Abstract
Cortexillins are actin-bundling proteins that play a critical role in regulating cell morphology and actin cytoskeleton reorganization in Dictyostelium. Here, we investigated dynamic subcellular localization of cortexillin I in chemotaxing Dictyostelium cells. Most of the cortexillin I was enriched on the lateral sides of moving cells. Upon chemoattractant stimulation, cortexillin I was rapidly released from the cortex followed by a transient translocation to the cell cortex with a peak at ~5 s and a subsequent decrease to basal levels, indicating that localization of cor-texillin I at the cortex in chemotaxing cells is controlled by two more signaling components, one for the initial delocalization from the cortex and another for the translocation to the cortex ~5 s after chemoattractant stimulation. Loss of cortexillins leads to reduced cell polarity and an in-creased number of lateral pseudopodia during chemotaxis, suggesting that cortexillins play an inhibitory role in producing pseudopodia along the lateral sides of the cell. Cells lacking cortexillins displayed extended chemoattrac-tantmediated Arp2/3 complex translocation kinetics to the cortex. Our present study provides a new insight into the function of cortexillins during reorganization of the actin cytoskeleton and cell migration.
Keywords: actin cytoskeleton, Arp2/3 complex, chemotaxis, cortexillin, Dictyostelium
INTRODUCTION
For efficient directional migration, cells must coordinate F-actinmediated protrusion at the leading edge and actomyosin contraction at the cell’s posterior. The differential polymerization of F-actin at the leading edge of the cell is regulated by numerous actin-modifying proteins, such as the Arp2/3 complex, WAVE/ SCAR, WASP, and ADF/cofilin (Firat-Karalar and Welch, 2011; Lee and Dominguez, 2010; Ridley et al., 2003; Sasaki and Firtel, 2006). The Arp2/3 complex directly nucleates actin assembly and forms new branch points. Efficient actin nucleation requires its interactions with WASP family members, which are Cdc42 and Rac effectors (Raftopoulou and Hall, 2004), and/or a pre-existing actin filament (Firat-Karalar and Welch, 2011; Raftopoulou et al., 2004; Rodal et al., 2005).
Cortexillins have been identified in Dictyostelium as actinbundling proteins that organize actin filaments preferentially into anti-parallel bundles and associate them into three dimensional meshworks (Faix et al., 1996). This activity is crucial for cytokinesis and cell morphology in Dictyostelium. Mutants lacking both cortexillin I and II display spread and flattened morphology and severely impaired cytokinesis, leading to the formation of large multinucleate cells (Faix, 2002; Faix et al., 1996; Weber et al., 1999). Recent reports have demonstrated that cortexillin I is involved in cell migration by negatively regulating cortical localization of RapGAP1 in Dictyostelium. Cells lacking both cortexillins exhibit constitutive and uniform RapGAP1 cortical localization during chemotaxis, leading to a reduced chemoattractantmediated Rap1 activation level (Jeon et al., 2007a; Lee et al., 2010). Cortexillin I is an F-actin bundling protein containing three domains (Faix, 2002; Faix et al., 1996; Weber et al., 1999). The N-terminal halves of cortexillin encompass two conserved actinbinding domains of the α-actinin/spectrin type (Faix, 2002; Faix et al., 1996). A coiled-coil domain required for forming dimmers is located at the central region. The C-terminal region is essential for targeting to the cleavage furrow during cytokinesis and for actin-bundling activity. The last positively charged nonapeptide motif has a phosphatidylinositol (4,5)bisphosphate (PIP2)- binding site (Stock et al., 1999). Cortexillin I forms a complex with RacI and the IQGAPs DGAP1 and GAPA (Faix et al., 2001; Lee et al., 2010; Mondal et al., 2010). The IQGAP/cortexillin complexes are involved in regulating cortical mechanics. It has been recently suggested that myosin II and IQGAP/cortexillin play important roles in regulating the ability of cells to restrict the F-actin assembly site and pseudopod formation at the leading edge of moving cells (Jeon et al., 2007b; Lee et al., 2010).
To further understand the functions of cortexillins in the regulation of actin cytoskeleton reorganization during cell migration, we have investigated the dynamic subcellular localization of cortexillin I in moving Dictyostelium cells. Our data suggest that the localization of cortexillin I at the lateral sides of moving cells is related to an inhibited production of lateral pseudopodia, and cortexillins are linked to the translocation of Arp2/3 complex to the cell cortex upon chemoattractant stimulation.
MATERIALS AND METHODS
Strains and plasmids
Dictyostelium wild-type KAx-3 cells and ctxA-/B- cells were obtained from the Dictyostelium stock center. All cell lines were cultured axenically in HL5 medium at 22℃. For expression of GFP-cortexillin I, the full coding sequence of the cortexillin I cDNA was generated by RT-PCR and cloned into the BglII-XhoI site of expression vector EXP-4(+) containing a GFP fragment. The expression plasmids for GFP-ArpD, GFP-coronin, and GFP-PhdA were described previously (Jeon et al., 2007a). The plasmids were transformed into KAx-3 or ctxA-/B- cells and the transformants were maintained in 20 μg/ml G418.
Development and chemotaxis analysis
Exponentially growing cells were harvested and washed twice with 12 mM Na/K phosphate buffer (pH 6.1) and plated on Na/K phosphate agar plates at a density of 4 × 106 cells/cm2 (Jeon et al., 2009). The developmental morphology of the cells was examined by photographing the developing cells at the time indicated in the figures.
The chemotaxis towards cAMP and changes in the subcellular localization of proteins in response to chemoattractant stimulation were examined as described previously (Jeon et al., 2007a; Sasaki et al., 2004). The aggregation competent cells were plated on glass-bottomed microwell plates, and then a micropipette filled with 150 μM cMP was positioned near the cells to stimulate them. The images of chemotaxing cells were taken at time-lapse intervals of 6 s for 30 min using an inverted microscope (IX71; Olympus, Japan) with a camera (DS-Fi1; Nikon, Japan).
Quantitation analysis of GFP fusion proteins
The quantitation of membrane or cortical localization of GFP fusion proteins was performed as described previously (Cha et al., 2010; Jeon et al., 2007a; Sasaki et al., 2004). The aggregation competent cells were allowed to adhere to the plate for 10 min. The cells were uniformly stimulated with cAMP by quickly pipetting 250 μl of 150 μM cAMP into the plate containing cells. The fluorescence images were taken at time-lapse intervals of 1 s for 1 min using an inverted microscope. The frames were captured using NIS-elements software (Nikon) and analyzed using ImageJ software (National Institutes of Health, USA). The intensity of cortical GFP was measured and the level of cortical GFP was calculated by dividing the intensity before stimulation (Eo) by the intensity at each time point (Et).
RESULTS
Cortexillins are required for formation of cell polarity, cell shape, and multicellular development
Cortexillins are actin-binding proteins containing three domains (Fig. 1A; Faix et al., 1999) whose activity is crucial for cytokinesis in Dictyostelium (Faix et al., 1996; Weber et al., 1999). Recent studies have shown that cortexillins are involved in directional cell migration by interacting with Rac and Rap1 signaling pathways (Jeon et al., 2007a; Lee et al., 2010). In this study, we examined the morphology and ability of mutant strains lacking cortexillins to polarize and chemotax up a chemoattractant gradient. Wild-type cells became elongated and polarized and chemotaxed to a micropipette emitting the chemoattractant cAMP with protrusions that were predominantly in the direction of the chemoattractant gradient (Figs. 1B and 1C). In contrast, cells lacking both cortexillin isoforms appeared in general to be slightly larger and more flat and exhibited reduced cell polarity (increased roundness) (Figs. 1B and 1C). Cell size measurement confirmed that ctxA/B null cells were much larger than wild-type cells (Fig. 1D). Mean sizes of wild-type and ctxA/B null cells were 11.4 ± 2.30 and 21.7 ± 5.95 μm, respectively. In addition, ctxA/B null cells showed increased production of pseudopodia around the cell, including the posterior and lateral sides of cells, and a slightly slower speed of movement toward the micropipette, suggesting that cortexillins might play some roles in establishing cell polarity and inhibiting protrusion formation at the posterior and lateral sides of moving cells.
Fig. 1. Chemotaxis and multicellular development of ctxA/B null cells. (A) Domain structure of cortexillin I showing three domains, two CH domains in the N-terminus, a coiledcoil domain at the central region, and an actin-bundling domain in the C-terminus. (B) Chemotaxis of wild-type KAx-3 and ctxA/B null cells. Representative frames from timelapse recordings of chemotaxis are shown. The micropipette filled with 150 μM cAMP was placed on the bottom and is visible in the images. Cells chemotax toward the cAMP gradient produced by diffusion of the cAMP from the micropipette. (C) Morphology of wild-type and ctxA/B null cells. A representative image from time-lapse recordings of chemotaxing wild-type and ctxA/B null cells is shown, respectively. (D) Cell size measurement. The sizes of the vegetative wild-type and ctxA/B null cells were measured by NIS-elements software (Nikon). The histogram graphs show the distribution of the sizes of 357 cells from wild-type and ctxA/B null cells each. (E) Development of wild-type and ctxA/B null cells. Cells were developed on non-nutrient agar plates. Photographs were taken at the times indicated after plating. Development at 12 h (wild-type tip forming stage) and 24 h (wild-type fruiting body stage) is shown.
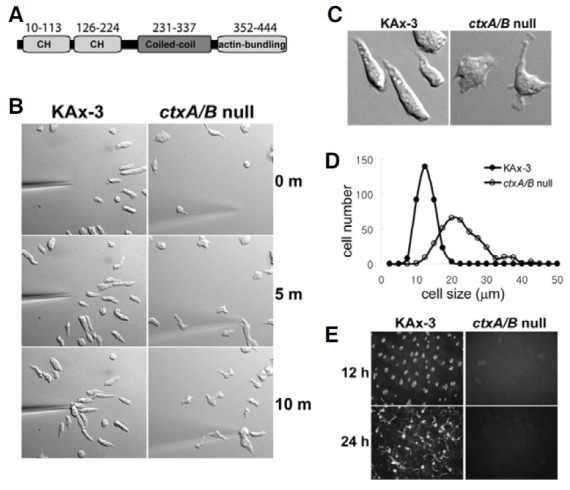
Dictyostelium cells undergo a multicellular developmental process upon starvation, eventually leading to the formation of a fruiting body within 24 h (Chisholm and Firtel, 2004). Individual cells aggregate to form a mound of 105 cells at approximately 10 h, primarily mediated by chemotaxis to cAMP. Cells within the mound then differentiate into several cell types and form a slug-shaped structure. Culmination follows, resulting in the formation of a mature fruiting body. Because both aggregation and morphogenesis require regulated cell movement, we examined the potential involvement of cortexillins in these processes. Wild-type cells aggregated and formed mounds approximately 12 h after initiating development, followed by the formation of mature fruiting bodies within 24 h (Fig. 1E). In contrast, cells lacking cortexillin I and II were unable to aggregate, and development did not proceed (Fig. 1E), indicating that cortexillins are essential for multicellular development of Dictyostelium.
Subcellular localization of cortexillin I
We examined the localization of GFP-cortexillin I in randomly moving vegetative cells to understand dynamic roles of cortexillin in cell migration. As shown in Fig. 2A, GFP-cortexillin I localized at the plasma membrane in the resting vegetative cells, and some cells showed enrichment at the regions of membrane ruffling where F-actin accumulates (Fig. 2A). To determine whether cortexillins are localized at the F-actin assembly sites, the localization of GFP-cortexillin I was compared with that of a newly formed F-actin marker protein GFP-coronin (Gerisch et al., 1995), which associates with the Arp2/3 complex on F-actin filaments and inhibits actin nucleation (Rodal et al., 2005). The majority of GFP-coronin was found at protruding regions as clearly accumulated forms (Fig. 2B), which was slightly different from the relatively even distribution of GFP-cortexillin I at the cell cortex. Another clear difference in the localization of the proteins was found on the bottom of the cells. When the bottoms of the cells expressing GFP-coronin were examined using a confocal microscope, small structures were found, which are known as F-actin-containing foci and may function in cellsubstratum adhesion (Jeon et al., 2007a; Uchida and Yumura, 2004). In contrast, such small structures were not found at the bottom of the cell expressing GFP-cortexillin I (Fig. 2C), indicating that cortexillin I does not localize to the actin foci at the bottom of the cell. These observations suggest that the sites where cortexillin I localizes do not match with regions of newly formed F-actin represented by GFP-coronin.
Fig. 2. Spatial localization of GFP-cortexillin I. (A) Localization of GFP-cortexillin I (GFP-CtxI) and GFP-coronin (B) in vegetative cells. Vegetative cells expressing GFP-cortexillin I or GFPcoronine were imaged. (C) Localization of GFPCtxI and GFP-coronin on the bottom of cells. Bottom sections of the cells expressing GFPctxI or GFP-coronin were imaged using a confocal microscope. (D) Spatial localization of GFPCtxI and GFP-coronin (E) in chemotaxing cells. The asterisk indicates the position of the micropipette containing cAMP. The arrow in (E) shows the direction of movement.
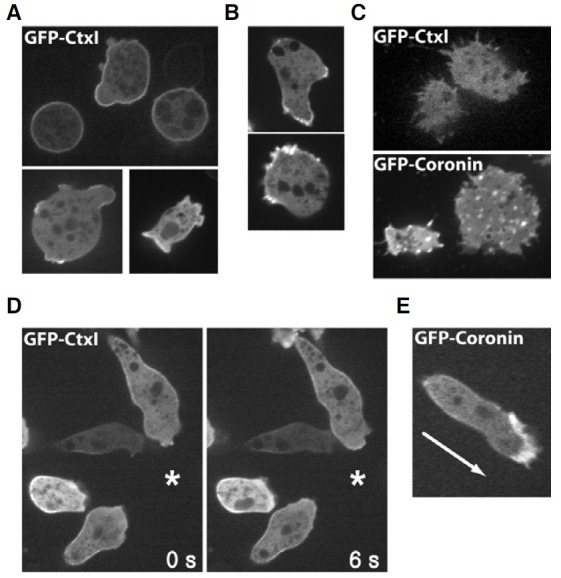
In chemotaxing cells, most coronins were enriched at the leading edge and only a small amount of protein was found in the cortex regions including the posterior and lateral sides of the cell, which is consistent with localization of the F-actin assembly (Fig. 2E, Jeon et al., 2007a; Uchida and Yumura, 2004). In contrast, GFP-cortexillin I exhibited no such enrichment at the leading edge of the moving cell as with coronin (Fig. 2D). Instead, most of the cortexillin I was found on the plasma membrane along the lateral sides of the cell and sometimes small amounts of cortexillin I localized to the leading edge, which was a rare case, and the level of accumulated GFP-cortexillin I was much less than that of GFP-coronin (Fig. 2D). The level of cortexillin I at the posterior of the cell was significantly reduced. This unique localization of cortexillin I in moving cells suggest that cortexillin I plays some roles in the lateral sides of the cell during migration, and that binding partners of cortexillin I or binding mechanisms of cortexillin I to F-actin are likely different from those of other general actin-binding proteins such as coronin and the Arp2/3 complex, which accumulate at the leading edge of moving cells.
Translocation kinetics of cortexillin I in response to chemoattractant stimulation
We analyzed the translocation kinetics of cortexillin I to the cell cortex in response to uniform chemoattractant stimulation to understand the temporal mechanism for the dynamic localization of cortexillins to the cell cortex in chemotaxing cells. Unstimulated cells displayed quite high levels of GFP-cortexillin I at the cortex. Upon uniform chemoattractant stimulation, GFP-cortexillin I at the cortex rapidly delocalized in 2-3 s shortly after stimulation and then transiently translocated to the cell cortex with a peak at ~5 s, followed by decrease of the protein amount at the cortex to the basal level within 30 s (Figs. 3A and 3B). The changes of the fluorescence intensity of GFP-cortexillin I at the cortex at 2 s and 5 s after stimulation were statistically significant, compared to that before stimulation (Fig. 3B, p < 0.005).
Fig. 3. Temporal localization of GFP-cortexillin I in response to chemoattractant stimulation. (A) Translocalization of GFP-CtxI to the cell cortex in response to uniform chemoattractant stimulation. Cells were stimulated with cAMP, and the images were taken every second for 1 min. Five representative frames from time-lapse recor-dings are shown. (B) Translocation kinetics of GFP-CtxI. The fluorescence intensity of membrane-localized GFPCtxI was quantified. The graphs represent the means of data on several cells from videos taken from 3 separate experiments. Error bars represent S.D. (n = 52). *, statistically significant difference compared to the fluorescence intensity at 0 s (t-test, p < 0.005). (C) Translocation kinetics of GFP-CtxI, GFP-ArpD, GFP-coronin and PhdA-GFP to the cortex were obtained from time-lapse recordings and quantified as described previously (Jeon et al., 2007a). The graphs are the means of data on several cells from at least three separate experiments.
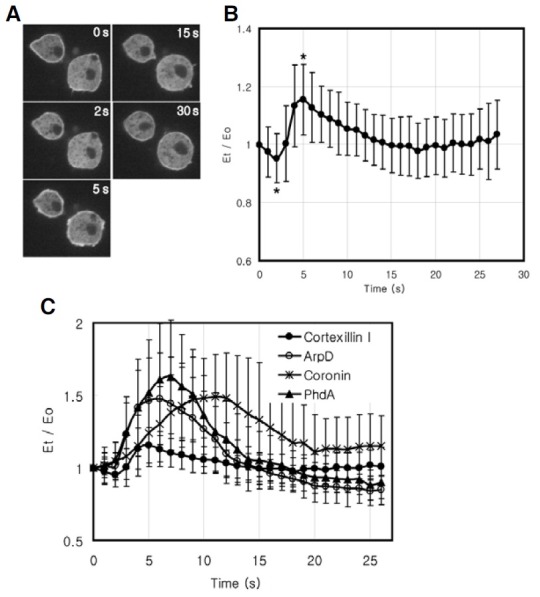
To understand temporal translocation kinetics of cortexillin I in more detail, the cell cortex translocation kinetics of cortexillin I were compared with those of other F-actin binding proteins such as ArpD, a subunit of Arp2/3 complex (Rodal et al., 2005), and coronin and PhdA-GFP (Funamoto et al., 2002), a pleckstrin homology domain containing PIP3 reporter (Fig. 3C). As shown in Fig. 3C, all the proteins examined in this study displayed similar rapid and transient translocation to the cell cortex with a peak at 5-10 s in response to chemoattractant stimulation. However, only GFP-cortexillin I exhibited unique initial delocalization from the cortex before transient translocation to the cortex upon stimulation (Figs. 3B and 3C), suggesting that localization of cortexillin I at the cortex is controlled by two more signaling components, one for the initial delocalization from the cortex and another for the translocation to the cortex ~5 s after chemoattractant stimulation.
Although the level of translocated cortexillin I to the cell cortex was much lower than those of other proteins and there was an initial delocalization of cortexillin I, the temporal translocation kinetics of cortexillin I upon chemoattractant stimulation was similar to that of ArpD with a maximal level of the proteins at the cortex ~5 s after stimulation, which was 1-2 s earlier than PhdA and 4-5 s earlier than coronin. The Arp2/3 complex, PhdA, and coronin are sequentially involved in regulating F-actin polymerization at the cell cortex upon chemoattractant stimulation (Firat-Karalar and Welch, 2011; Ridley et al., 2003). Our results indicate that, within the limits of our experiments, translocation of cortexillin I to the cortex is temporally correlated with that of the Arp2/3 complex.
Localization of GFP-ArpD in ctxA/B null cells
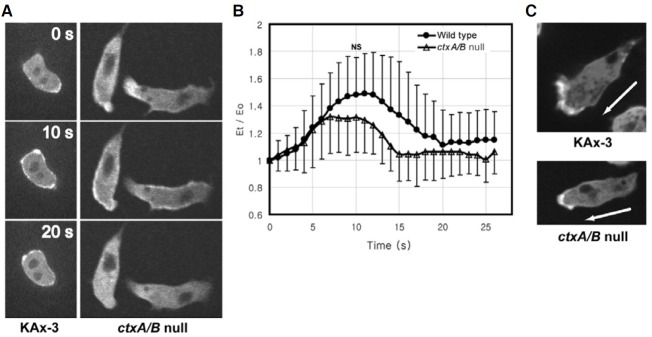
The experiments examining translocation kinetics of the actinbinding proteins upon chemoattractant stimulation showed that the temporal translocation kinetics of cortexillin I upon chemoattractant stimulation was similar to that of ArpD, a subunit of the Arp2/3 complex. We examined the localization of GFP-ArpD in ctxA/B null cells to examine the relationship between cortexillins and the Arp2/3 complex during translocation to the cell cortex in response to chemoattractant stimulation (Fig. 4).
Fig. 4. Localization of GFP-ArpD in ctxA/B null cells. (A) Translocation of GFP-ArpD in wildtype KAx-3 cells and ctxA/B null cells to the cell cortex in response to uniform chemoattractant stimulation. Cells expressing GFP-ArpD were stimulated with cAMP, and the images were taken every second. Four representative frames from time-lapse recordings are shown. (B) Translocation kinetics of the GFP-ArpD in KAx-3 and ctxA/B null cells to the cell cortex. The fluorescence intensity of membrane-localized GFPArpD was quantified and graphed as described in Fig. 3. Error bars represent S.D. (n = 30). The difference at 20 s after chemoattractant stimulation between KAx-3 and ctxA/B null cells is significant (*p < 0.001, t-test). (C) Spatial localization of GFP-ArpD in chemotaxing wild-type KAx-3 and ctxA/B null cells. The arrow indicates the direction of movement.
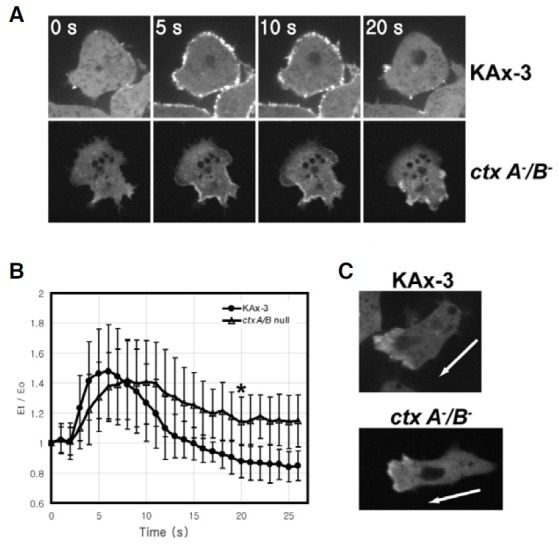
Both wild-type cells and ctxA-/B- cells exhibited a transient and rapid translocation of ArpD to the cell cortex in response to uniform cAMP stimulation. In chemotaxing cells, GFP-ArpD was highly accumulated at the leading edge in both wild-type and ctxA-/B- cells. One difference was found in the translocation kinetics of ArpD to the cell cortex upon chemoattractant stimulation. Wild-type cells have a maximal level of ArpD at the cortex approximately 5-6 s after stimulation and then decrease to the basal level within 20 s, whereas ctxA-/B- cells showed a slightly delayed and extended ArpD translocation kinetics. The maximal level of GFP-ArpD translocated to the cortex upon stimulation at approximately 8-9 s, which was 2-3 s slower than that in wild-type cells, and then the elevated level of ArpD at the cortex had a longer duration than that in wild-type cells, suggesting that cortexillins are involved in the delocalization of the Arp2/3 complex from the cortex following the transient translocation to the cortex upon chemoattractant stimulation.
Localization of GFP-coronin in ctxA/B null cells
We examined localization of coronin in ctxA/B double null cells to determine whether localization or translocation of the other proteins involved in F-actin polymerization is also affected by the loss of cortexillins. Coronin transiently translocated to the cell cortex in response to chemoattractant stimulation in both wild-type and ctxA-/B- cells (Fig. 5). The translocation kinetics of coronin in ctxA-/B- cells was similar to that in wild-type cells (Figs. 5A and 5B). However, ctxA-/B- cells displayed slightly lower levels of coronin translocated to the cortex upon stimulation compared to that in wild-type cells (Fig. 5B, p > 0.1, statistically not significant), probably resulting from the difference in coronin expression level in both cells and the flattened cell morphology of ctxA/B double null cells. In chemotaxing ctxA/B null cells, coronin localized at the leading edge as in wild-type cells. These results indicate that the localization of coronin, which binds to the Arp2/3 complex and inhibits nucleation activity of the complex (Rodal et al., 2005), is not affected by the absence of cortexillins.
Fig. 5. Localization of GFP-coronin in ctxA/B null cells. (A) Translocation of GFP-coronin in wild-type KAx-3 cells and ctxA/B null cells to the cell cortex in response to uniform chemoattractant stimulation. Cells expressing GFP-coronin were stimulated with cAMP, and the images were taken every second. Three representative frames from time-lapse recordings are shown. (B) Translocation kinetics of the GFP-coronin in KAx-3 and ctxA/B null cells to the cell cortex. The fluorescence intensity of membrane-localized GFP-coronin was quantified and graphed as described in Fig. 3. The difference between wild-type and ctxA/B null cells is not significant (t-test, p > 0.1, NS, not significant). (C) Spatial localization of GFP-coronin in chemotaxing wild-type KAx-3 and ctxA/B null cells. The arrow indicates the direction of movement.
DISCUSSION
Localization of cortexillin I in moving Dictyostelium cells
Here, we showed that most of the cortexillin I was enriched on the lateral sides of moving cells. The localization of cortexillin I was not common to other actin-binding proteins such as coronin and the Arp2/3 complex. These proteins were accumulated at the leading edge and only small amounts of the proteins were found on the posterior or lateral sides of moving cells, which largely matched the F-actin distribution. Consistent with this result, cortexillin I was not found in the actin foci at the bottom of the cells where other actin-binding proteins accumulate. These data suggest that cortexillins might play important roles in the lateral sides of moving cells. These findings support the suggestions that cortexillins are involved in establishing cell polarity and inhibiting the production of lateral pseudopodia. Loss of cortexillins caused severe defects in morphology and chemotaxis, such as a spread and flattened morphology and abnormal production of lateral pseudopodia during cell migration, suggesting that cortexillins provide cortical tension along the lateral sides of the cells and inhibit protrusions at the cell cortex, similar to myosin II.
In agreement with our view, recent studies have proposed that myosin II and IQGAP/cortexillin are important negative regulators of leading-edge function and restrict the site of pseudopod formation to the leading edge of moving cells (Jeon et al., 2007b; Lee et al., 2010; Ren et al., 2009). In response to chemoattractant stimulation, cells rapidly activate a series of signaling pathways including Ras/Rap1 proteins, PI3K and TORC2, and their effectors Akt/PKB and PKBR1 at the leading edge of the cell, leading to F-actin polymerization, pseudopod formation, and directional movement up the gradient (Charest et al., 2010; Kolsch et al., 2008; Kortholt and van Haastert, 2008). Disruption of myosin II or specific IQGAP/cortexillin complexes, which regulate cortical mechanics such as cortical tension, results in extended activation of PI3K and Akt/PKB, suggesting the negative roles of myosin II and IQGAP/cortexillin complexes in activating the signaling pathways (Lee et al., 2010). A distinct localization of cortexillin I different from other general actin-binding proteins was also found during cytokinesis. Cortexillins are much more strongly enriched in the cleavage furrow during cytokinesis rather than in the polar regions where F-actin and the potent actin-bundling protein P34 accumulate (Weber et al., 1999).
Translocation of cortexillin I to the cell cortex upon chemoattractant stimulation
Our analysis of the translocation kinetics of cortexillin I to the cell cortex in response to uniform chemoattractant stimulation suggests that the dynamic subcellular localization of cortexillin I is controlled by two more signaling components and that the cortexillin I translocation to the cortex upon stimulation temporally correlates with the Arp2/3 complex. As shown in the localization of cortexillin I in moving cells, cortexillin I displays unique translocation kinetics to the cell cortex in response to chemoattractant stimulation, compared with those of other actin-binding proteins. Upon chemoattractant stimulation, there is a rapid release of cortexillin I from the cortex followed by a transient translocation to the cell cortex with a peak at ~5 s and a subsequent decrease to the basal level. An initial delocalization from the cell cortex upon stimulation is also found in the translocation kinetics of several proteins including myosin II, PTEN, and PakA (Chung and Firtel, 1999; Funamoto et al., 2002; Jeon et al., 2007b), but the subsequent transient translocation of cortexillin I to the cortex within 10 s is not observed with other proteins. The transient translocation kinetics of cortexillin I, except the initial delocalization of the proteins, is similar to that of the Arp2/3 complex, a nucleator of F-actin assembly. These data indicate that two more signaling components govern the localization of cortexillin I in response to chemoattractant stimulation; one for the initial delocalization from the cortex and another for the translocation to the cortex ~5 s after chemoattractant stimulation.
Cortexillin I contains three domains, two actin-binding domains at the N-terminal, a coiled-coil domain at the central, and an actin-bundling activity containing domain at the C-terminal region. In addition, a PIP2-binding motif is located in the last nonapeptides of the protein (Faix et al., 1996; Stock et al., 1999; Weber et al., 1999). Several proteins have been identified as cortexillin binding partners. Cortexillin I interacts with RacI, IQGAPs, and GAPA forming a complex, which plays a key role in cytokinesis and cell migration (Lee et al., 2010; Mondal et al., 2010). Furthermore, an interaction with Rap- GAP1 has been demonstrated. RapGAP1 translocates to the cell cortex in response to chemoattractant stimulation and localizes to the leading edge of the moving cell in an F-actin dependent manner (Jeon et al., 2007a). We propose that intracellular localization of cortexillin I at the cortex is determined by a balance among the actin-binding activity of the N-terminal and the membrane-binding activity of the C-terminal region of cortexillins, as well as additional interactions with other binding proteins.
Extended translocation of the Arp2/3 complex to the cell cortex in cells lacking cortexillin I and II in response to chemoattractant stimulation
Our results indicate that cortexillins negatively regulate translocation of the Arp2/3 complex to the cell cortex upon chemoattractant stimulation. Loss of cortexillins resulted in an extension of the cortical localization of GFP-ArpD, a subunit of the Arp2/3 complex, upon chemoattractant stimulation. Coronin in ctxA/B null cells, an inhibitor of the Arp2/3 complex, displayed similar translocation kinetics to that in wild-type cells, although the amplitude of increased cortical fraction was smaller than that of wild-type cells. These data indicate that the effects of loss of cortexillins are limited to localization of the Arp2/3 complex, but not translocation of the inhibitor coronin, implicating that F-actin nucleation might increase in cells lacking cortexillins. Recent studies have shown a slightly increased amount of cortical Factin in ctxA/B null cells than that in wild-type cells (Lee et al., 2010).
The negative effect of cortexillins on the translocation of the Arp2/3 complex to the cortex is consistent with the findings of studies of propagating F-actin waves at the bottom of the cell (Schroth-Diez et al., 2009). Actin waves are formed at the substrate- attached surface in migrating Dictyostelium cells, particularly on strongly adhesive substrates (Heinrich et al., 2008). Actin waves separate two F-actin binding proteins, the Arp2/3 complex and cortexillins. The Arp2/3 complex is exclusively localized in the internal region of the F-actin waves, whereas cortexillins are localized at the external regions of the waves (Schroth-Diez et al., 2009). Taken together, our results suggest that cortexillins might play a role in restricting the Arp2/3 complex in the internal region of the F-actin waves.
Acknowledgments
We thank Richard A. Firtel and the DictyBase Stock Center for kindly providing strains and plasmids used in this study. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0065992 and 2009-0070924).
References
- 1.Cha I., Lee S.H., Jeon T.J. Chemoattractant-mediated Rap1 activation requires GPCR/G proteins. Mol. Cells. (2010);30:563–567. doi: 10.1007/s10059-010-0153-5. [DOI] [PubMed] [Google Scholar]
- 2.Charest P.G., Shen Z., Lakoduk A., Sasaki A.T., Briggs S.P., Firtel R.A. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev. Cell. (2010);18:737–749. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chisholm R.L., Firtel R.A. Insights into morphogenesis from a simple developmental system. Nat. Rev. Mol. Cell Biol. (2004);5:531–541. doi: 10.1038/nrm1427. [DOI] [PubMed] [Google Scholar]
- 4.Chung C.Y., Firtel R.A. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. (1999);147:559–576. doi: 10.1083/jcb.147.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faix J. The actin-bundling protein cortexillin is the downstream target of a Rac1-signaling pathway required for cytokinesis. J. Muscle Res. Cell Motil. (2002);23:765–772. doi: 10.1023/a:1024427712131. [DOI] [PubMed] [Google Scholar]
- 6.Faix J., Steinmetz M., Boves H., Kammerer R.A., Lottspeich F., Mintert U., Murphy J., Stock A., Aebi U., Gerisch G. Cortexillins, major determinants of cell shape and size, are actinbundling proteins with a parallel coiled-coil tail. Cell. (1996);86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- 7.Faix J., Weber I., Mintert U., Kohler J., Lottspeich F., Marriott G. Recruitment of cortexillin into the cleavage furrow is controlled by Rac1 and IQGAP-related proteins. EMBO J. (2001);20:3705–3715. doi: 10.1093/emboj/20.14.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firat-Karalar E.N., Welch M.D. New mechanisms and functions of actin nucleation. Curr. Opin. Cell Biol. (2011);23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funamoto S., Meili R., Lee S., Parry L., Firtel R.A. Spatial and temporal regulation of 3-phosphoinositides by PI 3- kinase and PTEN mediates chemotaxis. Cell. (2002);109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 10.Gerisch G., Albrecht R., Heizer C., Hodgkinson S., Maniak M. Chemoattractant-controlled accumulation of coronin at the leading edge of Dictyostelium cells monitored using a green fluorescent protein-coronin fusion protein. Curr. Biol. (1995);5:1280–1285. doi: 10.1016/s0960-9822(95)00254-5. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich D., Youssef S., Schroth-Diez B., Engel U., Aydin D., Blummel J., Spatz J.P., Gerisch G. Actin-cytoskeleton dynamics in non-monotonic cell spreading. Cell Adh. Migr. (2008);2:58–68. doi: 10.4161/cam.2.2.6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeon T.J., Lee D.J., Lee S., Weeks G., Firtel R.A. Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotaxing cells. J. Cell Biol. (2007a);179:833–843. doi: 10.1083/jcb.200705068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon T.J., Lee D.J., Merlot S., Weeks G., Firtel R.A. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. (2007b);176:1021–1033. doi: 10.1083/jcb.200607072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon T.J., Lee S., Weeks G., Firtel R.A. Regulation of Dictyostelium morphogenesis by RapGAP3. Dev. Biol. (2009);328:210–220. doi: 10.1016/j.ydbio.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Kolsch V., Charest P.G., Firtel R.A. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. (2008);121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kortholt A., van Haastert P.J. Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal. (2008);20:1415–1422. doi: 10.1016/j.cellsig.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Lee S.H., Dominguez R. Regulation of actin cytoskeleton dynamics in cells. Mol. Cells. (2010);29:311–325. doi: 10.1007/s10059-010-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S., Shen Z., Robinson D.N., Briggs S., Firtel R.A. Involvement of the cytoskeleton in controlling leading-edge function during chemotaxis. Mol. Biol. Cell. (2010);21:1810–1824. doi: 10.1091/mbc.E10-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mondal S., Burgute B., Rieger D., Muller R., Rivero F., Faix J., Schleicher M., Noegel A.A. Regulation of the actin cytoskeleton by an interaction of IQGAP related protein GAPA with filamin and cortexillin I. PLoS One. (2010);e15440. doi: 10.1371/journal.pone.0015440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. (2004);303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 21.Ren Y., Effler J.C., Norstrom M., Luo T., Firtel R.A., Iglesias P.A., Rock R.S., Robinson D.N. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr. Biol. (2009);19:1421–1428. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: integrating signals from front to back. Science. (2003);302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 23.Rodal A.A., Sokolova O., Robins D.B., Daugherty K.M., Hippenmeyer S., Riezman H., Grigorieff N., Goode B.L. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. (2005);12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki A.T., Firtel R.A. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur. J. Cell Biol. (2006);85:873–895. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki A.T., Chun C., Takeda K., Firtel R.A. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. (2004);167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroth-Diez B., Gerwig S., Ecke M., Hegerl R., Diez S., Gerisch G. Propagating waves separate two states of actin organization in living cells. HFSP J. (2009);3:412–427. doi: 10.2976/1.3239407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stock A., Steinmetz M.O., Janmey P.A., Aebi U., Gerisch G., Kammerer R.A., Weber I., Faix J. Domain analysis of cortexillin I: actin-bundling, PIP(2)-binding and the rescue of cytokinesis. EMBO J. (1999);18:5274–5284. doi: 10.1093/emboj/18.19.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchida K.S., Yumura S. Dynamics of novel feet of Dictyostelium cells during migration. J. Cell Sci. (2004);117:1443–1455. doi: 10.1242/jcs.01015. [DOI] [PubMed] [Google Scholar]
- 29.Weber I., Gerisch G., Heizer C., Murphy J., Badelt K., Stock A., Schwartz J.M., Faix J. Cytokinesis mediated through the recruitment of cortexillins into the cleavage furrow. EMBO J. (1999);18:586–594. doi: 10.1093/emboj/18.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]


