Abstract
The traditional focus on the central dogma of molecular biology, from gene through RNA to protein, has now been replaced by the recognition of an additional mechanism. The new regulatory mechanism, post-translational modifications to proteins, can actively alter protein function or activity introducing additional levels of functional complexity by altering cellular and sub-cellular location, protein interactions and the outcome of biochemical reaction chains. Modifications by ubiquitin (Ub) and ubiquitin-like modifiers systems are conserved in all eukaryotic organisms. One of them, small ubiquitin-like modifier (SUMO) is present in plants. The SUMO mechanism includes several isoforms of proteins that are involved in reactions of sumoylation and de-sumoylation. Sumoylation affects several important processes in plants. Outstanding among those are responses to environmental stresses. These may be abiotic stresses, such as phosphate deficiency, heat, low temperature, and drought, or biotic stressses, as well including defense reactions to pathogen infection. Also, the regulations of flowering time, cell growth and development, and nitrogen assimilation have recently been added to this list. Identification of SUMO targets is material to characterize the function of sumoylation or desumoylation. Affinity purification and mass spectrometric identification have been done lately in plants. Further SUMO noncovalent binding appears to have function in other model organisms and SUMO interacting proteins in plants will be of interest to plant biologists who dissect the dynamic function of SUMO. This review will discuss results of recent insights into the role of sumoylation in plants.
Keywords: Arabidopsis, SUMO, sumoylation
SMALL UBIQUITIN LIKE MODIFIER (SUMO)
The first identified SUMO homolog, suppressor of mif two 3 (Smt3), was originally discovered as a suppressor of the centromeric protein MIF2 in the budding yeast Saccharomyces cerevisiae (Meluh and Koshland, 1995). MIF2 is a kinetochore protein with homology to human CENP-C that is required for the structural integrity of the spindle during anaphase spindle elongation. Different human and mouse homologs of Smt3 have been identified and named hSmt3, Smt3A, Smt3B, and Smt3C (Chen et al., 1998; Mannen et al., 1996). This was followed by the detection of several other proteins, namely SUMO1, which was originally named ubiquitin like protein 1 (UBL1), PML interacting protein 1 (PIC1), and sentrin. Sentrin was found to interact with RAD51 (DNA repair protein RAD51 homolog)/RAD52, which are two components of the double-strand DNA break repair system, promyelocytic leukemia protein (PML), a RING finger protein involved in acute promyelocytic leukemia, and the Fas receptor (FasR, also known as Arp) involved in programmed cell death (apoptosis). (Boddy et al., 1966; Okura et al., 1996; Shen et al., 1996b). The covalent modification of RanGAP1, a GTPase-activating protein for nuclear Ran (Ras related nuclear protein, GTP binding protein), by SUMO1 triggers its association with cytoplasmic fibers of nuclear pore complexes (NPCs) (Matunis et al., 1996; 1998).
The three-dimensional structure of SUMO (12 kDa) can be superimposed onto that of ubiquitin (Ub) (9 kDa), despite their low amino acid sequence identity (~18%). This property is common for several ubiquitin-like modifiers, including RUB (related to ubiquitin) and ATG8/12 (autophagy-related protein 8/12), and SUMO. The characteristic structure of ubiquitin, termed the Ub-fold, consists of four β-strands surrounding an α helix that traverses the molecule diagonally (ββαββαβ) (Bayer et al., 1998; Vijay-Kumar et al., 1987). Similar to ubiquitin, SUMO is synthesized as a precursor with a C-terminal extension, and it is processed by a SUMO-specific protease to expose the C-terminus glycine that is essential for conjugation to target proteins. The SUMO conjugation pathway consists of a three enzyme cascade that includes an E1 SUMO activation enzyme, an E2 SUMO conjugation enzyme, and the E3 SUMO ligase. These are functionally the same as the enzymes that make up the ubiquitination system. However, a flexible N-terminal extension is present in SUMO and the surface charge of SUMO is different from that of ubiquitin, indicating different protein interactions (Bayer et al., 1998; Johnson, 2004).
The S. cerevisiae genome contains a single SUMO gene known as Smt3 that is essential for completing mitosis. The fission yeast Schizosaccharomyces pombe homolog Pmt3 is not essential for viability, but disruption of Pmt3 results in growth defects and compromised genome maintenance such as aberrant mitosis, longer telomeres and abnormal chromosome segregation. SUMO homologs are present in all eukaryotes. While yeasts, the fruit fry (Drosophila melanogaster), and Caenorhabditis elegans encode a single SUMO protein, four SUMO proteins have been identified in animals: SUMO1 (Smt3C, PIC1, UBL1, sentrin), SUMO2 (Smt3a, sentrin3), SUMO3 (Smt3b, sentrin2), and SUMO4. Human SUMO1 is a 101 amino acid polypeptide that shares 18% sequence identity with human ubiquitin. SUMO2 and SUMO3 from humans and animals share 87-95% protein sequence identity but they are only ~50% identical to SUMO1. The mRNA transcripts of SUMO1-3 are found in all tissues in humans and mice. However, the majority of the SUMO1 proteins can be found in conjugated forms, while SUMO2 and SUMO3 exist as free, unconjugated forms. The conjugation of SUMO2 and SUMO3 is strongly induced under various stress conditions, while SUMO1 conjugation is not inducible in animals (Bohren et al., 2004; Johnson, 2004; Saitoh and Hinchey, 2000). SUMO2 and SUMO3 have the consensus SUMO modification site, ψKXD/E (ψ, hydrophobic amino acid; K, lysine; X, any amino acid, D, aspartic acid; E, glutamic acid) in their N-terminal region. The sequence promotes poly SUMO chain formation in SUMO2 and SUMO3, but not in SUMO1 (Tatham et al., 2001). RanGAP1 is preferentially modified by SUMO1, but not SUMO2 and SUMO3 (Saitoh and Hinchey, 2000). These studies suggest a pattern in the functions of these proteins in animals by which irrespective of the similarities in structure, the differences in amino acid sequences between them reflect the specificity of each isoform for its own set of targets.
The first plant SUMO, T-SUMO, was found in tomato as an interacting partner of ethylene-inducing xylanase (EIX) from the plant pathogenic fungus Trichoderma viride (Hanania et al., 1999). In a search for Arabidopsis SUMO (AtSUM) orthologs in protein and DNA databases eight AtSUMOs (AtSUM1-AtSUM8) were identified. AtSUM9 is a pseudogene. Transcripts of AtSUM1, 2, 3 and 5 have so far been detected, suggesting that the other isoforms could be expressed in condition-, time- or space-specific manners. The detection of AtSUM1/2 and their conjugates in both cytoplasmic and nuclear compartments implies that both soluble and membrane-associated proteins may be targets of AtSUM1/2 (Kurepa et al., 2003). AtSUM1/2 and AtSUM3 have different conjugation dynamics and modify different sets of proteins: SUMO1 and SUMO2 conjugates, but not SUMO3 conjugates were dramatically increased by stress treatments including heat shock, H2O2, ethanol, and the amino acid analog canavanine. Especially AtSUM1 and AtSUM2 conjugates were induced within 2 min by heat shock (Kurepa et al., 2003) through reversible conjugation. Furthermore, heat-induced SUMO conjugates were detected in the nuclear fraction, suggesting that sumoylation is involved in the signaling and early response to heat stress (Saracco et al., 2007). In addition to Arabidopsis and tomato, SUMO gene families are present in other plants such as poplar, grape, sorghum, and rice (Kurepa et al., 2003; Reed et al., 2010; van den Burg et al., 2010). AtSUM1 is expressed from the early embryo stages (globule, heart, and torpedo stage) and its expression is constitutive in most organs (especially in the leaf and root), except for the vasculature, lateral root primordia and the root apex where AtSUM2 is highly expressed. AtSUM2 appears in the late heart stage in developing embryos. Although AtSUM1 and AtSUM2 are constitutively expressed, there is no overlap in their spatial expression (Saracco et al., 2007; van den Burg et al., 2010). AtSUM3 is not expressed in seeds or embryos, and its expression is restricted to hydathodes and the vasculature of mature leaves (van den Burg et al., 2010). AtSUM1 and AtSUM2 can form poly-SUMO chains for conjugation to ScPCNA, a known SUMO target in yeast in vitro. AtSUM3, on the other hand, can modify ScPCNA, but polymeric chains were not detected (Colby et al., 2006).
Single homozygous T-DNA inserted null mutants of AtSUM1 and AtSUM2, sum1-1 and sum2-1 were phenotypically similar to wild-type plants despite the reduction of heat shock-induced SUMO conjugation and free SUMO in these mutants. Similarly, heterozygous/homozygous (SUM1/sum1-1sum2-1/sum2-1) and homozygous/heterozygous (sum1-1/sum1-1sum2-1/SUM2) mutants looked similar to their wild-type counterparts, but heat shock-induced SUMO conjugation was significantly decreased. The double homozygous mutant, sum1-1sum2-1 is embryo lethal, suggesting that SUMO conjugation is essential for seed, seedling or early plant development (Saracco et al., 2007). Consistently, sum1sum2 knockdown mutants (sum1-1amiRSUM2) and transgenic plants overexpressing conjugationdeficient mutant SUM1 (ΔGG) or SUM2 (ΔGG) were smaller and flowered earlier in short days (8 h light/16 h dark) than wildtype plants. This phenotype is similar to that of the siz1-2 mutant, which is a null mutant of the SUMO E3 ligase, SIZ1 (see below) (van den Burg et al., 2010). Thus, although the spatial expression profile of AtSUM1and AtSUM2 shows some differences, these two isoforms appear to be functionally largely redundant.
PATHWAY OF SUMO CONJUGATION: MATURATION (PROTEASE), ACTIVATION (E1), CONJUGATION (E2), AND LIGATION (E3)
SUMO conjugation or sumoylation consists of the formation of a covalent bond between the C-terminal glycine carboxyl group of SUMO and the ε-amino group of the lysine residue in the sumoylation site of a target protein. The three-step enzyme pathway for the attachment of SUMO to its target shares similarity with the ubiquitin conjugation mechanism, but the enzymes involved in sumoylation are SUMO-specific.
Similar to ubiquitin, SUMO proteins are synthesized as longer precursors and processed by a SUMO-specific isopeptidase to expose the glycine at the C-terminus of mature SUMO. Mature SUMO is activated by a SUMO-specific E1 activating enzyme. Activated SUMO is transferred to the cysteine of a SUMO-conjugating enzyme E2, and then transferred from the E2 to the target lysine residue of substrates with the help of several E3 SUMO protein ligases (Johnson, 2004; Melchior, 2000). Although the target lysine residue is located in the short consensus sequence ψKXD/E, approximately 20-25% of all sumoylations occur at lysine residues in non-consensus sequences (Miller et al., 2010; Seeler and Dejean, 2003). Notably, recombinant E1, E2, and SUMO are sufficient for ATP-dependent sumoylation of targets in vitro (Bernier-Villamor et al., 2002). The enzymes involved in the SUMO conjugation system (Tables 1 and 2, Fig. 1) are described in detail in the following sections.
Table 1.
Sumoylation and desumoylation determinants in animals, yeasts, Arabidopsis and rice
| Mammals | S. cerevisiae | S. pombe | Arabidopsis | Arabidopsis locus # | Rice | |
|---|---|---|---|---|---|---|
| Modifier | SUMO-1 | Smt3 | Pmt3 | SUM1 | At4g26840 | Os01g0918300 |
| SUMO-2 | SUM2 | At5g55160 | Os01g0918200 | |||
| SUMO-3 | SUM3 | At5g55170 | Os07g0574500 | |||
| SUMO-4 | SUM4 | At5g48710 | ||||
| SUM5 | At2g32765 | |||||
| SUM6 | At5g48700 | |||||
| SUM7 | At5g55855 | |||||
| SUM8 | N.A | |||||
| SUM9 | N.A | |||||
| E1 | Sae1(Aos1) | Aos1 | Rad31 | SAE1a | At4g24940 | Os11g0497000 |
| SAE1b | At5g50580/At5g50680 | |||||
| Sae2(Uba2) | Uba2 | Fub2 | SAE2 | At2g21470 | Os07g0586500 | |
| E2 | Ubc9 | Ubc9 | Hus5 | SCE | At3g57870 | Os03g0123100 |
| Os10g0536000 | ||||||
| Os04g0580400 | ||||||
| E3-SP-RING | PIAS1 | Siz1(Ull1) | Pli1 | SIZ1 | At5g60410 | Os05g0125000 |
| PIAS3 | Siz2(Nfi) | Nse2 | SIZ2 | Os03g0719100 | ||
| PIASxα/ARIP3 | Mms21 | MMS21/HPY2 | At3g15150 | Os05g0563500 | ||
| PIASxβ/Miz1 | Zip3 | |||||
| PIASy | ||||||
| Mms21/Nsc2 | ||||||
| E3-others | RanBP2/Nup358 | |||||
| Pc2b | ||||||
| Protease | SENP1/SuPr-2 | Ulp1(Nib1) | Ulp1 | ULP1a | At3g06910 | Os01g0355900 |
| SENP2 | Ulp2(Smt4) | Ulp2 | ULP1b | At4g00690 | Os03g0410100 | |
| AXAM2, SMT3IP2 | ||||||
| SENP3 | ULP1c/OTS2 | At1g10570 | Os03g0344300 | |||
| SSP3, SMT31P1 | ||||||
| SENP5 | ULP1d/OTS1 | At1g60220 | ||||
| SENP6 | ESD4 | At4g15880 | ||||
| SUSP1, SSP1 | ||||||
| SENP7 | ULP2a | At4g33620 | Os05g0207900 | |||
| ULP2b | At1g09730 | Os01g0738100 | ||||
SUMO, small ubiquitin-related modifier; E1, two subunits of SUMO activation enzyme SAE1 and SAE2; E2, single subunit of SUMO-conjugating enzyme SCE1; E3, two types of SUMO ligase, SIZ and NSE2/MMS21; protease, two types of ubiquitin-like SUMO protease ULP1 and 2; NA, not annotated.
Table 2.
Arabidopsis proteins of the SUMO pathway
| Arabidopsis | ATG number | Location | Description |
|---|---|---|---|
| SUMO isoforms | |||
| SUM1 | At4g26840 | Nucleus | Single mutants look like wild type; double mutant is embryo lethal. |
| SUM2 | At5g55160 | Nucleus | |
| SUM3 | At5g55170 | ||
| SUM4 | At5g48710 | ||
| SUM5 | At2g32765 | ||
| SUM6 | At5g48700 | ||
| SUM7 | At5g55855 | ||
| SUM8 | N.A | ||
| SUM9 | N.A | ||
| SUMO-activating enzyme E1 | |||
| SAE1a | At4g24940 | ||
| SAE1b | At5g50580 | ||
| /At5g50680 | |||
| SAE2 | At2g21470 | sae2 is embryo lethal | |
| SUMO-conjugating enzyme E2 | |||
| SCE | At3g57870 | sce is embryo lethal | |
| E3 ligases | |||
| SIZ1 | At5g60410 | Nucleus | Dwarfism, elevated SA level with increased PR1 expression and resistance to bacterial pathogens, early flowering in short day, hypersensitivity to cold and phosphorus-limited condition (see the text). |
| MMS21/HPY2 | At3g15150 | Nucleus | Severe dwarf with short roots (fewer and shorter cells in the mature and elongation zones), premature transition from mitotic cycle to endocycle (see the text). |
| SUMO-specific isopeptidases | |||
| ULP1a/ELS1 | At3g06910 | Cytosolic compartment | Dwarfism, early flowering in short day. |
| ULP1b | At4g00690 | ||
| ULP1c/OTS2 | At1g10570 | Single mutants look like wild type; double mutant is hypersensitive to salt and plants flower early. | |
| ULP1d/OTS1 | At1g60220 | ||
| ESD4 | At4g15880 | Nucleus | Early flowering in short day. |
| ULP2a | At4g33620 | ||
| ULP2b | At1g09730 | ||
Fig. 1. The sumoylation/desumoylation cycle; Maturation, SUMO isoforms are encoded as precursor proteins and are processed by SUMO-specific cysteine proteases (ULP, ubiquitin-like protein-specific protease) with SUMO peptidase activity. Maturation consists of carboxyl terminal truncation to expose the di-glycine (GG) motif; Activation by E1, the SUMO carboxyl-terminal glycine is linked to AMP (SUMO-AMP) catalyzed by the heterodimeric E1 SUMO-activating enzymes 1 and 2 (SAE1 and SAE2) in an ATP-dependent reaction. Subsequently, the glycine of SUMO is coupled to a cysteine (C) residue in SAE2 via a high-energy thioester bond; Conjugation by E2, SUMO is transferred to a cysteine residue of the E2 SUMO-conjugating enzyme (SCE1) by transesterification catalyzed by SCE1; Ligation by E3, SUMO is transferred to the e-amino group of a lysine (K) side chain in the sumoylation consensus motif (ψKXE/D. ψ, a large hydrophobic residue; X, any amino acid; E/D, glutamic acid or aspartic acid) of the substrate protein, forming an isopeptide bond between the carboxyl of glycine in SUMO and the ε-amino of lysine in the substrate, a process that requires the E3 SUMO ligase in vivo; Deconjugation, SUMO-specific proteases (ULP) with isopeptidase activity cleave the isopeptide bond and SUMO is recycled through the conjugation system.
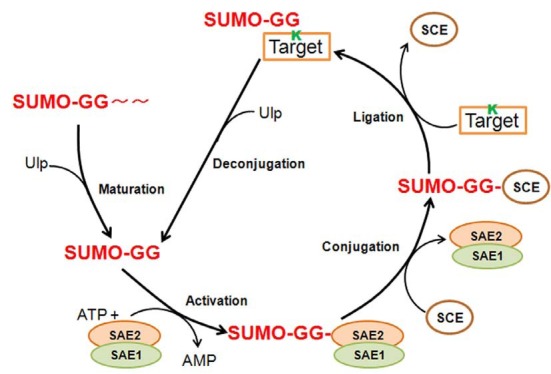
The SUMO-activating enzyme E1 and the SUMO-conjugating enzyme, E2
The SUMO-activating enzyme, E1, catalyzes a three-part reaction similar to that of ubiquitin E1 enzymes. In this reaction, the C-terminus carboxyl group of SUMO interacts with ATP (adenosine- 5′-triphosphate) to form adenylated SUMO and pyrophosphate. In next step, the thiol group (S) of an active site cysteine of E1 reacts with the SUMO adenylate, forming a thioester bond between the E1 and the SUMO C-terminus and releasing AMP (adenosine monophosphate). In the final step, activated SUMO is transferred to an active site cysteine of E2, a conjugation enzyme, forming a SUMO-E2 thioester intermediate that promotes the transfer of SUMO to a lysine residue in the target protein, mediated directly or indirectly by an E3 ligase (Johnson, 2004; Walden et al., 2003).
The SUMO E1 enzyme is a heterodimer consisting of a ~40 kDa Aos1 subunit (SAE1, Sua1) and a ~70 kDa Uba2 protein (SAE2, hUbs2) [yeast (Arabidopsis, human)], while the ubiquitin activating enzyme, Ub E1 is encoded by a single gene, Uba1. Aos1 and Uba2 show similarity to the N-terminus and C-terminus of Ub E1, respectively. The Arabidopsis E1 proteins are SAE1a (36 kDa) and SAE1b (35.7 kDa) as the small SAE1 proteins, and a single SAE2 (18 kDa). SAE1a/b and SAE2 are 28-33% identical to animal orthologs but share conserved domains including the active site cysteine, the Ub-activating enzyme C-terminal, UBA-CT (in SAE2), and the thiamine biosynthesis proteins F and ThiF (in SAE1a/b and SAE2) (Kurepa et al., 2003). SAE1a and SAE1b are 81% identical at the amino acid sequence level, suggesting functional redundancy (Saracco et al., 2007).
SUMO E2 is encoded by a single gene in yeast, humans, and Arabidopsis (Ubc9 in yeast and humans, SCE in Arabidopsis) whereas there are many Ub E2s. AtSCE shows sequence similarities of 64%, 59%, and 58% to HsUBC9, SpHUS5, and ScUBC9, respectively. AtSCE conjugates human SUMO1 to mouse RanGAP in vitro, and the E2 SUMO conjugation activity of AtSCE, which transfers SUMO from E1 to a consensus receptor lysine, was suppressed when the catalytic domain cysteine was mutated to a serine (Lois et al., 2003). SUMO conjugation was ATP-dependent, as suggested by the 32P-labeled SUMO produced in an ATP regenerating system but not in an ATP depleted system when labeled SUMO was incubated with a crude Arabidopsis protein extract (Kurepa et al., 2003).
In budding yeast, any single mutation in Aos1, Uba2 or Ubc9 leads to a loss of viability based on cell cycle defects (references in Saracco et al., 2007). In fission yeast, the corresponding mutants are viable but have defects in growth and mitotic processes, and alterations in genome maintenance (references in Saracco et al., 2007). The mRNA transcripts of SAE1a/b and SCE were detected in all Arabidopsis tissues including seedlings, cotyledons, roots, the stem, shoot tip, leaves, flower and siliques. In addition, AtSCE1 colocalized with AtSUM1/2 in the nucleus (Lois et al., 2003). Null T-DNA-inserted mutants of SAE2 and SCE1 (sae2 and sce1) were embryo lethal and arrested in an early embryonic stage (globular, heat, early torpedo), indicating that E1 and E2 are as essential for SUMO modification as AtSUM1/2 (Saracco et al., 2007).
The SUMO protein ligase E3
Unlike HECT type ubiquitin ligases, SUMO E3 ligases do not form a thioester bond with SUMO. SUMO E3s bind to the SUMO E2 conjugating enzyme Ubc9 and promote the transfer of SUMO to substrates, functioning as a bridge or adaptor between the E2 and the substrate (Gill, 2004).
SP-RING (Siz/PIAS RING) domain proteins {Siz (SAP and Miz finger domain) in yeast and PIAS [Protein Inhibitor of Activated STAT (signal transducer and activator of transcription-1)] in vertebrates} were identified as SUMO E3 ligases. The SPRING domain, which resembles the RING domain found in ubiquitination E3 ligases, binds to Ubc9 and is essential for SUMO E3 ligase activity. In addition, SP-RING proteins contain a SAP [Scaffold Attachment factor (SAF)-A/B, acinus, PIAS] motif (DNA binding), a PINIT motif (proline-isoleucine-asparagine- isoleucine-threonine, nuclear retention of SP-RING protein/ subcellular localization), a SIM (SUMO interacting motif) or SXS motif (interacting with SUMO, serine-X-serine, X is any amino acid), and a C-terminal domain. Fruit flies and worms have one E3 ligase named Su(var)2-10/zimp and gei-17, respectively, and loss of function mutants lead to embryonic lethality. In budding yeast, two ligases are present, namely Siz2/ Nfi1 and Siz1/Ull1, which are required for Smt3p conjugation and have different substrates. Siz1 and Siz2 in yeast are not essential for cell viability, unlike Aos1/Uba2 (E1 heterodimer) and Ubc9 (E2), as double siz1Δsiz2Δ mutants are viable and show low levels of SUMO conjugates probably derived from another SUMO E3, Mms21 (see below). Fission yeast has one SP-RING protein, Pli1, while mammals have five PIASs, namely PIAS1, PIAS3, α and β spliced forms of PIASx, and PIASy, which are encoded by four genes and are implicated in gene expression, signal transduction and genome maintenance (Johnson, 2004).
Arabidopsis has one AtSIZ1 and loss of SIZ1 function phenotypes include dwarfism, elevated SA level with increased PR1 (Pathogenesis Related gene 1) expression and resistance to bacterial pathogens, early flowering in short days, and hypersensitivity to cold and phosphorus limited conditions. These phenotypes led to the discovery of the SIZ1 targets PHR1, ICE1, FLD, and NIA1 and NIA2 (see “FUNCTIONS OF SUMOYLATION IN PLANTA”). AtSIZ1 has four conserved domains including SAP, PINIT, SP-RING, and SXS, and one plant PIAS protein-specific domain called PHD (plant homeodomain). PHD and SP-RING domains are required for binding to AtSCE and for sumoylation of GTE3 (global transcription factor group E3, Bromodomain-containing protein) (Garcia-Dominguez et al., 2008). Expression of the site directed-mutated domains named SIZ1sap, SIZ1phd, SIZ1pinit, SIZ1sp-ring, SIZ1sxs in siz1-2 mutants showed that the SP-RING domain was required for the punctuate subnuclear structure of SIZ1, and demonstrated that SUMO conjugation in vivo is involved in thermosensitivity, SAdependent resistance to bacterial pathogens, and SA-independent ABA sensitivity. The PHD and PINIT domains are likely to be involved in sumo-ylation and regulate the elongation of hypocotyls in response to sugar and light (Cheong et al., 2009).
Another SP-RING type protein characterized by its SP-RING domain, but with the loss of the SAP and PINIT domains, is methyl methanesulfonate sensitive 21 (Mms21). The fission yeast Pli3 protein and the Siz1 and Siz2 proteins in budding yeast are not essential, whereas deletion of Nse2 (fission yeast) and Mms21 (budding yeast) results in lethality. Human MMS21 is required for DNA repair (Potts and Yu, 2005). Arabidopsis has one AtMMS21/HPY2 (High ploidy 2) that is involved in cell proliferation and root meristem maintenance (see “FUNCTIONS OF SUMOYLATION IN PLANTA”).
Mammalian cells contain additional non-SP-RING type SUMO E3s that lack the SP-RING domain but have a distinct domain for binding to the E2 enzyme Ubc9, among them RAN Binding Protein 2 (RanBP2) and Polycomb group protein 2 (Pc2). RanBP2 is a nuclear-pore protein localized to cytoplasmic fibrils that mediates the sumoylation of certain substrates such as Ran GTPase activating protein (RanGAP). Pc2 is the specific SUMO E3 ligase for the transcriptional co-re-pressor C-terminal binding protein of adenoviruns E1A (CtBP) (Wang and Dasso, 2009). In animal cells, the localization of E3 ligases contributes to their functional specificity for a substrate or target: RanBP2 is associated with the nuclear pore complex, PIASs localizes to the nucleoplasm and nuclear bodies, and Pc2 is found in a subnuclear structure called the Polycomb body (Gill, 2004).
SUMO deconjugation, reversible SUMO conjugation (SUMO-specific protease)
Accumulation of heat shock-induced AtSUM1/2 conjugates was found to begin within 2 min of heat stress, and suspension of the heat stress stimulus resulted in the return of SUMO conjugation to a steady-state level and an increase in unconjugated free AtSUM1/2 levels (Kurepa et al., 2003). Rapid SUMO deconjugation was inhibited by N-ethyl-maleimide (NEM) (Lois et al., 2003). Plants have SUMO specific proteases that cleave the C-terminus of the nascent SUMO to expose the C-terminus glycine. Specific proteases also function to remove SUMO moieties from conjugated proteins and recycle free SUMO. Yeast has two SUMO-specific proteases called ScUlp1 (Ub like protein protease 1) and ScUlp2/Smt4. ScUlp1 null mutants are not viable due to defects in cell cycle progression, but ScUlp2 mutants are viable and show pleiotropic phenotypes such as abnormal cell morphology and decreased chromosome stability (references in Colby et al., 2006). The mammalian genome encodes six SUMO-specific proteases referred to as SENP1 (sentrin-specific protease 1), SENP2, SENP3, SENP5, SENP6, and SENP7. Arabidopsis has seven SUMO-specific proteases, namely ESD4, ULP1a/ELS1 (Ub like protein protease 1a/ESD4- like SUMO protease), ULP1b, and ULP1c/OTS2, which are homologous to yeast ScUlp1. ULP2a and ULP2b are homologous to yeast ScUlp2. Plants have an additional protease involved in salt stress tolerance named ULP1d/OTS1 (Colby et al., 2006; Hermkes et al., 2011; Kurepa et al., 2003; Miura and Hasegawa, 2010; Miura et al., 2007a). However, only four Arabidopsis SUMO-specific proteases have been characterized functionally: AtESD4, ULP1a/ELS1, ULP1c, and ULP1d.
AtESD4 and AtULP1c/d are located in the nucleus, as are the yeast proteases ScUlp1 and ScUlp2 (Conti et al., 2008; Murtas et al., 2003). AtESD4 was originally isolated from a deletion mutation in ESD4 (early in short days 4) causing extreme early flowering in short days and abnormal shoot development including inflorescence defects (Murtas et al., 2003; Reeves et al., 2002). Similarly, single T-DNA insertion mutants of OTS1 (AtULP1d) and OTS2 (AtULP1c) did not exhibit significant phenotypes, but the double mutant ots1ots2 (overly tolerant to salt 1 and 2) flowered earlier, was sensitive to salt, and had a short root phenotype (100 mM NaCl). By contrast, over-expression of OTS1 resulted in increased resistance to salt (Conti et al., 2008). Recently, ULP1a/ELS1 was found to have SUMO specific proteolytic activity but to locate to the cytosolic compartment or to endo-membrane associated structures (Hermkes et al., 2011).
Plant ULPs and ESD4 contain the conserved 200-amino acid ULP1-Catalytic (ULP1-C) domain surrounded by a catalytic triad of histidine (H), or aspartic acid (D), and cysteine (C) residues. This structure is similar to that of yeast ScUlp1 and ScUlp2/Smt4 (Chosed et al., 2006; Kurepa et al., 2003; Murtas et al., 2003). The protease activity of AtESD4 was lost when the 488th amino acid, an active site cysteine, was mutated to serine (Murtas et al., 2003). The dominant negative mutant OTS1 C526S (site-directed mutation of the active cysteine to serine) did not exhibit the same resistance to salt as OTS1 overexpressing transgenic plants (Conti et al., 2008). Similar to the effect of the N-terminal domain of ScUlp1 on its protease activity, localization, and target-binding properties (references in Conti et al., 2008), the N-terminal regulatory domain of AtULP1s plays a role in substrate-specificity and the activity of the catalytic domain. The N-terminus catalytic cores of AtULP1a and AtESD4 show 65% sequence identity and those of AtULP1c and AtULP1d show 72%. Consistent with their sequence identity/ similarity, AtULP1a and AtESD4 require the N-terminus regulatory domain for peptidase activity. Although all four peptidases (AtESD4 and AtULP1a/1c/1d) can cleave AtSUM1/2 conjugates, AtSUM3-HA conjugates are processed only weakly by AtULP1a and none of the proteases cleaves AtSUM5-HA (Chosed et al., 2006). AtULP1a has higher SUMO peptidase activity than isopeptidase activity, but AtESD4 is more active as a deconjugating enzyme than as a SUMO-processing peptidase (Chosed et al., 2006). Similarly, a separate study showed that the four Arabidopsis SUMO peptidases deconjugated AtSUM1/2 conjugates of yeast ScPCNA, which is a known target, but AtSUM3-ScPCNA conjugates were only cleaved by XopD, a bacterial virulence factor secreted by Xanthomonas campestris (see below). All four proteases processed AtSUM1/2 precursors to the mature AtSUM1/2, but none of the proteases produced mature AtSUM3 or AtSUM5 from their precursors (Colby et al., 2006).
FUNCTIONS OF SUMOYLATION IN PLANTA
Environmental (Abiotic) stress
The conjugation of AtSUM1/2 to protein substrates is significantly induced by heat shock, cold, oxidative stress, and drought. Loss and gain of function analyses have revealed that sumoylation is involved in plant responses to environmental signals and signaling pathways (Conti et al., 2008; Kurepa et al., 2003; Miura et al., 2007b; Yoo et al., 2006).
SIZ1 was originally isolated as a phenotype suppressor from NaCl sensitive sos3 (salt over sensitive 3) mutant seedlings (Miura et al., 2005). The siz1 mutants siz1-1/2/3 and siz1-1sos3-1 showed hypersensitive responses to Pi starvation characterized by short primary roots, increased number of lateral roots and root hairs and anthocyanin accumulation. These mutants showed an increase in the transcript levels of AtPT2, AtPS2, and AtPS3 under Pi-sufficient conditions and/or in the early stages of Pi starvation, but the induction of AtIPS1 and AtRNS2, which are controlled by PHR1 (phosphate starvation response 1), a MYB transcription factor, was reduced under Pi starvation conditions. PHR1 was sumoylated by AtSIZ1 in vitro. As PHR1 plays a central role in regulating many late Pi starvation genes, and AtSIZ1 is upregulated under Pi-deficient conditions, SIZ1-mediated sumoylation of PHR1 acts positively on Pi-starvation induced gene expression (Lin et al., 2009; Miura et al., 2005).
SIZ1 expedites basal thermotolerance independent of salicylic acid (SA) (Yoo et al., 2006). The siz1 T-DNA insertion mutants showed basal thermosensitivity with a lower germination rate at 45℃ for 4 h, but a similar capacity for acquired thermotolerance compared to wild type (basal heat tolerance is the tolerance of heat shock without prior heat acclimation, whereas acquired thermotolerance is acclimated heat tolerance, induced by prior exposure to moderately high temperatures, which enables the plant to cope with subsequent lethal heat exposure). SA-dependent signaling facilitates basal thermotolerance and plays a role in acquired thermotolerance by inducing the expression of HSPs (heat shock proteins). The siz1 mutants that accumulate higher levels of SA exhibit reduced basal thermotolerance independently of SA, as seen in the lower tolerance of NahGsiz1-2 compared to siz1 mutants and the lack of detectable changes in the gene expression of HSF (heat shock factors) and heat shock-induced genes in siz1 seedlings (Clarke et al., 2004; Larkindale et al., 2005) [NahG is the SA-degrading salicylate hydroxylase from Pseudomonas putida (Delaney et al., 1994)]. SIZ1-mediated basal thermotolerance in Arabidopsis seems to operate through a different signaling pathway because in other eukaryotes, sumoylation of HSFs regulates downstream expression of HSPs and acquired thermotolerance.
AtSIZ1-mediated sumoylation is essential for chilling and freezing tolerance (Miura et al., 2007b). The siz1-2 and siz1-3 mutants are less tolerant to cold and chilling stress than their wild-type counterparts, but transgenic plants overexpressing SIZ1 show normal tolerance. Cold-induced expression of CBF [CRT (C-repeat)/dehydration responsive element (DRE) binding protein]/dehydration responsive element binding protein (DREB1), especially CBF3/DREB1A, and their downstream genes (COR- 15A, CAR47, KIN1) was repressed in siz1-1 mutants, while MYB15 expression was induced. However, expression of siz1 did not change the constitutive transcript levels of transcription factor ICE1 (inducer of CBF/DREB1 expression), which is targeted and polyubiquitinated by the RING type ubiquitin E3 ligase HOS1 (High expression of osmotically responsive gene) leading to proteasome degradation. Instead, SIZ1 mediates SUMO1 conjugation to ICE1 and stabilizes the activity of ICE1. The K393R mutation in SIZ1 blocked the sumoylation of ICE1, which caused the inhibition of the expression of CBF/DREB1s and regulon genes, and the upregulation of MYB15, a negative regulator of CBF3/DREB1A. SIZ1-mediated sumoylation of ICE1 therefore not only stabilizes ICE1, but also facilitates downstream CBF expression and MYB15 repression during cold acclimation.
SIZ1 regulates drought responses, as revealed by the increased sensitivity of siz1-3 to drought stress and the repression of drought-induced SUMO conjugation in the siz1-3 mutant (Catala et al., 2007). Drought-induced SUMO conjugation appeared to be independent of ABA signaling, as accumulation of SUMO-protein conjugates in the ABA-deficient mutant aba2 was similar to that of wild-type plants and included enzymes of the anthocyanin synthesis pathway and jasmonate response.
SUMO is involved in salt responses. Two SUMO proteases, OTS1/ULP1d (Overly Tolerant to Salt1) and OTS2/ULP1c, show redundancy in their function in salt stress responses, as seen in the hypersensitivity of the double mutant ots1ots2, but not the single homozygous mutants, to salt (100 mM NaCl). In addition, root development and desumoylation of AtSUM1/2 conjugates were impaired in the double mutant (Conti et al., 2008).
Aabscisic acid (ABA) signaling
Sumoylation and desumoylation are implicated in ABA signaling in plants. Overexpression of AtSUM1 or AtSUM2 attenuated ABA-mediated root growth inhibition and siz1 mutants were hypersensitive to ABA, showing lower germination rates and shorter root length (Lois et al., 2003; Miura et al., 2009). Expression of ABA-responsive genes containing ABREs (ABA Responsive Elements) such as RD29A, RD29B, AtEm6, RAB18, and ADH1 was increased in siz1-2 and siz1-3 mutants, suggesting that SIZ1 functions as a negative regulator of ABA signaling. However, genetic crosses showed that although ABI5 (ABA Insensitive 5) was epistatic to SIZ1, the transcript levels of ABI5 were not affected in siz1 mutants. This was found to be due to the SIZ1-mediated SUMO conjugation of the 391th lysine of ABI5, which negatively regulated ABA signaling. Expression of ABI5 (K391R) in abi5-4 exhibited hypersensitive (increased) expression of ABA-response genes and hypersensitive phenotypes including lower germination rates and shorter root growth in response to ABA. It was proposed that SIZ1- mediated sumoylation of ABI5 protects this transcription factor from 26S proteasome degradation mediated by the ABI5- binding protein AFP and the RING type ubiquitin E3 ligase KEG (Keep on Going), and rapid desumoylation of ABI5 activates the protein, inducing ABA signaling (Miura and Hasegawa, 2010; Miura et al., 2009). In addition, mutations in the SP-RING and SXS domains of SIZ1 did not rescue the hypersensitivity of siz1-2 mutant to ABA, suggesting that the SP-RING and SXS domains are responsive to ABA signaling (Cheong et al., 2009) Rice OsSIZ1 and OsSIZ2 partially complemented the hypersensitivity of the siz1-2 mutant to ABA in Arabidopsis, indicating that OsSIZ1 and OsSIZ2 function as SUMO E3 ligases and are implicated in ABA signaling (Park et al., 2010).
Cell growth and development
Sumoylation and the components of the SUMO conjugation machinery are essential for cell viability, as shown by the embryo lethality of null mutations in SAE2 and SCE and the double mutant sum1-1sum2-1 (Saracco et al., 2007). The loss of function mutant siz1 is a dwarf with decreased number and size of cells compared to the wild type, but is still viable, probably due to the action of another SUMO E3 ligase, MM21/HPY2 (Catala et al., 2007; Huang et al., 2009; Ishida et al., 2009; Jin et al., 2008). The dwarfism and reduced growth of this mutant was suppressed by NahG, which restored the reduced expression of XTH8 and XTH31 (xyloglucan endotransglycosylase and hydrolase) in siz1-2, probably facilitating leaf cell expansion. It was suggested that SIZ1 regulates cell expansion and division by regulating SA accumulation (Miura et al., 2010). A T-DNA-inserted null mutant of Rice OsSIZ1, a functional ortholog of AtSIZ1, showed shorter primary roots, more and longer adventitious roots and root hairs, and dwarfism (Wang et al., 2011).
Recent studies showed that another SUMO E3 ligase, AtMMS21/HPY2, regulates cell proliferation and root meristem maintenance (Huang et al., 2009; Ishida et al., 2009). The loss of function T-DNA insertion mutant mms21-1 exhibited a short root phenotype with fewer and shorter cells in the mature and elongation zones. The cytokinin response genes, ARR1 ARR3, ARR4, ARR5 and ARR7 and the cytokinin receptor CRE1 were down-regulated in the mms21-1 mutant, but the expression of ARR6 was not affected (Huang et al., 2009; Zhang et al., 2010). Loss of HPY2 function resulted in a severe dwarf phenotype and premature transition from the mitotic cycle to the endocycle with additional rounds of endoreduplication. The involvement of HPY2 in the promotion of the mitotic cell cycle and/or the repression of the entry into the endocycle in the root meristem was demonstrated by a reduction in the cyclin dependent kinase B1 (CDKB1) and CDKB2 mRNA and protein levels, and the transcription level of the mitotic cyclin CYCB in hpy2. HPY2 is expressed in proliferating cells including the epidermis, cortex, endodermis, and pericycle in the root meristem and lateral root primordia, and their meristems and HPY2 expression and accumulation are positively regulated by PLT (plethora), an AP2 domain transcription factor (Ishida et al., 2009).
Flowering
Mutations in the SUMO protease ESD4 and its interacting partner NUA (nuclear pore anchor) result in early flowering under short and long days (Murtas et al., 2003; Reeves et al., 2002; Xu et al., 2007). The transcript level of the floral repressor FLC (Flowering Locus C) was reduced in esd4, and the expression of flowering-time genes repressed by FLC was increased, thus accelerating the transition from vegetative growth to flowering. The double mutant ots1ots2 flowered early compared with the wild type and single mutants (Conti et al., 2008). In addition, a knockdown mutant of ULP1a/ELS1 exhibited early flowering in short days (Hermkes et al., 2011), suggesting that the SUMO proteases ESD4 and AtULP1s negatively regulate the transition to flowering.
The siz1 mutant exhibited early flowering under short day conditions in Arabidopsis. This is partially due to the elevated SA level in siz1 mutants, as the flowering time of NahGsiz1-2 and NahG plants was similar to that of wild-type plants in the Columbia background. Another SA-independent pathway is the SIZ1-mediated SUMO modification of flowering locus D (FLD). SIZ1-mediated sumoylation of FLD suppresses FLD activity, which promotes acetylation of histone 4 in FLC and results in increased FLC expression and the subsequent suppressed expression of floral genes (Jin et al., 2008). Consistent with the phenotype of the siz1 mutants, sum1sum2 knockdown mutants exhibited early flowering in short days. By contrast, loss of SUM3 showed late flowering and overexpression of SUM3 caused early flowering (van den Burg et al., 2010). These results show that sumoylation and desumoylation are involved in different pathways that regulate flowering. Studies in rice showed that both siz1 mutants and SIZ1-RNAi rice transgenic plants exhibited defects in anther dehiscence, but pollen viability and flowering time were not affected. This finding is supported by the observation that mutant and transgenic plants expressed lower amounts of the rice orthologs of the Arabidopsis IRX and maize MADS2, which are known to be essential for endothelium development and anther dehiscence (Thangasamy et al., 2011). Rice SIZ1 is 50% and 41% identical to Arabidopsis SIZ1 and SIZ2, respectively. However, whether monocot rice SIZs function in a similar manner as dicot Arabidopsis SIZs remains to be elucidated.
Plant pathogen interactions
Sumoylation plays a role in the interaction between plants and plant pathogens and in the progression of pathogenesis. Tomato SUMO, or T-SUMO, was originally isolated as an interacting partner of ethylene-inducing xylanase (EIX) from the pathogenic fungus Trichoderma viride, and overexpression of T-SUMO in tobacco suppressed cell death and ethylene biosynthesis induced by EIX (Hanania et al., 1999). Tobacco NbSCE1 was found to interact with replication initiator protein (RepAC1/ Rep) from geminivirus tomato golden mosaic virus (TGMV) and Tomato yellow leaf curl Sardinia virus (TYLCSV) (Castillo et al., 2004).
Phytopathogenic bacteria use a type III secretion system (TTBS) to inject effector proteins into host cells, and some virulence factors possess SUMO specific protease activities even though prokaryotic bacteria do not have sumoylation and desumoylation systems. The pathogen applies TTBS effectors mimicking SUMO-specific proteases to disrupt cellular processes in the host and facilitate infection. The avirulence factor AvrBsT from the plant pathogen Xanthomonas campestris pv. campestris, a homolog of YopJ (Yersinia outer protein J), which is a virulent factor from Yersinia pestis (causative agent of Black Death) that shares the triad active sites (His154, Glu173, and Cys222), has SUMO protease activity. While wild-type AvrBsT protein induced a resistant, hypersensitive response (HR) in tobacco leaves (Nicotiana benthamiana), the AvrBsT protein containing mutations in the active site showed a disrupted signaling pathway and did not elicit a hypersensitive response (Orth et al., 2000). Another avirulent factor, AvrXV4 from Xanthomonas campestris pv vesicatoria 4, has SUMO-specific protease activity. Like AvrBsT, mutations in catalytic core residues of AvrXV4 resulted in a suppressed AvrXv4- dependent HR in tobacco (Roden et al., 2004). A unique virulent factor, XopD, which is secreted from X. campestris pv. vesicatoria was able to desumoylate/cleave not only AtSUM1/2 conjugates but also AtSUM3 conjugates of ScPCNA, which were not released by plant SUMO proteases such as AtULP1c, AtULP1d, and AtESD4 in vitro (Colby et al., 2006). XopD targets to the host nucleus and binds to host DNA via its Nterminus DNA-binding domain (DBD). XopD has the conserved active sites of a SUMO protease (H/E/C), and mutation of the 470th cysteine to alanine caused the loss of plant-specific SUMO protease activities. Interestingly, although XopD was required for bacterial growth, it reduced chlorophyll loss and SA levels in the host plant, suppressed senescence-associated host gene responses and plant defense-associated gene transcription through its two EFR-associated amphiphilic repression (EAR) motifs, and delayed plant disease symptoms in infected tomato leaves (Hotson and Mudgett, 2004; Kim et al., 2008).
Sumoylation is implicated in plant pathogen defenses. Null siz1 mutants exhibit constitutive expression of pathogenesisrelated (PR) genes and disease response genes (such as EDS1, PAD4, SID2, EDS5 but not NDR1 and NPR1), leading to increased resistance to the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000. These effects are due to elevated SA, as introducing NahG into siz1 mutants reduced SA levels to normal and restored the wild-type phenotype. Mutant crossing analysis showed that PAD4 is epistatic to SIZ1, suggesting that SIZ1 negatively regulates SA-mediated or - dependent defense signaling and responses through a PAD4- dependent pathway in non-infected plants (Lee et al., 2006). The sum1sum2 knockdown mutant sum1amiR-Sum2 displayed elevated SA levels, constitutive expression of PR1, subsequent resistance to Pst DC3000, and spontaneous cell death. In addition, it flowered early in short days similar to the siz1 mutant (Lee et al., 2006; van den Burg et al., 2010). Heat shock-induced AtSUM1/2 conjugation was suppressed in sum1sum2 knockdown mutants and siz1, suggesting that SIZ1 functions through conjugation to AtSUM1/2 (Cheong et al., 2009; van den Burg et al., 2010). Overexpression of AtSUM1-3 and their conjugation-deficient mutants AtSUM1-3ÄGG resulted in increased resistance to Pst DC3000, increased SA levels, and reduced cell death (HR). AtSUM3 transcript levels, but not those of AtSUM1/2, rapidly and transiently increased after treatment with the bacterial elicitor Flg22 in an SA-dependent manner (van den Burg et al., 2010). These results imply that plant defense systems inhibit the AtSUM1/2 dependent suppression of SA signaling and allow SA accumulation, which turns on AtSUM3 expression.
Metabolism: nitrogen assimilation
The phenotypes of siz1-2 including dwarf, early flowering, poor seed development, and high accumulation of SA leading strong resistance against bacterial pathogen result from loss of function of SUMO E3 ligase, and functional defects of proteins which are supposed to be sumoylated to be active. Interestingly supplementary ammonium resource complements the characteristic phenotype of siz1-2, but not other nitrogen source such as nitrate (Park et al., 2011b). Rather nitrate content is much higher in siz1-2 than wild type and nitrate reductase (NR) activity is reduced in siz1-2. Actually two NR, NIA1 and NIA2 could be SUMO conjugated. Sumoylation seems to regulate the NR activity by stabilizing the proteins.
SEARCHING ATSUMO TARGETS
In yeast and animal cells, SUMO attachment and desumoylation are involved in nuclear and subnuclear localization, DNA repair, mechanisms that antagonize ubiquitination, gene expression (transcriptional regulation), genome maintenance, protein-protein interaction, and signal transduction (Geiss-Friedlander and Melchior, 2007; Gill, 2004). Systematic screens have identified SUMO targets and the regulatory mechanisms of the target proteins related to sumoylation have been characterized (Golebiowski et al., 2009; Xu and Peng, 2006).
The identified targets of AtSUMOs are mostly transcription factors such as ICE1, FLD, and ABI5 that are involved in regulating or modulating different biological pathways, and are characterized through null loss of function siz1 mutant phenotypes (Jin et al., 2008; Miura et al., 2005; 2007b; Yoo et al., 2006). Yeast two-hybrid assays and immunoprecipitation experiments have been applied for screening or isolating SUMO conjugated proteins and/or SUMO-associating proteins (Budhiraja et al., 2009; Elrouby and Coupland, 2010; Garcia- Dominguez et al., 2008; Miller et al., 2010). GTE3, which belongs to the bromodomain and extra terminal (BET) domain protein family, was identified by a yeast two-hybrid screen using truncated SIZ1ΔC with SAP, PHD, and SP-RING domain as bait (Garcia-Dominguez et al., 2008). Affinity purification and mass spectrometric identification using tagged AtSUM1 (or 3 or 5) in plants with transient inducible expression added 15 potential SUMO targets involved in RNA-dependent and chromatinrelated processes (Budhiraja et al., 2009). Systemic yeast twohybrid screening using the SUMO E2 activating enzyme SCE and the SUMO specific protease ESD4, which interact with SUMO substrates for sumoylation and desumoylation, respectively, yielded 238 potential targets and 124 proteins were sumoylated experimentally. These proteins include sumoylation system components, transcription factors, proteins involved in RNA processing, chromatin and genome stability and maintenance, proteins for protein translation, folding, and stability, and stress responding genes (Elrouby and Coupland, 2010). Not only nucleus-localized proteins, but also chloroplast, mitochondrial, and cytoplasmic proteins were found, suggesting that SUMO conjugation occurs in other cell compartments. Affinity purification and enrichment with two rounds of Ni-NTA affinity chromatography and anti-SUMO antibody affinity chromatography, followed by LC-MS/MS with protein extracts from His- H89R SUMO1 sum1-1sum2-1-expressing transient plants allowed the identification of 357 putative SUMO targets (Miller et al., 2010). Protein extraction and subsequent purification included strong denaturants such as 7M guanidine or 8M urea to eliminate non-covalently bound conjugates and background from abundant ribulose-1,5-bisphosphate carboxylase oxygennase (RuBisCO) a nd c rosslink ed immunoglobulins from a ntibodies. The results showed that 76% of them were nuclearlocalizing proteins including transcription factors, components of RNA metabolism and processing, chromatin binding/remolding proteins, and cell cycle regulators. This list also contained components of the SUMO conjugation and deconjugation machinery, and of DNA repair/maintenance systems. In addition, transcription factors involved in hormonal signaling and stress responses were included. In this study, many proteins were present as sumoylated proteins both under normal conditions and stress-treated conditions such as heat and oxidative stresses suggesting stress increases fraction of SUMO conjugated form (Miller et al., 2010).
Our group followed two different approaches for screening sumoylated proteins: the first system included the overexpression of AtSUM1 fused to a 6His-3Flag tag in Arabidopsis. Total protein extracts were applied to Ni- NTA affinity chromatography columns followed by 2D SDSPAGE. Gel-excised putative SUMO-binding candidate proteins were identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry and were found to be involved in DNA/RNA-related processing, signaling pathways and metabolism (Park et al., 2011a). Another system was designed to utilize affinity chromatography with His-GSTAtSUM1 and total protein extracts from Arabidopsis under native conditions without strong denaturants to improve the probability of including SUMO binding conjugates. SUMO-binding protein complexes were then purified and identified by HPLC followed by tandem mass spectrometry (MS/MS). The list of 15 putative SUMO-binding proteins includes nucleus-localizing proteins such as the transcription factor MYB31 and the RNA processing-related genes DEAD helicase, as well as cytoplasmic proteins such as 1-amino-cyclopropane-1-carboxy-late synthase 7 (ACS7) (manuscript in preparation).
All studies for mining SUMO targets showed that many of them were transcription factors and genes involved in RNA/ DNA-dependent and/or chromatin-related processes. However, some candidate proteins were not nucleus-localized proteins, indicating that the nucleus is not the only place where sumoylation occurs. Although the øKXE/D motif is known as the canonical consensus sequence for covalent SUMO attachment, the motif is not essential for sumoylation because 20% of putatively sumoylated proteins isolated from His-H89R SUMO1 sum1-1sum2-1 plants or obtained by affinity purification do not contain this consensus sequence (Miller et al., 2010). This is consistent with the idea that the three-dimensional position or location of the target site lysine is important for SUMO conjugation (Bernier-Villamor et al., 2002). However, induced folding could also mask or expose a lysine that is not surrounded by the specific motif. Defining the putative SUMO targets will elucidate the biological and regulatory processes and mechanisms that govern sumoylation in planta.
SUMO BINDING
SUMO can be covalently conjugated to specific eukaryotic target proteins mainly through the consensus motif, ψKXE/D in the substrate proteins. Since the non-covalent interaction between human Rad51/52 and SUMO1 was found, additional substrates that interact with SUMO non-covalently have been identified using the yeast two-hybrid assay, which can differentiate between the covalent and non-covalent interactions with SUMO (Kroetz and Hochstrasser, 2009; Li et al., 2000; Shen et al., 1996a; 1996c). Non-covalent binding to SUMO occurs through SUMO-interacting motif (SIM) also known as SUMO-binding domain (SBD) and SUMO-binding motif (SBM) in the targets (Kerscher, 2007).
SUMO non-covalent binding is considered analogous to the ubiquitin system, where the covalently conjugated ubiquitin on substrates can recruit ubiquitin-binding proteins that bind noncovalently. Sumoylation probably provides the interaction platform for an effector protein, which has SUMO-binding activity or SIM (Grabbe and Dikic, 2009; Wilkinson and Henley, 2010).
The first study of SUMO-binding proteins and SIM was based on SUMO-interacting proteins from yeast in a two-hybrid assay using sumoylated human p73α, a member of the p53 family as the bait (Minty et al., 2000). It was proposed that an SXS motif (X is any amino acid and S is serine) surrounded by three or four acidic residues at the C-terminus and a few hydrophobic residues at the N-terminus is the requirement for SUMO interaction based on sequence comparisons. The importance of serine for SUMO binding was put in doubt by another group that instead suggested the involvement of a few amino acid sequences, V/I-X-V/I-V/I (Val/Ile-X-Val/Ile-Val/Ile) in the SIM in a study using nuclear magnetic resonance (NMR): a SIM forms an extended structure that is bound between the α-helix and β-strand of human SUMO1 (Song et al., 2004; 2005). Some SUMO-binding proteins indeed contain this motif, including the E3 ligase PIASx, the Ran binding protein2 (RanBP2/Nup358), and the SUMO activating enzyme unit Uba2/SAE2. From additional studies, SIMs are now defined by a hydrophobic consensus sequence that includes a cluster of valine, isoleucine, and leucine residues preceded or followed by a cluster of acidic (negatively charged) and/or phosphorylated (serine or threonine) residues. This region aligns as a â-strand with a matching region on the surface of SUMO (Hannich et al., 2005; Hecker et al., 2006). The hydrophobic groove in human SUMO1 and SUMO2, and yeast Smt3 is surrounded by basic residues that associate with negative charges in the SIM (Chupreta et al., 2005). The arrangement of hydrophobic and acidic/negative residues in SIMs appears to determine their specificity in binding to distinct SUMO isoforms (Hecker et al., 2006; Sekiyama et al., 2008; Zhu et al., 2008). The SIM and SUMO interaction also seems to impinge on sumoylation of target proteins or substrate recognition. This is based on the observation that SUMO loaded-Ubc9 (E2) (SUMO interaction with sumoylated Ubc9) could regulate the target discrimination of protein sumoylation by a mechanism involving the interaction between the substrate and the SIM in sumoylated Ubc9 (Knipscheer et al., 2007; 2008). In addition, the SIM-SUMO interaction plays a role in regulating the downstream effect of SUMO by recruiting SIM-containing proteins, and through intramolecular interactions and E3 ligase activity (Kerscher, 2007; Merrill et al., 2010; Yang and Sharrocks, 2010). For example, the recruitment of the anti-recombinogenic helicase Srs2 by sumoylated polymerase processing factor (PCNA) was reduced in the presence of truncated Srs2ÄN, from which the sequence ELLVID (Glu-Ile-Ile- Val-Ile-Asp) was deleted. This motif is considered a SIM in budding yeast (Kerscher, 2007). A conformational change of the thymine DNA glycosylase TDG caused the release of TDG from DNA, and the subsequent repair of the abasic site resulted from the binding of an internal SIM to the covalently attached SUMO moiety (Kerscher, 2007). Only one SIM was discovered in yeast and humans and there are no studies addressing the interaction between SIM and SUMO in plant proteins. Characterizing the mechanism by which SUMO-binding proteins attach to SUMO and how sumoylation of target protein is regulated will help us understand the biological function of SUMO proteins in cellular processes.
CROSSTALK BETWEEN UBIQUITINATION AND SUMOYLATION
The lysine residue in SUMO targets can be modified by other post-translational modifications such as acetylation or methylation. In addition, ubiquitination and sumoylation can also be altered by other modifications (Denuc and Marfany, 2010; Guo et al., 2007; Perry et al., 2008).
One renowned example of antagonistic relationships between sumoylation and ubiquitination is IκBα (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha), a binding partner of NF-κB (transcriptional activator nuclear factor- κB) (Ulrich, 2005). Sumoylation of IκBα protects itself from ubiquitination and proteasome-dependent degradation.
In animal and yeast cells, SUMO-conjugated and ubiquitynated proteins are accumulated in cells treated with proteasome inhibitors. This suggests that sumoylation could also work as a targeting signal for Ub-mediated proteasome-dependent degradation. The STUbL [SUMO-targeted Ubiquitin (Ub) ligase] family of RING finger domain ubiquitin E3 ligases recognizes sumoylated substrates and mediates their subsequent degradation. This unique ubiquitin E3 family includes budding yeast Hex3/Slx5 and Slx8, fission yeast Rfp1, Rfp2, and Slx8, Dictyostelium MIP1, and the mammalian RNF4/Snurf (RING finger protein4/small nuclear RING finger protein). STUbL proteins have two N-terminal SIMs and a C-terminal RING finger domain (Denuc and Marfany, 2010; Perry et al., 2008). In plants, however, orthologs of the STUbL family ubiquitin E3 ligase appear to be missing. Nonetheless, following heat stress treatment, ubiquitination of SUMO-conjugated proteins in Arabidopsis was found to increase substantially (Miller et al., 2010).
SUMO-UBIQUITIN PATHWAY INTERPLAY
SUMO also plays a role in the regulation of the ubiquitin pathway. Sumoylation of the animal deubiquitinating enzyme (DUB) ubiquitin-specific peptidase 25 (USP25) decreases its activity, whereas ubiquitination of the same lysine residue enhances the rescue of substrates from proteasomal degradation by USP25 (Denuc and Marfany, 2010).
Studies addressing the interactions between sumoylation and ubiquitination are in their beginning phases and we have not yet reached a detailed understanding of these mechanisms in yeasts or animals. As our knowledge about the two seemingly independent pathways accumulates in plants, information about the interplay between SUMO and Ub pathways will emerge as well.
CONCLUSION
In plants, the sumoylation system functions as a regulatory mechanism involved in cell expansion and division, flowering, hormonal signaling, and responses to abiotic and environmental stresses. Mining novel targets of SUMO proteins expands the field of sumoylation/desumoylation, revealing its unexpected complexity. SUMO binding identifies regulators and/or effectors that are involved in an increasing number of diverse downstream pathways.
Acknowledgments
This work was supported by grants from the World Class University Program (R32-10148) funded by the Ministry of Education, Science and Technology, and the Next-Generation BioGreen 21 Program (SSAC, grant #: PJ008025), Rural Development Administration, Republic of Korea.
References
- 1.Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R., Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. (1998);280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 2.Bernier-Villamor V., Sampson D.A., Matunis M.J., Lima C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. (2002);108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 3.Boddy M.N., Howe K., Etkin L., Solomon E., Freemont P. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. (1966);13:971–982. [PubMed] [Google Scholar]
- 4.Bohren K.M., Nadkarni V., Song J.H., Gabbay K.H., Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. (2004);279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 5.Budhiraja R., Hermkes R., Müller S., Schmidt J., Colby T., Panigrahi K., Coupland G., Bachmair A. substrates related to chromatin and to rna-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. (2009);149:1529–1540. doi: 10.1104/pp.108.135053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castillo A.G., Kong L.J., Hanley-Bowdoin L., Bejarano E.R. Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. (2004);78:1758–1769. doi: 10.1128/JVI.78.6.2758-2769.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catala R., Ouyang J., Abreu I.A., Hu Y., Seo H., Zhang X., Chua N. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. (2007);19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen A., Mannen H., Li S.S. Characterization of mouse ubiquitin-like SMT3A and SMT3B cNDAS and gene/pseudogenes. IUBMB Life. (1998);46:1161–1174. doi: 10.1080/15216549800204722. [DOI] [PubMed] [Google Scholar]
- 9.Cheong M.S., Park H.C., Hong M.J., Lee J., Choi W., Jin J.B., Bohnert H.J., Lee S.Y., Bressan R.A., Yun D. Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. (2009);151:1930–1942. doi: 10.1104/pp.109.143719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chosed R., Mukherjee S., Lois L.M., Orth K. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem. J. (2006);398:521–529. doi: 10.1042/BJ20060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chupreta S., Holmstrom S., Subramanian L., Iniguez-Lluhi J.A. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol. Cell. Biol. (2005);25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke S.M., Mur L.A.J., Wood J.E., Scott I.M. Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J. (2004);38:432–447. doi: 10.1111/j.1365-313X.2004.02054.x. [DOI] [PubMed] [Google Scholar]
- 13.Colby T., Matthäi A., Boeckelmann A., Stuible H. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. (2006);142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conti L., Price G., O’Donnell E., Schwessinger B., Dominy P., Sadanandom A. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. (2008);20:2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaney T.P., Uknes S., Vernooij B., Friedrich L., Weymann K., Negrotto D., Gaffney T., Gut-Rella M., Kessmann H., Ward E., et al. A central role of salicylic acid in plant disease resistance. Science. (1994);266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 16.Denuc A., Marfany G. SUMO and ubiquitin paths converge. Biochem. Soc. Trans. (2010);038:34–39. doi: 10.1042/BST0380034. [DOI] [PubMed] [Google Scholar]
- 17.Elrouby N., Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA. (2010);107:17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Dominguez M., March-Diaz R., Reyes J.C. The PHD domain of plant pias proteins mediates sumoylation of bromodomain GTE proteins. J. Biol. Chem. (2008);283:21469–21477. doi: 10.1074/jbc.M708176200. [DOI] [PubMed] [Google Scholar]
- 19.Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. (2007);8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 20.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Gene Dev. (2004);18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 21.Golebiowski F., Matic I., Tatham M.H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R.T. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. (2009);2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 22.Grabbe C., Dikic I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. (2009);109:1481–1494. doi: 10.1021/cr800413p. [DOI] [PubMed] [Google Scholar]
- 23.Guo B., Yang S., Witty J., Sharrocks A.D. Signalling pathways and the regulation of SUMO modification. Biochem. Soc. Trans. (2007);35(Pt 6):1414–1418. doi: 10.1042/BST0351414. [DOI] [PubMed] [Google Scholar]
- 24.Hanania U., Furman-Matarasso N., Ron M., Avni A. Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. (1999);19:533–541. doi: 10.1046/j.1365-313x.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- 25.Hannich J.T., Lewis A., Kroetz M.B., Li S., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. (2005);280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 26.Hecker C., Rabiller M., Haglund K., Bayer P., Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. (2006);281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 27.Hermkes R., Fu Y.-F., Nürrenberg K., Budhiraja R., Schmelzer E., Elrouby N., Dohmen R. J., Bachmair A., Coupland G. Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta. (2011);233:63–73. doi: 10.1007/s00425-010-1281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotson A., Mudgett M.B. Cysteine proteases in phytopathogenic bacteria: identification of plant targets and activation of innate immunity. Curr. Opin. Plant Biol. (2004);7:384–390. doi: 10.1016/j.pbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Huang L., Yang S., Zhang S., Liu M., Lai J., Qi Y., Shi S., Wang J., Wang Y., Xie Q., et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. (2009);60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 30.Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., Adachi S., Minamisawa K., Umeda M., Sugimoto K. SUMO E3 Ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell. (2009);21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin J.B., Jin Y.H., Lee J., Miura K., Yoo C.Y., Kim W., Oosten M.V., Hyun Y., Somers D.E., Lee I., et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. (2008);53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. (2004);73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 33.Kerscher O. SUMO junction-what’s your function? EMBO Rep. (2007);8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J., Taylor K.W., Hotson A., Keegan M., Schmelz E.A., Mudgett M.B. XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell. (2008);20:1915–1929. doi: 10.1105/tpc.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knipscheer P., van Dijk W.J., Olsen J.V., Mann M., Sixma T.K. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. (2007);26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knipscheer P., Flotho A., Klug H., Olsen J.V., van Dijk W.J., Fish A., Johnson E.S., Mann M., Sixma T.K., Pichler A. Ubc9 Sumoylation regulates SUMO target discrimination. Mol. Cell. (2008);31:371–382. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Kroetz M.B., Hochstrasser M. Identification of SUMOinteracting proteins by yeast two-hybrid analysis. Methods Mol. Biol. (2009);497:107–120. doi: 10.1007/978-1-59745-566-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurepa J., Walker J.M., Smalle J., Gosink M.M., Davis S.J., Durham T.L., Sung D., Vierstra R.D. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of sumo1 and -2 conjugates is increased by stress. J. Biol. Chem. (2003);278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 39.Larkindale J., Hall J.D., Knight M.R., Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. (2005);138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J., Nam J., Park H.C., Na G., Miura K., Jin J.B., Yoo C.Y., Baek D., Kim D.H., Jeong J.C., et al. Salicylic acidmediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. (2006);49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 41.Li W., Hesabi B., Babbo A., Pacione C., Liu J., Chen D.J., Nickoloff J.A., Shen Z. Regulation of double-strand break-induced mammalian homologous recombination by UBL1, a RAD51-interacting protein. Nucleic Acids Res. (2000);28:1145–1153. doi: 10.1093/nar/28.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin W., Lin S., Chiou T. Molecular regulators of phosphate homeostasis in plants. J. Exp. Bot. (2009);60:1427–1438. doi: 10.1093/jxb/ern303. [DOI] [PubMed] [Google Scholar]
- 43.Lois L.M., Lima C.D., Chua N. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell. (2003);15:1347–1350. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mannen H., Tseng H., Cho C., Li S.S. Cloning and expression of human homolog HSMT3 to yeast SMT3 suppressor of MIF2 mutations in a centromere protein gene. Biochem. Biophys. Res. Commun. (1996);222:178–180. doi: 10.1006/bbrc.1996.0717. [DOI] [PubMed] [Google Scholar]
- 45.Matunis M.J., Coutavas E., Blobel G. A novel ubiquitin- like modification modulates the partitioning of the Ran- GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. (1996);135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matunis M.J., Wu J., Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating Protein, Ran- GAP1, to the nuclear pore complex. J. Cell Biol. (1998);140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melchior F. SUMO-nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. (2000);16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 48.Meluh P.B., Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. (1995);6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merrill J.C., Melhuish T.A., Kagey M.H., Yang S., Sharrocks A.D., Wotton D. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLos one. (2010);5:e8794. doi: 10.1371/journal.pone.0008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2010);107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minty A., Dumont X., Kaghad M., Caput D. Covalent modification of p73α by SUMO-1. J. Biol. Chem. (2000);275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 52.Miura K., Hasegawa P.M. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. (2010);20:223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A.S., Raghothama K.G., Baek D., Koo Y.D., Jin J.B., Bressan R.A., et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA. (2005);102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miura K., Jin J.B., Hasegawa P.M. Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. (2007a);10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Hasegawa P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. (2007b);19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA. (2009);106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miura K., Lee J., Miura T., Hasegawa P.M. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. (2010);51:103–113. doi: 10.1093/pcp/pcp171. [DOI] [PubMed] [Google Scholar]
- 58.Murtas G., Reeves P.H., Fu Y., Bancroft I., Dean C., Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of small ubiquitin- related modifier conjugates. Plant Cell. (2003);15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okura T., Gong L., Kamitani T., Wada T., Okura I., Wei C.F., Chang H.M., Yeh E.T. Protection against Fas/ APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J. Immunol. (1996);157:4277–4281. [PubMed] [Google Scholar]
- 60.Orth K., Xu Z., Mudgett M.B., Bao Z.Q., Palmer L.E., Bliska J.B., Mangel W.F., Staskawicz B., Dixon J.E. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. (2000);290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 61.Park H.C., Kim H., Koo S.C., Park H.J., Cheong M.S., Hong H., Baek D., Chung W.S., Kim D.H., Bressan R.A., et al. Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant Cell Environ. (2010);33:1923–1934. doi: 10.1111/j.1365-3040.2010.02195.x. [DOI] [PubMed] [Google Scholar]
- 62.Park H.C., Choi W., Park H.J., Cheong M., Koo Y.D., Shin G., Chung W.S., Kim W.-Y., Kim M.G., Bressan R., et al. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol. Cells. (2011a);32:143–151. doi: 10.1007/s10059-011-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park B.S., Song J.T., Seo H.S. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. (2011b);2:400. doi: 10.1038/ncomms1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perry J.J.P., Tainer J.A., Boddy M.N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. (2008);33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 65.Potts P.R., Yu H. Human MMS21/NSE2 is a sumo ligase required for DNA repair. Mol. Cell. Biol. (2005);25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reed J.M., Dervinis C., Morse A.M., Davis J.M. The SUMO conjugation pathway in Populus: genomic analysis, tissue- specific and inducible Sumoylation and in vitro de-SUMOylation. Planta. (2010);232 doi: 10.1007/s00425-010-1151-8. [DOI] [PubMed] [Google Scholar]
- 67.Reeves P.H., Murtas G., Dash S., Coupland G. early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development. (2002);129:5349–5361. doi: 10.1242/dev.00113. [DOI] [PubMed] [Google Scholar]
- 68.Roden J., Eardley L., Hotson A., Cao Y., Mudgett M.B. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. (2004);17:633–643. doi: 10.1094/MPMI.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- 69.Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. (2000);275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 70.Saracco S.A., Miller M.J., Kurepa J., Vierstra R.D. Genetic analysis of sumoylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. (2007);145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seeler J., Dejean A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. (2003);4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 72.Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., Shirakawa M. Structure of the small ubiquitin-like modifier (SUMO)- interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. (2008);283:35966–35975. doi: 10.1074/jbc.M802528200. [DOI] [PubMed] [Google Scholar]
- 73.Shen Z., Pardington-Purtymun P.E., Comeaux J.C., Moyzis R.K., Chen D.J. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics. (1996a);37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- 74.Shen Z., Pardington-Purtymun P.E., Comeaux J.C., Moyzis R.K., Chen D.J. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. (1996b);36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 75.Song J., Durrin L.K., Wilkinson T.A., Krontiris T.G., Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA. (2004);101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song J., Zhang Z., Hu W., Chen Y. Small ubiquitinlike modifier (SUMO) recognition of a SUMO binding motif. J. Biol. Chem. (2005);280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 77.Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M.P., Botting C.H., Naismith J.H., Hay R.T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. (2001);276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 78.Thangasamy S., Guo C.L., Chuang M.-H., Lai M.-H., Chen J., Jauh G.-Y. Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence. New Phytol. (2011);189:869–882. doi: 10.1111/j.1469-8137.2010.03538.x. [DOI] [PubMed] [Google Scholar]
- 79.Ulrich H.D. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. (2005);15:525–532. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 80.van den Burg H.A., Kini R.K., Schuurink R.C., Takken F.L.W. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. (2010);22:1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vijay-Kumar S., Bugg C.E., Cook W.J. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. (1987);194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 82.Walden H., Podgorski M.S., Schulman B.A. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. (2003);422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y., Dasso M. Sumoylation and desumoylation at a glance. J. Cell Sci. (2009);122:4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang H., Makeen K., Yan Y., Cao Y., Sun S., Xu G. OsSIZ1 regulates the vegetative growth and reproductive development in rice. Plant Mol. Biol. Rep. (2011);29:411–417. [Google Scholar]
- 85.Wilkinson K.A., Henley J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. (2010);428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu P., Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim. Biophys. Acta. (2006);1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., Meier I. Nuclear pore anchor, the Arabidopsis Homolog of Tpr/Mlp1/Mlp2/Megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. (2007);19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang S., Sharrocks A.D. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol. Cell. Biol. (2010);30:2193–2205. doi: 10.1128/MCB.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoo C.Y., Miura K., Jin J.B., Lee J., Park H.C., Salt D.E., Yun D., Bressan R.A., Hasegawa P.M. SIZ1 small ubiquitin- like modifier E3 Ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. (2006);142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang S., Qi Y., Yang C. Arabidopsis SUMO E3 ligase AtMMS21 regulates root meristem development. Plant Signal. Behav. (2010);5:1–3. doi: 10.4161/psb.5.1.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu J., Zhu S., Guzzo C.M., Ellis N.A., Sung K.S., Choi C.Y., Matunis M.J. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. (2008);283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]


