Abstract
A moderate change in ambient temperature significantly affects plant physiology including flowering time. MiR399 and its target gene PHOSPHATE 2 (PHO2) are known to play a role in the maintenance of phosphate homeostasis. However, the regulation of flowering time by the miR399-PHO2 module has not been investigated. As we have previously identified miR399 as an ambient temperature-responsive miRNA, we further investigated whether a change in expression of the miR399-PHO2 module affects flowering time in response to ambient temperature changes. Here, we showed that miR399b-overexpressing plants and a loss-of-function allele of PHO2 (pho2) exhibited an early flowering phenotype only at normal temperature (23℃). Interestingly, their flowering time at lower temperature (16℃) was similar to that of wild-type plants, suggesting that alteration in flowering time by miR399 and its target PHO2 was seen only at normal temperature (23℃). Flowering time ratio (16℃/23℃) revealed that miR399b-overexpressing plants and pho2 mutants showed increased sensitivity to ambient temperature changes. Expression analysis indicated that expression of TWIN SISTER OF FT (TSF) was increased in miR399b-overexpressing plants and pho2 mutants at 23℃, suggesting that their early flowering phenotype is associated with TSF upregulation. Taken together, our results suggest that miR399, an ambient temperature-responsive miRNA, plays a role in ambient temperature-responsive flowering in Arabidopsis.
Keywords: ambient temperature, flowering time, MiR399, PHO2, TSF
INTRODUCTION
The initiation of flowering in Arabidopsis is affected by various environmental stimuli (Simpson and Dean, 2000), among which temperature plays an important role. Because plants are sessile organisms, they are continuously exposed to modest temperature changes and thus adjust their growth and development in response to moderate changes in ambient temperature. However, not much is known about how ambient temperature is sensed and triggers physiological response in plants. Recently, H2A.Z-containing nucleosomes mediate ambient temperature responses in plants (Kumar and Wigge, 2010). SHORT VEGETATIVE PHASE (SVP) was shown to control flowering time responsive to ambient temperature changes via direct binding to the FLOWERING LOCUS T (FT) locus (Lee et al., 2007). Moreover, it was also suggested that SVP is involved in small RNA-mediated flowering in response to ambient temperature changes (Lee et al., 2010). The integration of signals that promote or inhibit floral development in response to ambient temperature changes ultimately converge in the regulation of a few floral integrator genes including FT and TWIN SISTER OF FT (TSF) (Kardailsky et al., 1999; Kobayashi et al., 1999). Although some genetic evidence on the ambient temperatureresponsive flowering has been reported, obviously it is still an early stage and more data should be accumulated to better interpret ambient temperature signaling.
MicroRNAs (miRNA) are small non-coding RNAs (21-22 nucleotides) that have been implicated in various plant functions, including development, phase transitions, and responses to environmental stress (Lee et al., 2010; Palatnik et al., 2003; Wang et al., 2009). The mature miRNA associates with an RNA-induced silencing complex (RISC) and guides it to the target mRNA, resulting in the inhibition of the expression of the target gene. The association between miRNA and plant development is seen in the function of miR156 and miR172, which target SQUAMOSA PROMOTER BINDING PROTEIN (SBP)- box and AP2-like family genes, respectively, to control the expression of floral integrator genes and modulate flowering time and phase transitions (Wang et al., 2009; Wu et al., 2009). Constitutive expression of miR156 and miR172 resulted in delayed and accelerated flowering times, respectively, whereas their target mimicry lines show an opposite phenotype (Franco-Zorrilla et al., 2007; Todesco et al., 2010).
MiR399, which is generated from 6 loci (miR399a, b, c, d, e, and f) in the Arabidopsis genome, is known to play an important role in the maintenance of Pi homeostasis (Bari et al., 2006; Chiou et al., 2006; Pant et al., 2008). Overexpression of MiR399 results in overaccumulation of Pi (Chiou et al., 2006). PHO2 was identified as a target gene of miR399 (Aung et al., 2006; Bari et al., 2006) based on the reduction of PHO2 expression levels via miR399-mediated cleavage (Lin et al., 2008). The control of Pi homeostasis by miR399 is therefore mediated by the regulation of PHO2 expression. PHO2 encodes a ubiquitinconjugating E2 enzyme, which is a component in the ubiquitindependent protein degradation pathway (Sunkar and Zhu, 2004), suggesting that protein degradation is important in Pi homeostasis. Recently, miR399 was identified as an ambient temperature-responsive miRNA (Lee et al., 2010). Mature miR399 is more abundant in plants grown at 23℃ than at 16℃. The expression levels of PHO2 are negatively correlated with miR399 expression at different temperatures, suggesting that miR399 is involved in the response to ambient temperature changes in plants. However, the role of miR399 and its target gene PHO2 in the regulation of ambient temperature-responsive flowering time is not known.
In the present study, we studied the roles of miR399 and PHO2 in the regulation of ambient temperature-responsive flowering time. We analyzed p35S:miR399b plants and a lossof- function allele of PHO2 (pho2). Both p35S:miR399b plants and pho2 mutants showed early flowering only at normal temperature, thus exhibiting increased sensitivity to ambient temperature changes. TSF transcript levels were increased in both p35S:miR399b and pho2 plants, which may explain their early flowering phenotype. Collectively, the present results suggest that the miR399-PHO2 module plays a role in the regulation of flowering time in response to ambient temperature changes in plants.
MATERIALS AND METHODS
Plant materials and measurement of flowering time
Wild-type and transgenic Arabidopsis plants were grown on MS medium or soil at 23℃ or 16℃ under long day (16 h light/8 h dark) conditions at a light intensity of 120 μmol m-2 s-1. Construction of the p35S:miR399b was described previously (Chiou et al., 2006). Flowering time was determined either by counting the total number of leaves (rosette + cauline) on the main shoot of plants grown in soil or by measuring the number of days elapsed between the time of germination and the time of bolting. Flowering time was determined by scoring at least 10 plants.
Small RNA Northern hybridization
Plants were harvested at zeitgeber time (ZT) 8. Total RNA was extracted with Plant TRIzol® Reagent (Invitrogen) from 8-dayold whole seedlings grown at 23℃ or 16℃ under LD conditions. For small RNA Northern blots, 10 μg of total RNA was separated on a denaturing 17% polyacrylamide gel (8 M urea) in TBE buffer and transferred to an N+ Hybond membrane (Amersham). Hybridization was carried out at 42℃ using PerfectHyb™ Plus hybridization buffer (Sigma). DNA oligonucleotide probes specific to miR399b were end-labeled with γ32P-ATP using Optikinase™ (USB). The bands’ intensities were quantified using Image Gauge. Expression was normalized against U6 RNA (Yoo et al., 2011).
Gel-based RT-PCR
The reverse transcriptase-mediated PCR (RT-PCR) procedure has been described previously (Yoo et al., 2005). Total RNA was isolated from whole seedlings using Trizol® reagent (Invitrogen), according to the manufacturer’s instructions. The cDNA was synthesized from 1 μg total RNA treated with DNaseI (New England Biolab). PCR cycle numbers of each gene were determined as an amplicon exponentially amplified by PCR. Resulting amplicons were separated using 1.2% agarose gel electrophoresis. UBQ10 (At4G05320) was used as an internal positive control (Lee et al., 2010).
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
The cDNA was synthesized from extracted total RNA using oligo dT primers and the First Strand cDNA Synthesis Kit for RT-PCR (Roche) and analyzed by real-time (qRT-PCR). Quantitative RT-PCR analysis was carried out according to the ‘Eleven Golden Rules for Quantitative RT-PCR’ (Udvardi et al., 2008). The qRT-PCR reaction was performed in a 384-well plate with a LightCycler 480 Real Time PCR system (Roche) using the KAPA SYBR® FAST qPCR Kit (Kapa Biosystems) (Li et al., 2010). LingRegPCR was used for calculating threshold cycle (Ct) and PCR efficiency of the primers used (Ramakers et al., 2003). Relative expression of the transcripts was calculated using PCR efficiency and Ct value according to the instruction of geNorm (Vandesompele et al., 2002). In order to quantify more precisely, we used two genes (At3G01150 and At4G26410) that were identified as stably expressed genes at 23℃ and 16℃ for reference genes during quantification (Hong et al., 2010), instead of conventional housekeeping genes. All qRTPCR experiments were carried out in biological duplicates with technical triplicates for each. The oligonucleotide sequences of the hybridization probes and PCR primers used in this study are presented as Supplementary Table S1.
RESULTS
MiR399b overexpression caused ambient temperaturesensitive flowering
To elucidate the role of miR399 in the regulation of flowering time in response to different ambient temperatures, we analyzed the phenotype of transgenic plants overexpressing miR399b (p35S:miR399b). Before we measured flowering time of p35S:miR399b plants, we first confirmed the overproduction of miR399b in transgenic plants at 23℃ and 16℃ (Fig. 1A). Small RNA hybridization analysis indicated that mature miR399b was highly expressed at both temperatures (8.0-fold increase at 23℃ and 7.2-fold increase at 16℃) in transgenic plants grown in normal nutrient media under Pi-sufficient conditions. This indicated that the transgenic plants we used could be used to test the effect of miR399 activity on flowering time. In addition, we found that in wild-type (WT) plants expression levels of miR399b at 23℃ was higher than at 16℃, which indicated that upregulation of miR399 at 23℃ was reproducible (Lee et al., 2010).
Fig. 1. Overexpression of miR399 caused an early flowering phenotype only at normal temperature. (A) Small RNA northern blot analysis of mature miR399b in 8-day-old p35S:miR399b plants grown at 23℃ and at 16℃ in P i-sufficient M S medium. U 6 RNA was used to show an equal amount of loading. Numbers below each blot indicate fold change relative to the miR399 level in WT plants grown at 23℃. (B, C) Phenotype and flowering time presented as total leaf number (rosette and cauline) and bolting day of p35S: miR399b plants grown at 23℃ and 16℃ under LD conditions. The vertical T-bars indicate standard deviation. Numbers above each genotype indicate leaf number ratio (16℃/ 23℃) or bolting-days ratio (16℃/23℃). (D) Quantitative RT-PCR analysis of PHO2 transcript levels in 8-day-old p35S:miR399b plants grown at 23℃ and 16℃ in Pi-sufficient MS medium. PHO2 expression levels were normalized against the expression levels of At3G01150 and At4G26410 and PHO2 expression level in wild-type plants was arbitrarily set to 1.0.
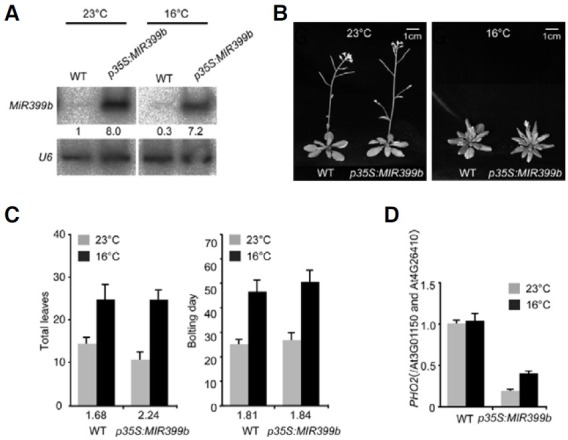
Flowering time measurement showed that transgenic p35S:miR399b plants grown at 23℃ under LD conditions flowered with 11 leaves (WT plants = 14.5 leaves), showing that p35S:miR399b plants were slightly early flowering (Figs. 1B and 1C). However, the leaf number of p35S:miR399b plants grown at 16℃ was similar to that of WT plants (24.6 leaves versus 24.4 leaves). This observation indicated that p35S: miR399b plants were early flowering only at 23℃. We also measured a flowering time ratio (defined as the proportion of the total number of leaves at 16℃/23℃) to determine the ambient temperature sensitivity. We used this ratio as an indicator of ambient temperature-responsive flowering. In miR399boverexpressing plants, flowering time ratio was 2.24, which was greater than that of WT plants (1.68). This indicated that in p35S:miR399b plants flowering time variation in response to ambient temperature changes was greater than in WT plants, revealing that miR399b overexpression caused ambient temperature- sensitive flowering. However, p35S:miR399b plants bolted almost at the same time as WT plants. The bolting time ratio, which is the number of days elapsed between the time of germination and the time of bolting, was similar in miR399b-overexpressing plants and in WT plants (1.84 versus 1.81). Thus, comparison of leaf numbers and bolting days indicated that the plastochron length of miR399b-overexpressing plants was decreased only at 23℃, whereas the plastochron length of p35S:miR399b plants was not altered at 16℃. These results demonstrated that miR399b overexpression affects ambient temperature-responsive flowering.
To assess the effect of miR399b overexpression on PHO2 expression, we analyzed the transcript levels of PHO2 in p35S:miR399b plants at both temperatures. For this experiment, we used a set of PCR primers that amplify a region containing miR399’s target sites in the 5′ UTR of PHO2. qRT-PCR analysis showed that PHO2 transcript levels were decreased in p35S:miR399b plants at both temperatures (Fig. 1D), indicating that miR399 overexpression significantly reduced PHO2 expression.
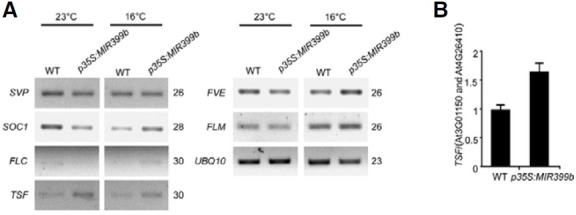
Because miR399b-overexpressing plants showed an early flowering phenotype, the flowering time gene affected by miR399 overexpression was investigated. The expression levels of flowering time genes were analyzed in 8-day-old whole seedlings of WT and p35S:miR399b plants grown under LD conditions in Pi-sufficient MS medium at 23℃ and 16℃. RT-PCR analysis showed that TSF expression was slightly upregulated in p35S:miR399b plants (Fig. 2A). Among the flowering time genes analyzed in this study, the expression of FVE, SVP, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), FLOWERING LOCUS M (FLM), and FLOWERING LOCUS C (FLC) was not obviously affected by miR399 overexpression (Fig. 2A) (Ausin et al., 2004; Jang et al., 2009; Moon et al., 2003; Scortecci et al., 2003; Sheldon et al., 2000). To further confirm the upregulation of TSF expression, a quantitative assessment of TSF expression levels was performed by qRT-PCR. We found that TSF expression was increased by 1.67-fold in p35S:miR399b plants (Fig. 2B), consistent with the RT-PCR results. These results suggested that the weak early flowering phenotype of p35S:miR399b plants was associated with the increased TSF expression at 23℃. Considering that TSF is a strong floral promoter (Yamaguchi et al., 2005), the upregulation of TSF explained the early flowering phenotype of miR399-overexpressing plants.
Fig. 2. Upregulation of TSF was observed in miR399-overexpressing plants at 23℃. (A) Expression of flowering time genes in 8-day-old p35S:miR399b plants grown at 23℃ and 16℃ under LD conditions analyzed by RT-PCR. UBQ10 was used as an internal control. PCR cycle numbers are indicated on the right of each gel. (B) Relative expression level of TSF in 8-day-old p35S:miR399b plants grown at 23℃ under LD conditions determined via qRTPCR. The TSF expression level was normalized against the expression levels of At3G01150 and At4G26410. TSF expression level in wild-type plants at 23℃ was arbitrarily set to 1.0.
Loss-of-PHO2 function caused an early flowering phenotype via increased TSF transcript level
As we observed an ambient temperature-sensitive flowering phenotype in p35S:miR399b plants, we analyzed a loss-of-function allele of PHO2 to test whether a similar flowering phenotype was obtained from the mutant. We used pho2 mutants, in which a point mutation in the sixth exon (from G2539 to A, relative to the translational start codon) causing an early termination at the 671 amino acid (Fig. 3A). The site of the early termination was at the beginning of the UBC domain and thus the mutation therefore caused the loss of the ubiquitin-conjugating activity of UBC24 in pho2 mutants (Aung et al., 2006). We first confirmed the down-regulation of PHO2 in pho2 mutants by qRT-PCR. The qRT-PCR analysis showed an approximately 6.25-fold decrease in PHO2 transcript levels (Fig. 3B). Thus, it seemed that pho2 mutants produced truncated (possibly non-functional) PHO2 protein with low abundance.
Fig. 3. pho2 mutants showed an early flowering phenotype only at 23℃. (A) Gene structure of PHO2 and the site of mutation in pho2. The translation initiation/termination sites (ATG/TAA) and the ubiquitin- conjugating conserved domain (UBC) are indicated. The asterisks indicate five putative miR399 target sites in the 5′ UTR. The single nucleotide change in the sixth exon in the pho2 mutant leading to early termination is indicated by a circle below W (TGG). (B) Analysis of PHO2 transcript levels in 8-day-old pho2 mutants grown at 23℃ and 16℃ in Pisufficient MS medium by qRTPCR. PHO2 expression level was normalized against the expression levels of At3G01150 and At4G 26410 and PHO2 expression level in WT plants grown at 23℃ was set to 1.0. (C, D) Phenotype and flowering time presented as total leaf number and bolting day of pho2 mutants grown at 23℃ and 16℃ under LD conditions. Numbers above each genotype indicate leaf number ratio (16℃/23℃) or bolting-days ratio (16℃/23℃). The vertical T-bars indicate standard deviation.
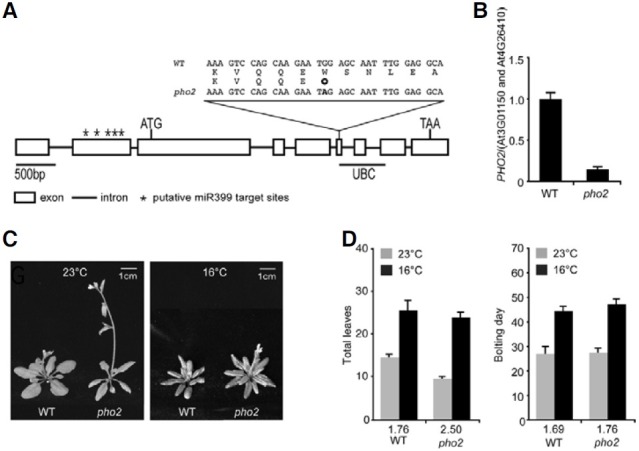
Flowering time measurement showed that the pho2 mutants grown at 23℃ under LD conditions showed an early flowering phenotype (9.3 leaves) in comparison to WT plants (WT plants = 14.3 leaves) (Figs. 3C and 3D). However, the flowering time in pho2 mutants at 16℃ was not significantly different from that of WT plants (23.4 leaves versus 25.1 leaves). This result showed that early flowering was observed only at 23℃. We then measured a flowering time ratio to determine how ambient temperature-responsive flowering was altered. The flowering time ratio of pho2 mutants was 2.5 (WT plants = 1.76). This indicated that in pho2 mutants flowering time variation in response to ambient temperature changes was greater than in WT plants, indicating that loss-of-function of PHO2 led to ambient temperature-sensitive flowering, as seen in p35S:miR399b plants (Figs. 1C and 3D). However, the bolting time ratio of pho2 mutants was similar to that of WT plants (1.76 versus 1.69), revealing that these mutants also displayed a decreased plastochron length only at 23℃. The decreased plastochron length of pho2 mutants at 23℃ was consistent with the phenotype of p35S:miR399b plants (Fig. 1C).
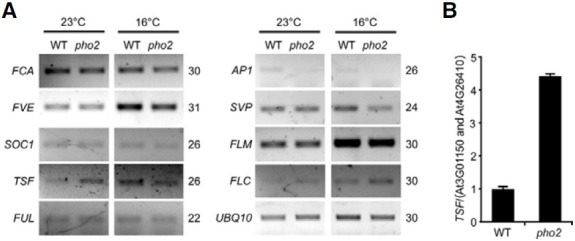
As pho2 mutants showed an early flowering phenotype, we investigated the expression levels of flowering time genes in pho2 mutants under LD conditions to determine which flowering time gene’s expression was altered. RT-PCR analysis indicated that TSF expression was significantly elevated at 23℃ in pho2 mutants (Fig. 4A). Among the flowering time genes analyzed, the transcript levels of FCA, FVE, SVP, SOC1, FRUIT-FULL (FUL), APETALA 1 (AP1), FLM, and FLC remained unaltered in pho2 mutants (Ausin et al., 2004; Jang et al., 2009; Macknight et al., 1997; Moon et al., 2003; Scortecci et al., 2003; Sheldon et al., 2000; Wang et al., 2009). To further confirm the upregulation of TSF expression in pho2 mutants, a quantitative assessment of TSF expression levels was performed via qRTPCR. We found that TSF expression was increased by 4.5-fold in pho2 mutants at 23℃ (Fig. 4B), consistent with the RT-PCR results. Interestingly, TSF expression was not upregulated in pho2 mutants grown at 16℃. Since pho2 mutants showed an early flowering only at 23℃, these results suggested that an increase in TSF levels downstream of PHO2 is responsible for the ambient temperature-responsive flowering of pho2 mutants.
Fig. 4. Upregulation of TSF was observed in pho2 mutants at 23℃. (A) Expression of flowering time genes in 8-day-old pho2 mutants grown at 23℃ and 16℃ under LD conditions via RT-PCR. UBQ10 was used as an internal control. PCR cycle numbers are indicated on the right of each gel. (B) Relative expression level of TSF in 8-day-old pho2 mutants grown at 23℃ under LD conditions via qRT-PCR. TSF expression level in wild-type plants at 23℃ was arbitrarily set to 1.0.
DISCUSSION
In this study, we analyzed the flowering time of miR399-overexpressing plants and a loss-of-function allele of PHO2, which is a target of miR399. We identified that alteration of miR399 and PHO2 activity caused early flowering only at normal temperature, suggesting that the miR399-PHO2 module regulates ambient temperature-responsive flowering in Arabidopsis.
Previous studies have described the close relationship between plant miRNAs and flowering time regulation. GI-regulated miR172 was shown to function in a genetic pathway that controls photoperiodic flowering by inducing FT in a CONSTANS (CO)-independent manner (Jung et al., 2007). A mutant, in which TARGET OF EAT 1 (TOE1), the target gene of miR172, is constitutively activated by the 35S enhancer, responded to vernalization and gibberellic acid treatment but not to day length changes. FT expression levels in this mutant were significantly reduced without affecting the expression of other flowering time genes. MiR172-overexpressing plants showed an early flowering phenotype under both LD and SD conditions, even in the absence of functional CO. In the present study, the overexpression of miR399b and a loss-of-function allele of PHO2 caused early flowering by increasing TSF expression. MiR399-mediated PHO2 cleavage may regulate photoperiodic flowering in a CO-independent manner based on our preliminary qRT-PCR results revealing that CO expression was not altered in these mutants (data not shown). Further analysis on miR399-overexpressing plants and pho2 mutants under both LD and SD conditions is required to precisely define the role of miR399-PHO2 in the photoperiod pathway.
The regulation of plastochron length by miRNA and its target genes has recently been reported. Down-regulation of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (SPL9) expression by miR156 overexpression resulted in reduced plastochron length and increased leaf numbers in Arabidopsis. Conversely, increased levels of SPL9 as a result of decreased miR156 expression caused an increase in plastochron length (Wang et al., 2008). Our study reveals that miR399-mediated PHO2 downregulation has an opposite effect against miR156- SPL in the regulation of plastochron length. Both p35S:miR399b plants and pho2 mutants showed a reduced number of rosette leaves but they bolted at the same time as WT plants at both 23℃ and 16℃ (Figs. 1C and 3D), suggesting that the miR399- targeted PHO2 module may also play a role in the regulation of plastochron length.
Devaiah and his colleagues demonstrated the relationship between phosphate starvation responses and gibberellic acid biosynthesis through studies with MYB62-overexpressing plants (Devaiah et al., 2009). MYB62 is an R2R3-type transcriptional factor that is induced in response to Pi deficiency. Overexpression of MY-B62 resulted in a GA-deficient phenotype characterized by delayed germination, bolting of the inflorescence stalk and rosette leaf initiation, and an increased number of rosette leaves. MYB62-overexpressing plants suppressed the transcript level of SOC1 and SUPERMAN (SUP), the transcription factors associated with floral homeotic genes. In the present study, 35S:miR399b plants and pho2 mutants showed early flowering via increased TSF expression. Overexpression of miR399 and loss-of-function of PHO2 were reported to induce overaccumulation of Pi in the shoot and result in Pi toxicity symptoms such as necrosis or chlorosis on the margins of mature leaves. Pi overaccumulation was found to be the result of increased Pi uptake (Aung et al., 2006; Chiou et al., 2006). Taken together, our results suggest that miR399 and its target gene PHO2 not only affect Pi homeostasis but also impact ambient temperature-responsive flowering. Although genetic evidence that the miR399-PHO2 module regulates flowering time was presented, the possibility that the alteration of flowering time seen in p35S:miR399b and pho2 plants was an indirect effect of Pi toxicity cannot be ruled out. Further detailed study is required to provide further evidence connecting the miR399-PHO2 module to flowering time regulation. A possible approach would be the measurement of flowering time and expression levels of floral integrator genes in miR399-overexpressing plants and pho2 mutants grown under high and low Pi conditions.
In conclusion, our study suggests that miR399 and its target gene PHO2, the genes that were known to function in the maintenance of Pi homeostasis in plants, plays a role in regulating flowering time in response to ambient temperature changes. We believe that this information will help improve the understanding of the molecular mechanism underlying the regulation of flowering time by small RNAs in response to ambient temperature changes.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
W. Kim was supported by the Brain Korea 21 program and National Research Foundation of Korea Grant funded by the Korea Government (Ministry of Education, Science and Technology) [NRF-2011-355-C00040]. This research was funded by the Creative Research Initiatives (R16-2008-106-01001-0) of the Ministry of Education, Science and Technology/National research foundation of Korea to J.H. Ahn.
References
- 1.Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. (2006);141:1000–1011. doi: 10.1104/pp.106.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausin I., Alonso-Blanco C., Jarillo J.A., Ruiz-Garcia L., Martinez- Zapater J.M. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. (2004);36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 3.Bari R., Datt Pant B., Stitt M., Scheible W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. (2006);141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell. (2006);18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devaiah B.N., Madhuvanthi R., Karthikeyan A.S., Raghothama K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant. (2009);2:43–58. doi: 10.1093/mp/ssn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., Garcia J.A., Paz- Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. (2007);39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 7.Hong S.M., Bahn S.C., Lyu A., Jung H.S., Ahn J.H. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. (2010);51:1694–1606. doi: 10.1093/pcp/pcq128. [DOI] [PubMed] [Google Scholar]
- 8.Jang S., Torti S., Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. (2009);60:614–625. doi: 10.1111/j.1365-313X.2009.03986.x. [DOI] [PubMed] [Google Scholar]
- 9.Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. (2007);19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. Activation tagging of the floral inducer FT. Science. (1999);286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. (1999);286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S.V., Wigge P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. (2010);140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. (2007);21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H., Yoo S.J., Lee J.H., Kim W., Yoo S.K., Fitzgerald H., Carrington J.C., Ahn J.H. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. (2010);38:3081–3093. doi: 10.1093/nar/gkp1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S., Zhou X., Chen L., Huang W., Yu D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells. (2010);29:475–483. doi: 10.1007/s10059-010-0059-2. [DOI] [PubMed] [Google Scholar]
- 16.Lin S.I., Chiang S.F., Lin W.Y., Chen J.W., Tseng C.Y., Wu P.C., Chiou T.J. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. (2008);147:732–746. doi: 10.1104/pp.108.116269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macknight R., Bancroft I., Page T., Lister C., Schmidt R., Love K., Westphal L., Murphy G., Sherson S., Cobbett C., et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. (1997);89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 18.Moon J., Suh S.S., Lee H., Choi K.R., Hong C.B., Paek N.C., Kim S.G., Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. (2003);35:613–623. doi: 10.1046/j.1365-313x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- 19.Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. Control of leaf morphogenesis by microRNAs. Nature. (2003);425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 20.Pant B.D., Buhtz A., Kehr J., Scheible W.R. Micro-RNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. (2008);53:731–738. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramakers C., Ruijter J.M., Deprez R.H., Moorman A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. (2003);339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 22.Scortecci K., Michaels S.D., Amasino R.M. Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Biol. (2003);52:915–922. doi: 10.1023/a:1025426920923. [DOI] [PubMed] [Google Scholar]
- 23.Sheldon C.C., Rouse D.T., Finnegan E.J., Peacock W.J., Dennis E.S. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA. (2000);97:3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson G.G., Dean C. Environmental-dependent acceleration of a developmental switch: the floral transition. Sci. STKE. (2000);pe1. doi: 10.1126/stke.2000.18.pe1. [DOI] [PubMed] [Google Scholar]
- 25.Sunkar R., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. (2004);16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. (2010);6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udvardi M.K., Czechowski T., Scheible W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell. (2008);20:1736–1737. doi: 10.1105/tpc.108.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. (2002);3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH 0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J.W., Schwab R., Czech B., Mica E., Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. (2008);20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J.W., Czech B., Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. (2009);138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. (2009);138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. (2005);46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 33.Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. (2005);139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo S.K., Hong S.M., Lee J.S., Ahn J.H. A genetic screen for leaf movement mutants identifies a potential role for AGAMOUS-LIKE 6 (AGL6) in circadian-clock control. Mol. Cells. (2011);31:281–287. doi: 10.1007/s10059-011-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


