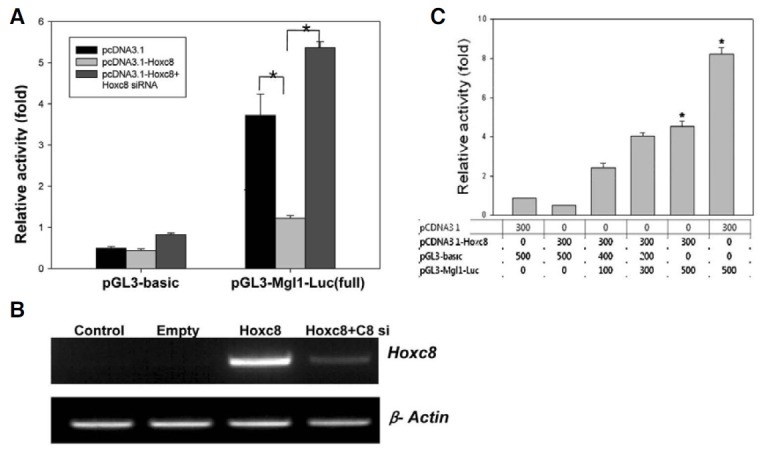
Since Hoxc8 binding to the promoter region of Mgl1 does not mean that Hoxc8 regulates Mgl1 gene expression, the upstream ~2 kbp region of Mgl1 was cloned into the luciferase reporter plasmid, pGL3-basic vector, and the reporter activity was analyzed in the presence or absence of Hoxc8. The cells transfected with pGL3-Mgl1-Luc vector showed strong reporter activity, while this activity was dramatically reduced (about 4- fold) by co-tranfection with the effector plasmid (pcDNA3.1- Hoxc8) expressing Hoxc8 (Figs. 4A and 4C). To confirm that this reduction was really due to expression of Hoxc8, siRNA against Hoxc8 was treated. As shown in Fig. 4A, the reduction of luciferase activity was fully recovered. Induction of Hoxc8 expression and depletion by siRNA in the same set of cells used for Fig. 4A was confirmed by RT-PCR (Fig. 4B). These findings strongly suggest that Hoxc8 represses Mgl1 promoter activity by binding to the promoter region.
Fig. 4. Hoxc8 negatively regulates Mgl1 promoter activity. C3H10T1/2 Cells were plated at a density of 4 × 104 cells per well in a 24-well plate for transfection. (A) pGL3-basic and pGL3-Mgl1-promoter construct (pGL3-Mgl1-Luc) were co-transfected with effec-tor plasmids, pcDNA 3.1 (empty in B) or pcDNA 3.1-Hoxc8 (Hoxc8 in B) with or without siRNA as indicated. Transfected cells were harvested after 48 h of transfection. Luciferase activity was measured and normalized to that of Renilla luciferase used as an internal control. Data are shown with the mean ± SEM of triplicates. The data shown are representative of at least two independent experiments. * represents significant difference (P < 0.05). (B) mRNA level of Hoxc8 and β-Actin in different transfection conditions. Control means no transfection. (C) Effect of Hoxc8 on a different titration of Mgl1 promoter.
Hoxc8 negatively affects Mgl1’s function on cell adhesion
Since it has been reported that the expression of Hugl1 increased cell adhesion, thus decreasing cell migration (Schimanski et al., 2005), we tested whether the mouse homologue Mgl1 had the same function. In order to do this, cell adhesion analysis was performed in C3H10T1/2 cells after transfection with different combinations of effector plasmid and siRNA (see “Materials and Methods”). As shown in Fig. 5, the number of adherent cells decreased by about 15% in the presence of Hoxc8. Since Hoxc8 did not completely shut down expression of Mgl1 (Fig. 2), we further treated siRNA against Mgl1 and counted the adherent cells after 60 h of transfection. Interestingly, a synergic inhibitory effect was achieved by knockdown of Mgl1 with Hoxc8 expression; that is, the fraction of adherent cells decreased another 35% compared to those overexpressing Hoxc8.
Fig. 5. Cell adhesion analysis upon Hoxc8 expression. C3H10T1/2 cells were transfected with either pcDNA3.1 or pcDNA3.1-Hoxc8 in the presence or absence of siRNA against Mgl1 (Mgl1 siRNA). Cells were trypsinized after 60 h of transfection and counted using a Hemocytometer. The adherent cells were plotted as a percentage of the untreated controls. Each bar indicates the mean ± SEM of three replicates of one experiment. All the experiments were performed at least in duplicate and obtained a similar pattern of results. * represents significant difference (P < 0.05).

DISCUSSION
Hox genes are implicated in several cancer types and altered patterns of Hox gene expression are considered to be an indicator of tumor progression or suppression (Alami et al., 1999; Chen et al., 2005; Miller et al., 2003). Since Mgl1 h as been proposed as a direct target gene of Hoxc8 and has a role as a tumor suppressor (Tomotsune et al., 1993), we aimed to clarify the binding capacity of Hoxc8 in the promoter region of Mgl1 and to evaluate the functional significance of Hoxc8 in Mgl1 gene expression and action.
First, we analyzed the expression pattern of Mgl1 and Hoxc8 in embryonic and adult tissues to understand the relationship between Hoxc8 and Mgl1 gene expression in mice. The expression pattern of Hoxc8 in developing mouse embryos has been intensively studied by our group and others (Awgulewitsch and Jacobs, 1990; Breier et al., 1988; Kwon et al., 2005; Le Mouellic et al., 1988). Our current data verified the distinct expression patterns of Hoxc8 in early embryos, in accordance with the previous results (Fig. 1). Hoxc8 is also expressed in some adult tissues, with considerably high levels in the spleen and kidney but rarely in the brain and heart. Although comprehensive expression profiles of Hox genes have not been previously reported for a variety of mouse adult tissues, the differential expression pattern of Hoxc8 in the several tissues we examined is partly in agreement with the previous studies showing Hox gene expression in human adult tissues (Takaha-shi et al., 2004; Yamamoto et al., 2003). Compared to the Hoxc8 expression pattern, Mgl1 was expressed relatively constantly in embryo, but has strong expression in the brain, tail and trunk regions harboring the forelimb, showing slight tendency toward negative correlation with Hoxc8 expression (Fig. 1); these results were similar to those of another group (Kim et al., 2003). The opposite pattern of expression between Hoxc8 and Mgl1 was more prominent in the adult brain, suggesting a possible regulatory mechanism for Mgl1 expression by Hoxc8. The high expression of Mgl1 in both embryonic and adult mouse brains implies that Mgl1 plays a critical role in brain development. It is interesting to emphasize the fact that Mgl1 knockout mice displayed brain dysplasia and died neonatally from severe hydrocephalus (Klezovitch et al., 2004). This opposing pattern of expression of Hoxc8 and Mgl1 was confirmed not only in vivo but also in the transient transfection experiment (Fig. 2).
Mgl1 was originally identified as one of the in vivo targets of Hoxc8 (Tomotsune et al., 1993). Through the combination of immunoenrichment and library screening procedures, they analyzed the genomic sequence of Mgl1 containing 21 exons and determined the sequence corresponding to the immunopurified clones at the 17th intron. However, it had not been verified at that time whether this genomic region was essential for the regulation of Mgl1 expression by Hoxc8. Here we demonstrated that the upstream promoter region of Mgl1 contains putative Hox binding sites and acts as a docking site for Hoxc8 binding (Fig. 3). On the contrary, no Hox binding site was found in the 17th intronic region. In addition, the binding of Hoxc8 into this intronic sequence was much weaker than that of the promoter region (data not shown). The luciferase reporter assay also clearly showed that ~2 kb promoter region of Mgl1 h as transcription enhancing activity, which is negatively regulated by Hoxc8 (Fig. 4). Taken together, our results suggest that Hoxc8 effectively binds to the promoter region and downregulates the transcriptional level of Mgl1.
Lgl is highly conserved in eukaryotes. Tumor-like characteristics, including rapid growth, invasiveness, loss of cell polarity and disorganization of the cell-cell adhesion structures, observed in Drosophila lgl mutants, re-emphasizes the important role of Lgl as a tumor suppressor (De Lorenzo et al., 1999; Vasiokhin, 2006). Considering that loss of cell polarity is one of the hallmarks of human cancer, the conserved function of Lgl in the cell polarity and asymmetric cell division pathway in different model organisms raises a lot of questions about the mechanisms responsible for Lgl functions in cancer. A recent study showed that downregulation of Hugl1 contributes to the progression of human colorectal cancer (Schimanski et al., 2005). In addition, they characterized the biological effects of Hugl1 using a functional assay with a Hugl1-inducible cell line and found that Hugl1 expression increased cell adhesion and decreased cell migration (Schimanski et al., 2005). In this experiment, when Hoxc8 was expressed, the number of adherent cells was reduced via knockdown of Mgl1 expression (Figs. 2 and 5), and further knockdown of Mgl1 with siRNA in Hoxc8- expressing cells exacerbated the Hoxc8-induced negative effect on cell adhesion (Fig. 5).
In conclusion, this study demonstrated that Hoxc8 downregulates Mgl1 expression at a transcriptional level by directly binding to its 5′ upstream promoter region, eventually affecting Mgl1 function on cell adhesion. Considering the tumor suppressor function of Mgl1, the aberrant expression of Hoxc8 in cancer and the subsequent effects on the function of Mgl1 will provide a plausible mechanism for how the alteration of normal Hox expression could promote tumorigenesis.
Acknowledgments
Ms. K. Ruthala is a graduate student supported by the Brain Korea 21 scholarship. This work was supported by grants from 2008-0058561 (2010-0000155) and 2010-0026759 from National Research Foundation (NRF), and in part by 20070401- 034-030 from the BioGreen21 Program, Rural Development Administration (RDA), Korea.
References
- 1.Alami Y., Castronovo V., Belotti D., Flagiello D., Clausse N. HOXC5 and HOXC8 expression are selectively turned on in human cervical cancer cells compared to normal keratinocytes. Biochem. Biophys. Res. Commun. (1999);257:738–745. doi: 10.1006/bbrc.1999.0516. [DOI] [PubMed] [Google Scholar]
- 2.Awgulewitsch A., Jacobs D. Differential expression of Hox 3.1 protein in subregions of the embryonic and adult spinal cord. Development. (1990);108:411–420. doi: 10.1242/dev.108.3.411. [DOI] [PubMed] [Google Scholar]
- 3.Breier G., Dressler G.R., Gruss P. Primary structure and developmental expression pattern of Hox 3.1, a member of the murine Hox 3 homeobox gene cluster. EMBO J. (1988);7:1329–1336. doi: 10.1002/j.1460-2075.1988.tb02948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K.N., Gu Z.D., Ke Y., Li J.Y., Shi X.T., Xu G.W. Expression of 11 Hox genes is deregulated in esophageal squamous cell carcinoma. Clin. Cancer Res. (2005);11:1044–1049. [PubMed] [Google Scholar]
- 5.Chung H.J., Lee J.Y., Deocaris C.C., Min H., Kim S.H., Kim M.H. Mouse homologue of the Schizophrenia susceptibility gene ZNF804A as a target of Hoxc8. J. Biomed. Biotechnol. (2010);2010:231708. doi: 10.1155/2010/231708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Lorenzo C., Mechler B.M., Bryant P.J. What is Drosophila telling us about cancer? Cancer Metastasis Rev. (1999);18:295–311. doi: 10.1023/a:1006381526008. [DOI] [PubMed] [Google Scholar]
- 7.Grifoni D., Garoia F., Schimanski C.C., Schmitz G., Laurenti E., Galle P.R., Pession A., Cavicchi S., Strand D. The human protein Hugl-1 substitutes for Drosophila lethal giant larvae tumour suppressor function in vivo. Oncogene. (2004);23:8688–8694. doi: 10.1038/sj.onc.1208023. [DOI] [PubMed] [Google Scholar]
- 8.Kang M., Bok J., Deocaris C.C., Park H.W., Kim M.H. Hoxc8 represses BMP-induced expression of Smad6. Mol. Cells. (2010);29:29–33. doi: 10.1007/s10059-010-0007-1. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y.S., Song J., Kim Y., Kim I.O., Kang I., Baek K.H. Functional and expression analyses of mgl-1, a mouse orthologue of lethal giant larvae recessive oncogene. Int. J. Oncol. (2003);23:1515–1519. [PubMed] [Google Scholar]
- 10.Klezovitch O., Fernandez T.E., Tapscott S.J., Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. (2004);18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuphal S., Wallner S., Schimanski C.C., Bataille F., Hofer P., Strand S., Strand D., Bosserhoff A.K. Expression of Hugl-1 is strongly reduced in malignant melanoma. Oncogene. (2006);25:103–110. doi: 10.1038/sj.onc.1209008. [DOI] [PubMed] [Google Scholar]
- 12.Kwon Y., Ko J.H., Kim B.-G., Kim M.H. Analysis of plausible downstream target genes of Hoxc8 in F9 teratocarcinoma cells. Mol. Biol. Rep. (2003);30:141–148. doi: 10.1023/a:1024920418148. [DOI] [PubMed] [Google Scholar]
- 13.Kwon Y., Shin J., Park H.W., Kim M.H. Dynamic expression pattern of Hoxc8 during mouse early embryogenesis. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. (2005);283:187–192. doi: 10.1002/ar.a.20160. [DOI] [PubMed] [Google Scholar]
- 14.Lee G.S., Kim B.S., Sheih J.H., Moore M. Forced expression of HoxB4 enhanced hematopoietic differentiation by human embryonic stem cells. Mol. Cells. (2008);25:487–493. [PubMed] [Google Scholar]
- 15.Lei H., Wang H., Juan A.H., Ruddle F.H. The identification of Hoxc8 target genes. Proc. Natl. Acad. Sci. USA. (2005);102:2420–2424. doi: 10.1073/pnas.0409700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei H., Juan A.H., Kim M.S., Ruddle F.H. Identification of a Hoxc8-regulated transcriptional network in mouse embryo fibroblast cells. Proc. Natl. Acad. Sci. USA. (2006);103:10305–10309. doi: 10.1073/pnas.0603552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei H., Juan A.H., Kim M.S., Ruddle F.H. Mouse Naked Cuticle 2 (mNkd2) as a direct transcriptional target of Hoxc8 in vivo. J. Exp. Zool. (2007);307A:1–6. doi: 10.1002/jez.a.327. [DOI] [PubMed] [Google Scholar]
- 18.Le Mouellic H., Condamine H., Brulet P. Pattern of transcription of the homeo gene Hox-3.1 in the mouse embryo. Genes Dev. (1988);2:125–135. doi: 10.1101/gad.2.1.125. [DOI] [PubMed] [Google Scholar]
- 19.Miller G.J., Miller H.L., van Bokhoven A., Lambert J.R., Werahera P.N., Schirripa O., Lucia M.S., Nordeen S.K. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. (2003);63:5879–5888. [PubMed] [Google Scholar]
- 20.Min H., Lee J.Y., Bok J., Chung H.J., Kim M.H. Proliferating cell nuclear antigen (Pcna) as a direct downstream target gene of Hoxc8. Biochem. Biophys. Res. Commun. (2010);392:543–547. doi: 10.1016/j.bbrc.2010.01.059. [DOI] [PubMed] [Google Scholar]
- 21.Morgan R. Hox genes: a contimuation of embryonic patterning? Trends Genet. (2006);22:67–69. doi: 10.1016/j.tig.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. (2005);6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 23.Schimanski C.C., Schmitz G., Kashyap A., Bosserhoff A.K., Bataille F., Schäfer S.C., Lehr H.A., Berger M.R., Galle P.R., Strand S., et al. Reduced expression of Hugl-1, the human homologue of Drosophila tumour suppressor gene lgl, contributes to progression of colorectal cancer. Oncogene. (2005);24:3100–3109. doi: 10.1038/sj.onc.1208520. [DOI] [PubMed] [Google Scholar]
- 24.Shashikant C.S., Ruddle F.H. Combinations of closely situated cis-acting elements determine tissue-specific patterns and anterior extent of early Hoxc8 expression. Proc. Natl. Acad. Sci. USA. (1996);93:12364–12369. doi: 10.1073/pnas.93.22.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi Y., Hamada J., Murakawa K., Takada M., Tada M., Nogami I., Hayashi N., Nakamori S., Monden M., Miyamoto M., et al. Expression profiles of 39 HOX genes in normal human adult organs and anaplastic thyroid cancer cell lines by quantitative real-time RT-PCR system. Exp. Cell Res. (2004);293:144–153. doi: 10.1016/j.yexcr.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Tomotsune D., Shoji H., Wakamatsu Y., Kondoh H., Takahashi N. A mouse homologue of the Drosophila tumoursuppressor gene l(2)gl controlled by Hox-C8 in vivo. Nature. (1993);365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- 27.Vasiokhin V. Lethal giant puzzle of Lgl. Dev. Neurosci. (2006);28:13–24. doi: 10.1159/000090749. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto M., Takai D., Yamamoto F., Yamamoto F. Comprehensive expression profiling of highly homologous 39 hox genes in 26 different human adult tissues by the modified systematic multiplex RT-pCR method reveals tissue-specific expression pattern that suggests an important role of chromosomal structure in the regulation of hox gene expression in adult tissues. Gene Exp. (2003);11:199–210. doi: 10.3727/000000003108749071. [DOI] [PMC free article] [PubMed] [Google Scholar]