Abstract
Respiratory tract exposure to viruses, air pollutants, or bacterial pathogens can lead to pulmonary diseases. The molecular mechanism of mucous overproduction increased by these pathogens provides the knowledge for developing new therapeutic strategies. There is established in vitro data demonstrating that the overexpression of MUC5AC is induced by peptidoglycan (PGN) derived from Staphylococcus aureus. However, the mechanisms by which PGN activates MUC5AC gene expression in the airway remain unclear. The aim of this study was to identify the mechanism of PGN-induced MUC5AC gene expression. We found that PGN could induce MUC5AC gene expressions in a time- and dose-dependent manner. Moreover, activations of ERK1/2 and JNK increased after treatment of cells with PGN, whereas phosporylation of p38 was undetected. Of these MAPKs, pharmacologic inhibition of ERK1/2 decreased PGN-induced MUC5AC gene expression. In addition, we checked the activation of p90 ribosomal S6 kinase 1 (RSK1) as a downstream signaling target of ERK1/2 in PGN signaling. The activation of RSK1 was prevented by pretreatment with PD98059. We also found that RSK1 mediated the PGN-induced phosphorylation of cAMP response element-binding protein (CREB) and the transcription of MUC5AC. Furthermore, the cAMPresponse element (CRE) in the MUC5AC promoter appears to be important for PGN-induced MUC5AC gene expression in NCI-H292 cells.
Keywords: CREB, inflammation, MUC5AC, mucous overproduction, peptidoglycan
INTRODUCTION
Mucus plays an integral role in the innate immune system as a defense against pathogenic organisms. Mucus hyperproduction and hypersecretion by airway epithelium are seen frequently in a number of respiratory diseases, including asthma, chronic bronchitis, COPD, and cystic fibrosis (Voynow et al., 2006). Thus, insights into the intracellular signaling mechanisms that induce airway mucus hyperproduction and hypersecretion are necessary for understanding inflammation control. Since bacteria interact with host epithelial cells to activate intracellular signaling pathways, which, in turn, cause the selective increase of specific mucin gene expression and mucin production (Song et al., 2009), it is important to clarify the mechanism by which mucin is increased in the airway by major pathogens such as Staphylococcus aureus, nontypable Haemophilus influenza, and Pseudomonas aeruginosa (Rylander, 2002; 2006; Wang et al., 2002).
Mucins are a family of large, heavily glycosylated proteins that comprise a significant portion of secreted mucus and secretory granules (Rose et al., 2006). The heavily glycosylated structures are believed to contribute to the highly viscoelastic property of secreted airway mucus. MUC genes encode the protein backbones of mucins. Twenty mucin genes have been identified, but it remains unclear which mucins are secreted in the various airway diseases. Mucins are usually subdivided into two groups based on domain: membrane-bound (MUC1, MUC3, MUC4, MUC11, MUC12, MUC13, MUC17, MUC18, and MUC20) and secreted (MUC2, MUC5AC, MUC5B, MUC6, MUC7, MUC9, and MUC19). The other mucin genes, including MUC8, have not yet been fully characterized (Thai et al., 2008).
Peptidoglycan (PGN) is an important and unique component of bacteria (Schleifer and Kandler, 1972). PGN is found as a thick exposed layer comprising the bacterial cell wall of Gram-positive bacteria in association with lipoteichoic acid, whereas it is present in Gram-negative bacteria as a thin layer overlaid by a thick layer of lipopolysaccharide (LPS) (McDoland et al., 2005). PGN stimulates the production of inflammatory cytokines, such as IL-6, IL-1α/β, and TNF-α in monocytes, macrophages, neutrophils, and epithelial cells (McDoland et al., 2005). The recognition of toll-like receptor (TLR) 2 by PGN leads to activation of intracellular signaling pathways such as NF-κB (Wang et al., 2001). MAPK signaling also activates to induce proinflammatory cytokines or related proteins (Schorey and Cooper, 2003). In addition, PGN also activates caspase-1 to induce proIL-1β (Martinon et al., 2004).
Cyclic AMP response element (CRE)-binding protein (CREB) is a member of the ATF-1 family of transcription factors and is involved in the regulation of many cytokine genes including IL-1, IL-6, and COX2 (Mayr and Montminy, 2001). It can be activated via multiple pathways by various upstream kinases, including protein kinase A, protein kinase C, MAPK-activated protein-2, Akt. In addition, ribosomal S6 kinase (p90RSK) 1 and mitogenand stress-activated kinase (MSK) are important upstream activator of CREB to induced MUC5AC gene expression. RSK1 was regulated by MAPK and translocated to nucleous to transfer the signaling in the airway epithelium cells (Kim et al., 2009). We therefore examined the role of RSK1-CREB pathway in signal transduction facilitated by PGN to induce mucus overproduction.
Because MUC5AC overproduction during bacterial infection plays an important feature in the pathogenesis of respiratory system, we hypothesized that bacterial product, PGN, upregulates MUC5AC gene expression by activating specific signaling mechanism(s) in the airway. Finding signaling mechanism will provide new insights into the interaction bacterial infection and innate immunity in human being.
MATERIALS AND METHODS
Materials
Peptidoglycan (#77140), lipopolysaccharide (#L9143), and exotoxin A (#P0184) were purchased from Sigma-Aldrich (USA). PD98059, MEK1 inhibitor, U0126, MEK1/2 inhibitor, and SP600125, JNK inhibitor, were purchased from Calbiochem (USA). All phospho-specific antibodies were purchased from Cell Signaling (USA). All primers and siRNAs were synthesized by Bioneer (Korea): RSK1, CUGAUGACACCUUCUACUU (dTdT), CREB, UCAAGGAGGCCUUCCUACA (dTdT) and negative control CCUACGCCACCAAUUUCGU (dTdT).
Real-time PCR
Real-time PCR was performed using a BioRad iQ iCycler Detection System (BioRad Laboratories; USA) with iQ SYBR Green Supermix. Reactions were performed in a total volume of 20 μl which included 10 μl 2× SYBR Green PCR Master Mix, 300 nM of each primer and 1 μl of previously reverse-transcribed cDNA template. The following primers were used: MUC5AC, forward 5′-CAGCCACGTCCCCTTCAATA-3′ and reverse 5′-ACCGCATTTGGGCATCC-3′; and β2-microglobulin, used as a reference for normalization, forward 5′-CGCTCCG TGGCCTTAGC-3′ and reverse 5′-GAGTACGCTGGATAGCC TCCA-3′. Real time RT-PCR was performed on a MiniOption Real-Time PCR Detection System (Bio-Rad). Parameters were 95℃ for 10 m, followed by 40 cycles of 95℃ for 15 s, 60℃ for 30 s, and 72℃ for 30 s. All reactions were performed in triplicate. The relative quantity of MUC5AC mRNA was obtained using the comparative cycle threshold method and was normalized using β2-microglobulin as an endogenous control.
Western blot analysis
For Western blot analysis, NCI-H292 cells were grown to confluence in six-well plates. After PGN treatment, the cells were lysed with 2× lysis buffer [250 mM Tris-Cl (pH 6.5), 2% SDS, 4% β-mercaptoethanol, 0.02% BPB, 10% glycerol]. Equal amounts of whole cell lysates were resolved using 10-15% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (PVDF; Millipore, USA). The following antibodies were used: anti-phospho ERK1/2 1:3000, anti-phospho p38, phospho-JNK, phosphor-RSK1, phospho-MSK1, phospho-CREB 1:2000, and secondary antibody 1:2000 were diluted.
Plasmids, transient transfection, and luciferase assay
Cells were transiently transfected with plasmids using the FuGENE 6 transfection reagent (Roche) and Dharmafect (Thermo Scientific; USA) reagent for siRNA according to the manufacturer's instructions. Deletion mutants covering the promoter regions of MUC5AC were generated by PCR as previously descrived (Song et al., 2003a). Cells were incubated for 48 h, harvested, and assayed for luciferase activity using a luciferase assay system (Promega; USA) according to the manufacturer's instructions. Luciferase values were normalized to β-galactosidase. Transfection experiments were performed in duplicate and repeated at least three times.
Statistical analysis
The data are presented as the mean ± S.D. of at least three independent experiments. When appropriate, statistical differences were assessed using Wilcoxon Mann-Whitney tests. A p value less than 0.05 was considered statistically significant.
RESULTS
Peptidoglycan derived from S. aureus can induce secretory mucin gene expression in NCI-H292 cells
To determine which mucin genes are regulated by bacterial products, PGN derived from S. aureous and lipopolysaccaride (LPS) and exotoxin A from Pseudomonas aeruginosa were used in the present study. Twenty-four hours after the treatment of exotoxins, we examined the alterations of secretory mucin gene expression in NCI-H292 cells (Fig. 1). PGN and LPS could induce MUC5AC and MUC8 gene expressions, but not MUC5B, MUC7, and MUC19 in airway epithelial cells. Because MUC5AC and MUC8 are highly expressed in airway epithelium, these mucins may be susceptible for bacterial infection in the airway. No changed β2-microgubulin as an internal control. Because the molecular mechanism of LPS-induced MUC5AC gene expression has been reported and investigating the signal pathway of MUC8 has several experimental limitations (Song et al., 2003b; 2009), we focused on the molecular mechanism by which PGN induces MUC5AC gene expression in the present study. NCI-292 cells were treated with various doses of PGN for 24 hr. Increased MUC5AC gene expression occurred in a dose-dependent manner with an EC50 value of 6.4 ± 0.19 μM (Fig. 1B), and, as a result, 10 μM of PGN was used for all the subsequent experiments. In addition, MUC5AC gene expression increased in a time-dependent manner for up to 12 hr after PGN treatment and then reached a plateau (Fig. 1C). These results show that extracellular PGN induces MUC5AC gene expression in a dose- and time-dependent manner in NCI-H292 cells.
Fig. 1. Effect of exotoxins on mucin gene expression. Confluent cells were rendered quiescent in RPMI-1640 with 0.2% FBS for 24 h prior to treatment with PGN (10 μg/ml), exotoxin A (500 ng/ml), or LPS (10 μg/ml) respectively for 24 h, and cell lysates were harvested for RTPCR. β-2-microglobulin (β2M) was employed as an internal control. The figures shown are representative of three independent experiments. Densitometry results are presented as the means ± SD (Fig. 1A, right panel). NCI-H292 Cells were treated for 24 h with exogenous PGN at the indicated concentrations (B) and times (C). Cell lysates were harvested for real-time quantitative PCR. *p < 0.05 compared to control (vehicle). Data are presented as mean ± SD values of three independent experiments. The figures are representative of three independent experiments.
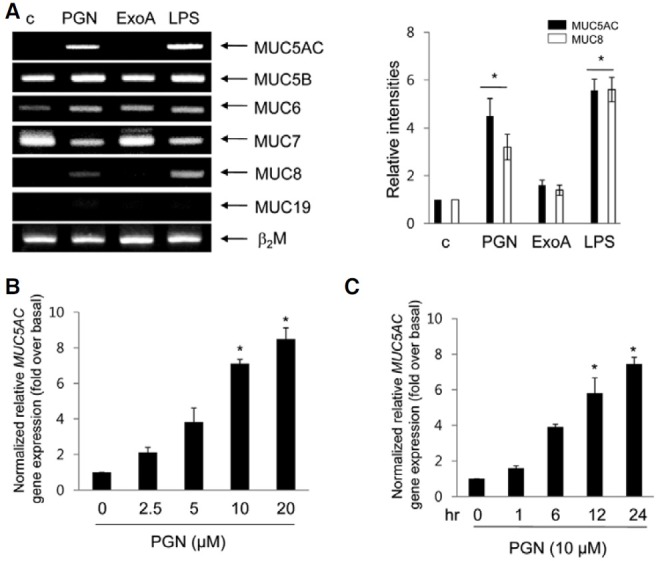
ERK1/2 MAPK is mediated in PGN-induced MUC5AC gene expression, but not JNK MAPK
To investigate possible mechanisms through which PGN induces MUC5AC gene expression, we started with the mitogenactivated protein kinase (MAPK) pathway. Because MAPKs are involved in many signal pathways crucial for adaptation to various physiological changes, it is likely this pathway may play a role in PGN-induced transcriptional responses. After the treatment of PGN, the phosphorylations of ERK1/2 and JNK increased in a time-dependent manner, while that of p38 MAPK did not (Fig. 2A). To examine whether these MAPKs are essential for PGN-induced MUC5AC gene expression, PD98059, a specific MEK1 inhibitor, SP600125, a selective JNK inhibitor, and U0126, MEK1/2 inhibitor were used. Both PD98059 and U0126 inhibited the phosphorylation of ERK1/2 MAPK in a dose-dependent manner, and SP600125 also inhibited the phosphorylation of JNK MAPK in a dose-dependent manner (Fig. 2B). Interestingly, PD98059 inhibited PGN-induced MUC5AC gene expression, but SP600125 did not affect PGN-induced MUC5AC gene expression (Fig. 2C), suggesting that ERK1/2 might be involved in PGN-induced MUC5AC gene expression in NCI-H292 cells.
Fig. 2. Effect of ERK1/2 on PGN-induced MUC5AC gene expression. (A) Confluent cells were treated with PGN for different times (min), and cell lysates were harvested for Western blot analysis using phospho-specific antibodies. Total ERK1/2 was used as a loading control. (B) Cells were treated with different dose of PD98059, U0126, or SP600125 for 10 min, and cell lystes were performed Western blot analysis. (C) Cells were treated with PD98059 (20 μM) or SP600125 (20 μM) for 2 h, and then stimulated for 24 h with PGN prior to collection of total RNA for real-time RT-PCR analysis of MUC5AC mRNA expression. *p < 0.05 compared to control (vehicle); **p < 0.05 compared to PGN treatment only. The figures shown are representative of three independent experiments.
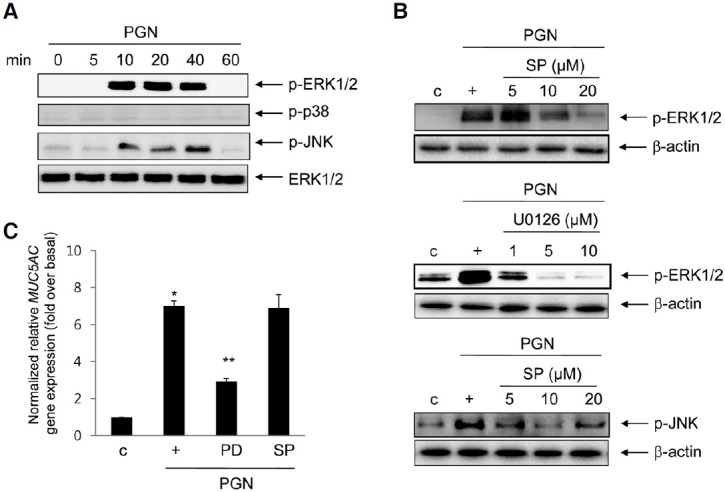
p90 ribosomal S6 kinase (RSK) 1 signaling functions as a down-stream mediator of the ERK1/2 MAPK pathway for PGN-induced MUC5AC gene expression
We examined the activations of RSK1 and mitogen- and stress-activated protein kinase (MSK) 1 as a down-stream effect of PGN signaling. MSK1 and RSK1 have been shown to be downstream players in the MAPK signal cascade in our system (Song et al., 2003b; 2008). The phosphorylation of RSK1, but not MSK1, by PGN was picked at 10 min and decreased at 60 min (Fig. 3A). Pretreatment with PD98059 (20 μM) inhibited PGN-activated RSK1 phosphorylation (Fig. 3B), suggesting that RSK1 is regulated by ERK1/2 MAPK. Moreover, to examine whether RSK1 is essential for PGN-induced MUC5AC gene expression, cells were transfected with a DNA expression construct encoding wild-type RSK1 and siRNA-RSK1. Endogenous RSK1 expression was increased in cells transfected with the construct expressing wild-type RSK1 and was decreased in cells transfected with siRNA-RSK1 (Fig. 3C, upper panel). Whereas MUC5AC gene expression was increased in cells transfected with the construct expressing wild-type RSK1 compared with that in treatment with PGN alone, it was significantly inhibited by siRNA-RSK1, suggesting that RSK1 appears to function closely in PGN-induced MUC5AC gene expression and RSK1 functions the downstream of ERK1/2 MAPK in NCIH292 cells.
Fig. 3. Effect of RSK1 on PGN-induced MUC5AC gene expression. (A) Cells were stimulated for the indicated times with PGN, and then total proteins were collected for Western blot analysis using phospho-RSK1 or MSK1 antibody. Total β-actin was used as a loading control. (B) The cells were pretreated for 2 h with 20 μM PD98059 and then stimulated with PGN for 10 min prior to Western blot analysis. (C) The cells were transiently transfected with RSK1 wild-type construct or siRNA-RSK1. The cells were treated for 24 h prior to real-time RT-PCR. *p < 0.05 compared to control (vehicle); **p < 0.05 compared to PGN treatment only. The figures shown are representative of three independent experiments.
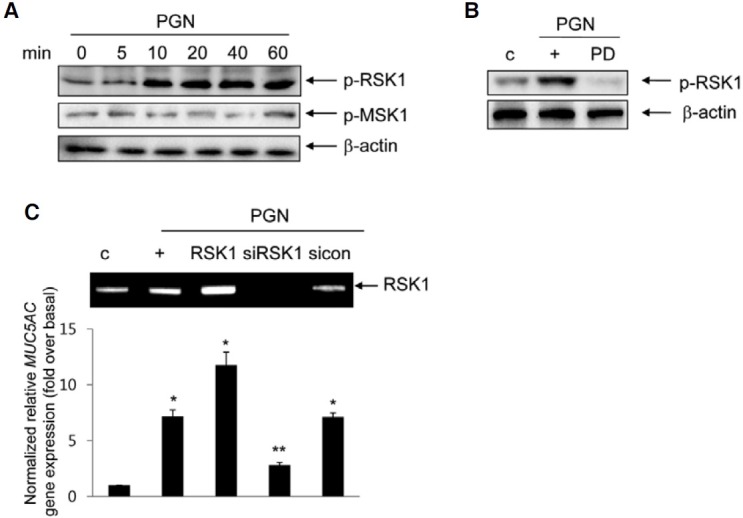
cis-acting regulatory element, CRE, mediates PGN-induced MUC5AC transcriptional activity
To investigate whether CRE-binding protein (CREB) participates in PGN-induced MUC5AC gene expression, we investigated CREB activation. The phosphorylation of CREB was increased after treatment with PGN (Fig. 4A). PGN-induced CREB activation and MUC5AC gene expression were dramatically suppressed in cells pretreated with PD98059 and transfected with siRNA-RSK1 (Fig. 4B), indicating that the activation of CREB may be essential for PGN-induced MUC5AC gene expression through the ERK1/2 MAPK via RSK1 pathway in NCI-H292 cells. To examine whether CREB is essential for PGN-induced MUC5AC gene expression, cells were transfected with DNA expression construct encoding wild-type CREB or siRNA-CREB. CREB expression increased in cells transfected with the construct expressing wild-type CREB and decreased in cells transfected with siRNA-CREB (Fig. 4C, upper panel). Whereas MUC5AC gene expression increased in cells transfected with the construct expressing wild-type CREB compared with that of treatment with PGN alone, it was significantly inhibited by siRNA-CREB. Furthermore, to determine whether the CREB binding site (CRE) may be critical for PGN-induced MUC5AC transcription, cells were transfected with several deletion constructs containing with CRE or not and treated with PGN for 24 h, respectively. PGN selectively increased luciferase activity of CRE element of MUC5AC promoter (Fig. 4D). In addition, in order to examine whether the activation of CRE is essential for PGN-induced MUC5AC transcription, we carried out site-directed mutagenesis to generate CRE mutant constructs. The luciferase activities of the mutant constructs, Mut1 and Mut2, were dramatically decreased (Fig. 4E), suggesting that CRE functions as a cis-element to regulate MUC5AC transcription by PGN.
Fig. 4. Involvement of CREB on PGN-induced MUC5AC transcription. (A) Cells were treated for the different times with PGN, and then total proteins were collected for Western blot. Total β-actin was used as a loading control. (B) The cells were pretreated for 2 h with 20 μM PD98059 or were transiently transfected with either siRNA-RSK1 or siRNA-CREB (C). The cells were harvested for Western blot analysis (upper panel) and the cells were treated for 24 h prior to real-time RT-PCR (lower panel). *p < 0.05 compared to control (vehicle); **p < 0.05 compared to PGN treatment only. (D) Cells were transiently transfected with several MUC5AC promoter luciferase reporter constructs which did or did not contain the CRE motif and were stimulated with PGN for 24 h. Cell lysates were harvested, and reporter assays were performed according to the manufacturer’s instructions (see “Materials and Methods”). *p < 0.05 compared to -776/-1 reporter construct. (E) Site-directed selective mutagenesis was performed to construct CRE-binding site mutants as indicated. Luciferase activities are shown after correction for transfection efficiency using the β-galactosidase activity of the cell lysates. Values shown are means ± S.D. of experiments performed three or more times. *p < 0.05 compared to wild-type reporter construct. The figures shown are representative of three independent experiments.
DISCUSSION
PGN derived from Staphylococcus aureus, a major cell wall compound in Gram (+) bacteria, can trigger inflammatory responses in t he human b ody. In this a rticle, we suggest that PGN can increase MUC5AC gene expression via MAPK and CREB. Although LPS functions have been established well in the airway, the immunogenicity of PGN from Gram (+) bacteria in pulmonary diseases has not been well documented.
Thai et al. (2008) have reported that PGN could elevate MUC5AC expression independent of human calcium-dependent chloride channel 1 in primary human tracheobronchial epithelial cells and Evans and Koo (2009) showed that the TLR2 or 6 signaling activated by PGN plays a crucial role in PGN-induced MUC5AC gene expression. Hypothesized that the TLR2-MyD88-NF-κB pathway plays a crucial role in PGNinduced MUC5AC expression (Thai et al., 2008). Interestingly, S. pyogenes PGN could not induce MUC2 gene expression, whereas lipoteichoic acid (LTA) was able to do so in HM3 cells (Lemjabbar et al., 2002). The discrepancy of PGN signaling in S. aureus versus S. pyogenes may derive from the difference in pathogeneses of these two bacteria in the airway. PGN induced MUC5AC and MUC8 gene expressions which have been considered the major mucins in the airway (Fig. 1) (Song et al., 2003a). Involvement of ERK1/2 MAPK is consistent with data from our previous results indicating that ERK1/2 MAPK activated by bacterial products and proinflammatory mediators increases mucin production (Song et al., 2003a; 2003b; 2008). However, it is unclear why the numbers of signaling pathways have shared ERK1/2 MAPK protein for mucin production. It may be responding to a receptor and its downstream signaling. Our previous study showed that TLR4 activated by LPS increased the activity of ERK1/2 MAPK via the P2Y2 G-protein coupled receptor (Song et al., 2008). Based on these results, ERK1/2 MAPK may have crucial roles in pathogeneses of both Gram (+) and Gram (-) bacteria in the airway.
We further investigated how PGN can induce MUC5AC gene expression in the nucleus. We examined MSK1 and RSK1 in the downstream signaling of ERK1/2 involved in cytokineinduced mucin gene expression (Song et al., 2003a; 2003b). Because MSK1 and RSK1 are regulated by MAPK in cells, their effects on PGN signaling also need to be investigated. The ERK1/2-RSK1 pathway was involved in the PGN-mediated signaling pathway, but MSK1 was not (Fig. 3), suggesting that the signaling pathways leading to MUC5AC gene expression are distinct, depending on the types of stimulant and cells used. However, the results shown account for the action of PGN on mucosal epithelial cells. The determination of either MSK1 or RSK1 as a downstream signaling pathway protein depends heavily on MAPK upstream proteins. MSK1 was activated by both ERK1/2 and p38 MAPK phosphorylated by proinflammatory cytokine, whereas RSK1 was regulated by only ERK1/2 MAPK (Song et al., 2003a; 2003b). Therefore, the possibility of intra-cellularly different expression level between MSK1 versus RSK1 can be ruled out, upstream signaling protein, however, may be responsible for this phenomenon in the cells.
Until now, the signaling pathway related to the downstream signaling of RSK1 for PGN-induced MUC5AC gene expression has been unknown. Previously, we demonstrated that CREB may be essential for cytokine-induced mucin gene expression (Song et al., 2003a; 2003b; 2008). We also found that CREB was responsible for PGN-induced MUC5AC gene expression through RSK1 signaling (Fig. 4). This finding is consistent with our previous studies in which we have shown involvement of the ERK1/2 pathway in the transcriptional activation of MUC5AC gene expression by several cytokines and bacterial product (Song et al., 2003a; 2003b), which in turn, CREB may play as a key factor of inflammation in the airway, much like NF-κB.
However, the CREB-binding partner(s) for the elevated transcriptional activity by PGN has been yet unclear. Our results showed that c-Ets1 interacted with CREB to regulate mucin production induced by cytokines or bacterial products (Our unpublished data). However, c-Ets1 may be unrelated to PGNinduced MUC5AC gene expression (data not shown). These findings are consistent with our hypothesis that CREB may interact directly or indirectly with other transcription factor(s) that function as binding partner between, which form a transcriptional complex for diverse signaling pathways in the regulation of MUC5AC gene expression.
In summary, our results showed that ERK1/2 is essential for PGN-induced MUC5AC gene expression in airway epithelial cells. In addition, the phosphorylation of RSK1-CREB is related to MUC5AC gene expression through the activity of PGN. We show that the mucus overproduction seen during bacterial infection may be a result of PGN-mediated increased MUC5AC gene expression within the inflamed airway. Further analysis of the signaling mechanisms activated by various cytokines or bacterial products may provide deeper insights into the pathogenesss of pulmonary diseases.
Acknowledgments
The authors have no conflicting financial interests. This study was supported by a grant from Kosin University College of Medicine (2010).
References
- 1.Evans C.M., Koo J.S. Airway mucus: the good, the bad, the sticky. Pharmacol. Ther. (2009);182:2349–2356. doi: 10.1016/j.pharmthera.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim C.H., Kim K.E., Yoon J.H., Song K.S. Upregulation of MUC5AC gene expression by IL-4 through CREB in human airway epithelial cells. J. Cell. Biochem. (2009);108:974–981. doi: 10.1002/jcb.22330. [DOI] [PubMed] [Google Scholar]
- 3.Lemjabbar H., Basbaum C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat. Med. (2002);8:41–46. doi: 10.1038/nm0102-41. [DOI] [PubMed] [Google Scholar]
- 4.Martinon F., Agostini L., Meylan E., Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. (2004);14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 5.Mayr B., Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. (2001);2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 6.McDonald C., Inohara N., Nuñez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J. Biol. Chem. (2005);280:20177–20180. doi: 10.1074/jbc.R500001200. [DOI] [PubMed] [Google Scholar]
- 7.Rose MC., Voynow J.A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. (2006);86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 8.Rylander R. Endotoxin in the environment-exposure and effects. J. Endotoxin. Res. (2002);8:241–252. doi: 10.1179/096805102125000452. [DOI] [PubMed] [Google Scholar]
- 9.Rylander R. Endotoxin and occupational airway diseases. Curr. Opin. Allergy Clin. Immunol. (2006);6:62–66. doi: 10.1097/01.all.0000202356.83509.f7. [DOI] [PubMed] [Google Scholar]
- 10.Schleifer K.H., Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. (1972);36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schorey J.S., Cooper A.M. Macrophage signalling upon mycobacterial infection: the MAP kinases lead the way. Cell. Microbiol. (2003);5:133–142. doi: 10.1046/j.1462-5822.2003.00263.x. [DOI] [PubMed] [Google Scholar]
- 12.Song K.S., Lee W.J., Chung K.C., Koo J.S., Yang E.J., Choi J.Y., Yoon J.H. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J. Biol. Chem. (2003a);278:23243–23250. doi: 10.1074/jbc.M300096200. [DOI] [PubMed] [Google Scholar]
- 13.Song K.S., Seong J.K., Chung K.C., Lee W.J., Kim C.H., Cho K.N., Kang C.D., Koo J.S., Yoon J.H. Induction of MUC8 gene expression by interleukin-1 beta is mediated by a sequential ERK MAPK/RSK1/CREB cascade pathway in human airway epithelial cells. J. Biol. Chem. (2003b);278:34890–34896. doi: 10.1074/jbc.M303911200. [DOI] [PubMed] [Google Scholar]
- 14.Song K.S., Lee T.J., Kim K., Chung K.C., Yoon J.H. cAMP-responding element-binding protein and c-Ets1 interact in the regulation of ATP-dependent MUC5AC gene expression. J. Biol. Chem. (2008);283:26869–26878. doi: 10.1074/jbc.M802507200. [DOI] [PubMed] [Google Scholar]
- 15.Song K.S., Kim H.J., Kim K., Lee J.G., Yoon J.H. Regulator of G-protein signaling 4 suppresses LPS-induced MUC5AC overproduction in the airway. Am. J. Respir. Cell Mol. Biol. (2009);41:40–49. doi: 10.1165/rcmb.2008-0280OC. [DOI] [PubMed] [Google Scholar]
- 16.Thai P., Chen Y., Dolganov G., Wu R. Differential regulation of MUC5AC/Muc5ac and hCLCA-1/mGob-5 expression in airway epithelium. Am. J. Respir. Cell Mol. Biol. (2005);33:523–530. doi: 10.1165/rcmb.2004-0220RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thai P., Loukoianov A., Wachi S., Wu R. Regulation of airway mucin gene expression. Annu. Rev. Physiol. (2008);70:405–429. doi: 10.1146/annurev.physiol.70.113006.100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voynow J.A., Gendler S.J., Rose M.C. Regulation of mucin genes in chronic inflammatory airway diseases. Am. J. Respir. Cell Mol. Biol. (2006);34:661–665. doi: 10.1165/rcmb.2006-0035SF. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q., Dziarski R., Kirschning C.J., Muzio M., Gupta D. Micrococci and peptidoglycan activate TLR2 → MyD88 → IRAK → TRAF → NIK → IKK → NF-κB signal transduction pathway that induces transcription of interleukin-8. Infect. Immun. (2001);69:2270–2276. doi: 10.1128/IAI.69.4.2270-2276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B., Lim D.J., Han J., Kim Y.S., Basbaum C.B., Li J.D. Novel cytoplasmic proteins of nontypeable Haemophilus influenzae up-regulate human MUC5AC mucin transcription via a positive p38 mitogen-activated protein kinase pathway and a negative phosphoinositide 3-kinase-Akt pathway. J. Biol. Chem. (2002);277:949–957. doi: 10.1074/jbc.M107484200. [DOI] [PubMed] [Google Scholar]


