Abstract
MicroRNAs (miRNAs) are recently discovered, noncoding, small regulatory RNA molecules that negatively regulate gene expression. Although many miRNAs are identified and validated in many plant species, they remain largely unknown in Brassica rapa (AA 2n =, 20). B. rapa is an important Brassica crop with wide genetic and morphological diversity resulting in several subspecies that are largely grown for vegetables, oilseeds, and fodder crop production. In this study, we identified 186 miRNAs belonging to 55 families in B. rapa by using comparative genomics. The lengths of identified mature and pre-miRNAs ranged from 18 to 22 and 66 to 305 nucleotides, respectively. Comparison of 4 nucleotides revealed that uracil is the predominant base in the first position of B. rapa miRNA, suggesting that it plays an important role in miRNA- mediated gene regulation. Overall, adenine and guanine were predominant in mature miRNAs, while adenine and uracil were predominant in pre-miRNA sequences. One DNA sequence producing both sense and antisense mature miRNAs belonging to the BrMiR 399 family, which differs by 1 nucleotide at the, 20th position, was identified. In silico analyses, using previously established methods, predicted 66 miRNA target mRNAs for 33 miRNA families. The majority of the target genes were transcription factors that regulate plant growth and development, followed by a few target genes that are involved in fatty acid metabolism, glycolysis, biotic and abiotic stresses, and other cellular processes. Northern blot and qRT-PCR analyses of RNA samples prepared from different B. rapa tissues for 17 miRNA families revealed that miRNAs are differentially expressed both quantitatively and qualitatively in different tissues of B. rapa.
Keywords: Brassicaceae, in silico, Small RNAs
INTRODUCTION
Gene expression of higher eukaryotes is regulated either at a transcriptional or posttranscriptional level. Various factors regulating gene expression at both the stages have been identified and demonstrated with experimental results (Depicker and Montagu, 1997; Vaucheret and Fagard, 2001). One class of such regulators at the posttranscriptional level is small, endogenous, noncoding RNA molecules known as microRNAs (miRNAs). miRNAs are generally 18-24 nucleotides (nt) long, and are abundant in plants and animals. miRNAs regulate target gene expression by degrading complementary mRNA sequences or by reducing the translation of the target gene (Bartel, 2004; Voinnet, 2009). miRNAs are transcribed by RNA polymerase II in plants, and the primary transcripts of mature miRNAs fold back into stable hairpin loop structures that form precursor miRNA (pre-miRNA). The processing of primary miRNA sequences, including capping, splicing, and polyadenylation, is completed in the nucleus by RNase III-like endonuclease and Dicer-like-1 enzyme (DCL1) in plants (Bartel, 2004; Dugas and Bartel, 2004). The HASTY5 gene exports the processed methylated miRNA/miRNA* to the cytoplasm (Bollman et al., 2003; Park et al., 2005; Voinnet, 2009). The matured miRNAs are then incorporated into argonauts containing RNA-induced silencing complex; they interact with the complementary sites of the target gene transcripts, which then negatively regulate the target gene expression by degrading or repressing the mRNA transcripts (Carthew and Sontheimer, 2009; Voinnet, 2009).
At present, miRNAs are mainly identified using 2 methods: (a) computational identification and structure prediction because a majority of known mature miRNAs are conserved within and between different plant species, allowing for comparative analysis using presently available bioinformatics tools to search for putative miRNAs (Wang et al., 2004a; 2004b); (b) the construction and sequencing of a small RNA library (Griffiths-Jones, 2006; Zhang et al., 2006a). These 2 methods have been used to identify hundreds of miRNAs in several plant species, including the model plant Arabidopsis thaliana (Alves-Junior et al., 2009; Buhtz et al., 2008; Jagadeeswaran et al., 2009; Jin et al., 2008; Klevebring et al., 2009; Song et al., 2009; Sunkar et al., 2005; 2008; Szittya et al., 2008; Xie et al., 2007; Zhang et al., 2006b; 2007a; 2008). Recently, many miRNAs have been shown to regulate a wide range of biological functions of plants. A growing amount of experimental evidence shows that miRNAs are involved in the regulation of transcription factors and other genes that play important roles in developmental timing, including the transitions from juvenile to adult and from the vegetative to reproductive phase (Lauter et al., 2005; Schwab et al., 2005; Wu and Poethig, 2006), floral development (Chen, 2004; Rhoades et al., 2002), leaf formation (Palatnik et al., 2003), stem development (Mallory et al., 2004), root development (Guo et al., 2005), signal transduction (Rhoades et al., 2002), male and female reproductive development (Wu and Poethig, 2006), and many other cellular processes (Ambros and Chen, 2007; Carrington and Ambros, 2003; Zhang et al., 2007b). Changes in the expression levels of miRNAs in plant tissues subjected to biotic stresses such as viral, fungal, and bacterial infection are reported (Bazzini et al., 2007; Lu et al., 2007; Navarro et al., 2006). Recently, changes in the expression of miRNAs due to abiotic stresses such as cold, drought, salinity, oxidative stress, mechanical strain, and UV-B radiation have also been identified in plants (Lu et al., 2005; Sunkar and Zhu, 2004; Sunkar et al., 2006; Zhao et al., 2007; Zhou et al., 2007) apart from nutrient stresses, such as phosphate or sulfate starvation (Chiou et al., 2006; Fujii et al., 2005; Jones-Rhoades and Bartel, 2004), and other nutrient responses (Sunkar et al., 2007).
However, despite a growing number of informations regarding miRNAs for specific gene expression regulation, miRNAs in Brassica species - especially B. rapa - are largely unknown despite a previous study reporting few miRNAs in this species (He et al., 2008). The systematic identification and characterization of miRNAs in B. rapa would help to elucidate the background of a large amount of genetic and morphological diversity that might have resulted in part to the differential gene expression regulated by miRNAs within and between B. rapa subspecies. B. rapa is one of the economically important crop species and exhibits a wide range of morphological diversity, forming different subspecies grown mainly for their leafy vegetables (Chinese cabbage, Pakchoi), oilseeds (Yellow sarson, Brown sarson, and Toria) and fodder (Turnip). Although these subspecies have the same genome complement, they exhibit variations in leaf morphology, head formation, plant type, and other morphological traits. Due to the global economic importance of B. rapa and the fact that it is a diploid progenitor parent that contributes its genome to widely cultivated amphidiploid Brassica oilseed crops, such as B. napus (AACC) and B. juncea (AABB), the Multinational B. rapa Genome Sequencing Project was started in, 2003 (Yang et al., 2005). This project, which is near completion, has generated a large amount of genome survey sequences (GSS, including BAC sequences) in addition to a large number of expressed sequence tags (ESTs) and messenger mRNAs (mRNAs) taken from different tissues and developmental stages of B. rapa; these were deposited in the National Center for Biotechnology Information (NCBI) database. In this study, we comprehensively investigated the presence of miRNAs in B. rapa that could be involved in a variety of functions, using genome survey sequences (GSS), ESTs, mRNAs, and complementary DNA (cDNA) presently available in a public database (NCBI).
MATERIALS AND METHODS
Reference set of miRNAs and B. rapa sequences
Two thousand eighty-five previously known mature plant miRNA sequences from A. thaliana, Brassica napus, Glycine max, Oryza sativa, Populus trichocarpa, Triticum aestivum, Vitis vinifera, and Zea mays were downloaded from the miRBase (Griffiths-Jones, 2006; Griffiths-Jones et al., 2008). From these 1408 repeated miRNA sequences were removed using Perl script (http://www.perl.org/), and the remaining 977 miRNAs were used as final reference set. A total of 397,600 GSSs, ESTs, and mRNAs of B. rapa sequences were downloaded from the NCBI GenBank database (ftp://ftp.ncbi.nih.gov/genbank/, http://www.brassica-rapa.org, and http://www.brassica.info). In addition, we used 12,098 full-length cDNAs and 5× NGS whole-genome sequence data of the B. rapa cultivar “Chiifu,” which is available in the laboratory. The 5× NGS wholegenome sequence data of the Chiifu cultivar were developed in our laboratory in collaboration with the Macrogen Company, Seoul, Korea (http://www.macrogen.com).
Software used to identify B. rapa miRNAs and their targets
We used BLAST-2.2.20 (Camacho et al., 2009) to identify conserved miRNAs from B. rapa sequences (GSSs, ESTs, mRNAs, and cDNAs), Mfold-3.1.2 (Zuker, 2003; Zuker et al., 1998) for predicting the secondary structure and stability of RNA, miRU (Zhang, 2005) to identify and confirm the targets genes of identified miRNA, and Perl scripts to remove overlapping and repeated mature miRNA sequences.
Computational identification of miRNAs
The computational method developed by Meyers et al. (2008) was used to search the conserved miRNAs in the B. rapa genome sequence. The nonredundant mature miRNAs were implemented as query sequence, using BLASTN to search for homologues from the downloaded B. rapa genome sequences, because they are highly conserved among plant species compared to precursor miRNA sequences. BLASTX was performed using the UniProt protein database (http://www.uniprot.org); putative B. rapa candidate miRNA nucleotide sequences with < 4 mismatches were compared to previously identified plant miRNAs; and all protein-coding sequences were removed. The hairpin structure for the remaining non-protein-coding putative B. rapa miRNA candidate sequences was predicted by the Mfold program (Zuker et al., 1998). The following criteria reported by Meyers et al. (2008) were used: (a) < 4 mismatches allowed for the identification of B. rapa mature miRNAs compared to previously known plant miRNAs; (b) precursor miRNAs folded into stem-loop hairpin secondary structures; (c) mature miRNAs fall on the stem part of the hairpin secondary structure; (d) mature miRNAs allowed < 6 mismatches with the other arm of the structure; and (e) predicted precursor miRNAs secondary structures have high negative minimal folding free energy (MFE), adjusted minimum folding free energy index (AMFE), and higher minimal folding free energy indexes (MFEIs). MFE, MFEI, and AFME values were determined using the methods outline by Zuker (2003) and Zhang et al. (2006c; 2008), respectively. To avoid redundancy and to confirm different members of the gene, we compared the predicted whole and precursor miRNA sequence-containing gene by using B. rapa unigenes and A. thaliana genes (http://www.arabidopsis.org).
Expression analyses of putative B. rapa miRNAs
Northern blot analyses
Total RNA was isolated from, 20-day-old B. rapa cultivar Kenshin seedling plants by using Trizol reagent (Invitrogen, USA) according to the procedure previously described by Kim et al. (2005). Total RNA was separated in 15% urea-polyacrylamide gel and electrically transferred to Hybond-N+ membranes (Amersham Bioscience, UK). Membranes were UV cross-linked and baked for 30 min at 80℃. Blots were hybridized with the ULTRA-Hyb Oligo solution (Ambion, USA) and oligonucleotide probes that were end-labeled using 32P-γATP and T4 polynucleotide kinase (New England Biolabs). After hybridization, the blots were washed with washing solution (2× SSC + 0.5% SDS) and briefly air dried. Images were obtained by exposing the blots to X-ray film. EtBr-stained 5S rRNA was used as a loading control. The 5 miRNA probes used to test for their expression analysis in B. rapa were as follows: (5′-3′, antisense) BrMiR 159 TAGAGCTCCCTTCAATCCAAA; BrMiR 160, TGGCATACAGGGAGCCAGGCA; BrMiR 167, TAGAT CATGCTGGCAGCTTCA; BrMiR 398, AGGGGTGACCTGAG AACACA; and BrMiR 408, GCCAGGGAAGAGGCAGTGCAT.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from B. rapa cultivar Chiifu plants as described in previous paragraph. Shoot apex (unopened floral buds and primary shoot apex), stems, old leaves (source, green tissues of old leaves larger than 13 cm length and 8 cm width), young leaves (smaller than 3 cm length and 2.5 cm Width), midribs (sink, white tissue of old leaves larger than 13 cm length and 8 cm width) and roots were collected for RNA extraction. Real time PCR was performed six times for each miRNAs following Shi and Chiang (2005). One microgram of total RNA polyadenylated with ATP by poly(A) polymerase according to the manufacturer’s protocol. After phenol-chloroform extraction, the RNAs were reverse-transcribed using Reverse Transcription Kit (Promega, USA) with poly(T) adaptor primer (Table 1). For real-time PCR, 1 ul cDNA was mixed with 12.5 ul of 2× SYBR Green Mix (Qiagen, USA) and 5 pmol each of the miRNA specific primer and reverse primers in a final volume of 25 ul. PCR runs were 45 cycles each cycles at 95℃ for 10 s, 65℃ for 15 s and 72℃ for 20 s. The relative microRNA expression was quantified using comparative ΔΔCT method (Livak and Schmittgen, 2001). All expression profiles are normalized with expression levels at shoot apex. B. rapa Actin gene (BrACT) was used as an internal control and each primer sequences were described in Table 1.
Table 1.
Primer sequences used for qRT-PCR analyses
| Name | Sequence (5′→ 3′) |
|---|---|
| BrMiR 158b | CCCAAATGTAGACAAAGCA |
| BrMiR 162b | TCGATAAACCTGTGCATCCAG |
| BrMiR 164c | TGGAGAAGCAGGGCACGTGCA |
| BrMiR 170b | TGATTGAGCCGTGCCAATATC |
| BrMiR 171f | TGATTGAGCCGCGCCAATATC |
| BrMiR 172g | GAATCTTGATGATGCTGCAT |
| BrMiR 396b | TTCCACAGCTTTCTTGAACTG |
| BrMiR 400b | TATGAGAGTATTATACGTCAC |
| BrMiR 837 | ATCAGTTTCTTGTTCTTTTC |
| BrMiR 842 | ATGGTCAGATCGGTCATC |
| BrMiR 1132b | ATTATGAAACGGAAGGAG |
| BrMiR 1521b | CTGTTGATGGAAAATGTT |
| Poly(T) adaptor | GCGAGCACAGAATTAATACGACTCACT ATAGG(T)12VaNb |
| Reverse primer | GCGAGCACAGAATTAATACGAC |
| BrACT Forward primer | GAACCGGGTGCTCCTCAGGA |
| BrACT Reverse primer | ATGGTACCGGAATGGTCAAGGC |
aV = A,G,C
bN = A,T,G,C
Prediction of identified miRNA targets
miRNAs bind to targets with perfect or near-perfect complementary; this nature motivates the prediction of potential targets through a computational comparative approach. To find the target genes, we compared the complementary sequences of the predicted miRNAs with all the available B. rapa sequences downloaded from the NCBI database. Less than 4 mismatches without gaps were allowed between predicted complementary miRNAs and their target sequences. We used miRU (http://bioinfo3.noble.org/psRNATarget; Zhang, 2005) and TAIR BLAST (http://www.arabidopsis.org/Blast/index.jsp) software to identify the targets in A. thaliana. To validate the identified targets, we compared miRNA as well as whole-target gene sequences with those of A. thaliana genes and identified the corresponding target genes in A. thaliana.
RESULTS
Identification of B. rapa miRNAs
miRNAs are conserved between plant species due to their important regulatory mechanisms. Their highly conserved nature allows the identification of miRNAs in different plant species for which genome sequence information is partially or fully available, or previously unidentified. The summary of steps followed in the present study is provided in Supplementary Fig. 1. Initially 977 known mature miRNA sequences were analyzed against B. rapa sequences (GSS, EST, and mRNA) in BLASTN, of which 780 probable conserved sequences appeared to contain miRNAs. Further screening removing the repeats and protein- coding sequences narrowed down the probable miRNA candidates to 234. Structure prediction using the Mfold program finally gave 186 mature miRNAs with reasonable stem-loop formation. The 186 identified B. rapa miRNAs belong to 55 families, and, 20 miRNAs belonging to 9 miRNA families were derived from EST sequences. Varying numbers of miRNAs from the 55 families were identified (See Table 2 and Fig. 1 for details). A maximum of 19 miRNAs and minimum of 1 miRNA were detected in a single family, and the length of matured ranged from 18 to 22 nt long (Table 2 and Fig. 1), consistent with the findings in B. napus (Xie et al., 2007) and cotton (Qiu et al., 2007). The majority of miRNAs were 21 nt long (54.84%) followed by, 20 (21.51%), 19 (10.22%) 18 (9.14%), and 22 (2.69%) (Fig. 2). It is suggested that the difference in size of the identified miRNAs within different families might offer unique functions for the regulation of miRNA biogenesis or gene expression in plants. It is reported that uracil is more represented at the 5′ side of mature miRNAs; the percentage of representation of mature miRNAs having U at the 5′ side of the stem loop is 66.67%, followed by A (14.52%), C (9.68%), and G (9.14%); this is consistent with previous findings (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008; Zhang et al., 2008). It is speculated that the strong bias of uracil in the first 5′ nucleotide position is due to its important role in the recognition of the miRNA by argonaute1 (Zhang et al., 2006b). The analyses of nucleotide contents in the mature miRNAs show a majority of G [26.96% (10.13%)], followed by A [26.61% (10.07%)], U [23.71% (10.87%)], and C [22.72% (10.61%)] nucleotides. The magnitude of diversity of the identified miRNAs was also found in the location of mature miRNAs sequences (Fig. 3). Of the 186 newly identified mature miRNAs, 93 were located toward the 5′ side of the stem loop of the precursor sequences, while the other 93 toward the 3′ side.
Fig. 1. Number of miRNAs identified in 55 miRNA families in Brassica rapa.

Table 2.
Details of identified Brassica rapa (Br) miRNAs, their structure information, and corresponding Arabidopsis thaliana (At) genes
| miRNA family | Br-GSS/EST | At-gene | Mature miRNA | ML | MAS | Len(Pre) | GC% | MFE | AMFE | MFEI |
|---|---|---|---|---|---|---|---|---|---|---|
| BrMiR 156a | ED524353 | AT5G55835.1 | UUGACAGAAGAAAGAGAGCAC | 21 | 5′ | 109 | 48.62 | 42.9 | 39.36 | 0.809 |
| BrMiR 156b | AJ858639 | AT4G30972.1 | UGACAGAAGAGAGUGAGCAC | 20 | 5′ | 151 | 46.36 | 64.8 | 42.91 | 0.925 |
| BrMiR 156c | DU829396 | AT4G30972.1 | UGACAGAAGAGAGUGAGCAC | 20 | 5′ | 92 | 47.83 | 30.9 | 33.59 | 0.702 |
| BrMiR 156d | DU831758 | AT4G30972.1 | UGACAGAAGAGAGUGAGCAC | 20 | 5′ | 98 | 45.92 | 39 | 39.8 | 0.866 |
| BrMiR 156e | DU121870 | AT5G10945.1 | UGACAGAAGAGAGUGAGCAC | 20 | 5′ | 111 | 36.94 | 39.5 | 35.59 | 0.963 |
| BrMiR 156f | AJ859176 | GACAGAACAGAGUGAGCAC | 19 | 3′ | 211 | 33.65 | 50.8 | 24.08 | 0.715 | |
| BrMiR 156g | ED529145 | GACAGAAUAGAGUGAGCAC | 19 | 3′ | 209 | 32.5 | 47.7 | 22.82 | 0.701 | |
| BrMiR 156h | DX011343 | UGACAGAAGAGAGAGAGC | 18 | 3′ | 241 | 47.72 | 75.4 | 31.29 | 0.655 | |
| BrMiR 156i | contig182998 | AT4G31877.1 | UGACAGAAGAGAGUGAGCAC | 20 | 3′ | 118 | 40.68 | 48.1 | 40.76 | 1.002 |
| BrMiR 156j | CV432746.1 | AT4G31877.1 | UGACAGAAGAGAGUGAGCAC | 20 | 5′ | 151 | 45.03 | 53.2 | 35.23 | 0.782 |
| BrMiR 156k | contig156839 | AT5G26147.1 | UGACAGAAGAGAGUGAGCAC | 20 | 3′ | 158 | 41.77 | 54.5 | 34.49 | 0.825 |
| BrMiR 156l | contig067399 | AT5G10945.1 | UGACAGAAGAGAGUGAGCAC | 20 | 5′ | 144 | 38.89 | 51.4 | 35.69 | 0.917 |
| BrMiR 156m | contig024357 | AT4G31877.1 | UGACAGAAGAGAGUGAGCAC | 20 | 3′ | 95 | 40 | 15.9 | 16.74 | 0.418 |
| BrMiR 156n | contig162829 | AT2G19425.1 | CGACAGAAGAGAGUGAGCAC | 20 | 5′ | 104 | 49.04 | 45.1 | 43.37 | 0.884 |
| BrMiR 156o | contig010293 | AT5G10945.1 | UGACAGAAGAGAGCGAGCACA | 21 | 5′ | 114 | 38.6 | 43.8 | 38.42 | 0.995 |
| BrMiR 157a | contig117388 | AT1G66783.1 | UUGACAGAAGAUAGAGAGCAC | 21 | 3′ | 136 | 38.97 | 45.5 | 33.46 | 0.858 |
| BrMiR 157b | contig126773 | UUGACAGAAGAAAGAGAGCAC | 21 | 5′ | 126 | 46.83 | 44.4 | 35.24 | 0.752 | |
| BrMiR 158a | CT015463 | AT3G10745.1 | CCAAAUGUAGACAAAGCA | 18 | 3′ | 88 | 37.5 | 25.8 | 29.32 | 0.781 |
| BrMiR 158b | contig145627 | AT3G10745.1 | UCCCAAAUGUAGACAAAGCA | 20 | 5′ | 120 | 41.67 | 37.1 | 30.92 | 0.741 |
| BrMiR 159a | ED518706 | AT1G73687.1 | UUUGGAUUGAAGGGAGCUCUA | 21 | 3′ | 71 | 36.62 | 15.1 | 21.27 | 0.58 |
| BrMiR 159b | EX050542.1 | AT1G73687.1 | UUUGGAUUGAAGGGAGCUCUA | 21 | 3′ | 71 | 36.62 | 15.1 | 21.27 | 0.58 |
| BrMiR 159c | EX048967.1 | AT1G73687.1 | UUUGGAUUGAAGGGAGCUCUA | 21 | 3′ | 71 | 36.62 | 15.1 | 21.27 | 0.58 |
| BrMiR 159d | EX047628.1 | AT1G73687.1 | UUUGGAUUGAAGGGAGCUCUA | 21 | 3′ | 71 | 36.62 | 15.1 | 21.27 | 0.58 |
| BrMiR 159e | ED525625 | AT1G18075.1 | UUGGAUUGAAGGGAGCUC | 18 | 3′ | 73 | 35.62 | 15.6 | 21.27 | 0.6 |
| BrMiR 159f | CT016164 | AT5G61730.1 | UUGCAUGCCCCAGGAGCU | 18 | 3′ | 70 | 48.57 | 16.6 | 23.71 | 0.488 |
| BrMiR 159g | EX039355.1 | AT1G73687.1 | UUUGGAUUGAAGGGAGCUCUA | 21 | 3′ | 194 | 36.08 | 58.9 | 30.36 | 0.841 |
| BrMiR 159h | contig176646 | AT1G73687.1 | UUUGGAUUGAAGGGAGCUCUA | 21 | 3′ | 206 | 38.35 | 65.8 | 31.94 | 0.832 |
| BrMiR 160a | DX045892 | AT2G39175.1 | UGCCUGGCUCCCUGUAUGCCA | 21 | 5′ | 94 | 51.06 | 39.9 | 42.45 | 0.831 |
| BrMiR 160b | EX044968.1 | AT2G39175.1 | UGCCUGGCUCCCUGUAUGCCA | 21 | 5′ | 94 | 51.06 | 39.9 | 42.45 | 0.831 |
| BrMiR 160c | DX016569 | AT1G77850.1 | UGCCUGGCUCCCUGCAUGCCA | 21 | 3′ | 186 | 51.61 | 54 | 29.03 | 0.562 |
| BrMiR 160d | contig053638 | AT4G17788.1 | UGCCUGGCUCCCUGUAUGCCA | 21 | 5′ | 126 | 46.03 | 48.7 | 38.65 | 0.839 |
| BrMiR 160e | contig136837 | AT5G46845.1 | UGCCUGGCUCCCUGUAUGCCA | 21 | 5′ | 134 | 41.79 | 49.6 | 37.01 | 0.885 |
| BrMiR 160f | contig152673 | AT4G17788.1 | UGCCUGGCUCCCUGUAUGCCA | 21 | 3′ | 121 | 41.32 | 48.2 | 39.83 | 0.964 |
| BrMiR 160g | contig133622 | AT5G46845.1 | UGCCUGGCUCCCUGUAUGCCA | 21 | 5′ | 105 | 43.81 | 36 | 34.29 | 0.782 |
| BrMiR 160h | EX025484.1 | AT2G39175.1 | UGCCUGGCUCCCUGUAUGCCA | 21 | 5′ | 126 | 43.65 | 44.7 | 35.48 | 0.812 |
| BrMiR 161 | contig045367 | UCAACGCAUUGAAAGUGACUA | 21 | 5′ | 128 | 35.94 | 56 | 43.75 | 1.21 | |
| BrMiR 162a | ED527680 | AT5G08185.3 | UCGAUAAACCUGUGCAUCCAG | 21 | 3′ | 108 | 42.59 | 31.1 | 28.8 | 0.676 |
| BrMiR 162b | EX133401.1 | AT5G08185.3 | UCGAUAAACCUGUGCAUCCAG | 21 | 3′ | 108 | 42.59 | 31.1 | 28.8 | 0.676 |
| BrMiR 162c | EX071919.1 | AT5G08185.3 | UCGAUAAACCUGUGCAUCCAG | 21 | 3′ | 107 | 42.99 | 33.3 | 31.12 | 0.723 |
| BrMiR 162d | EX071254.1 | AT5G08185.3 | UCGAUAAACCUGUGCAUCCAG | 21 | 3′ | 108 | 42.59 | 31.1 | 28.8 | 0.676 |
| BrMiR 162e | EX069816.1 | AT5G08185.3 | UCGAUAAACCUGUGCAUCCAG | 21 | 3′ | 108 | 42.59 | 31.1 | 28.8 | 0.676 |
| BrMiR 162f | AC189332.2 | AT5G08185.3 | UCGAUAAACCUGUGCAUCCAG | 21 | 3′ | 99 | 43.43 | 30.4 | 30.71 | 0.707 |
| BrMiR 162g | contig177394 | AT5G23065.1 | UCGAUAAACCUGUGCAUCCAG | 21 | 3′ | 118 | 44.92 | 43.5 | 36.86 | 0.82 |
| BrMiR 164a | CW984396 | AT2G47585.1 | UGGAGAAGCAGGGCACGUGCA | 21 | 5′ | 108 | 50 | 51.1 | 47.31 | 0.944 |
| BrMiR 164b | DX051151 | AT2G47585.1 | UGGAGAAGCAGGGCACGUGCA | 21 | 5′ | 81 | 48.15 | 30.8 | 38.02 | 0.789 |
| BrMiR 164c | contig142689 | AT5G01747.1 | UGGAGAAGCAGGGCACGUGCA | 21 | 3′ | 151 | 44.37 | 52.6 | 34.83 | 0.785 |
| BrMiR 164d | contig167289 | AT2G47585.1 | UGGAGAAGCAGGGCACGUGCA | 21 | 5′ | 109 | 49.54 | 39.1 | 35.87 | 0.724 |
| BrMiR 164e | contig016289 | AT2G47585.1 | UGGAGAAGCAGGGCACGUGCA | 21 | 3′ | 110 | 51.82 | 57.5 | 52.27 | 1.008 |
| BrMiR 164f | contig181636 | AT5G27807.1 | UGGAGAAGCAGGGCACGUGC | 20 | 3′ | 112 | 42.86 | 25.9 | 23.13 | 0.539 |
| miRNA family | Br-GSS/EST | At-gene | Mature miRNA | ML | MAS | Len(Pre) | GC% | MFE | AMFE | MFEI |
| BrMiR 165 | contig026374 | UCGGACCAGGCUUCAUCCCCC | 21 | 3′ | 122 | 41.8 | 38.5 | 31.56 | 0.754 | |
| BrMiR 166a | DX911364 | AT3G61897.1 | UCGGACCAGGCUUCAUUCCCC | 21 | 3′ | 140 | 41.43 | 33.9 | 24.21 | 0.584 |
| BrMiR 166b | EX063968.1 | AT2G46685.1 | UCGGACCAGGCUUCAUUCCCC | 21 | 3′ | 140 | 41.43 | 33.9 | 24.21 | 0.584 |
| BrMiR 166c | EX063242.1 | AT2G46685.1 | UCGGACCAGGCUUCAUUCCCC | 21 | 3′ | 140 | 41.43 | 33.9 | 24.21 | 0.584 |
| BrMiR 166d | contig073929 | UCGGACCAGGCUUCAUUCCCC | 21 | 3′ | 136 | 42.65 | 43.7 | 32.13 | 0.753 | |
| BrMiR 167a | CT022223 | AT3G22886.1 | UGAAGCUGCCAGCAUGAUCUA | 21 | 5′ | 93 | 41.94 | 23.1 | 24.84 | 0.592 |
| BrMiR 167b | DX028242 | AT3G22886.1 | UGAAGCUGCCAGCAUGAUCUA | 21 | 5′ | 124 | 39.52 | 30.8 | 24.84 | 0.628 |
| BrMiR 167c | DX025819 | AT1G31173.1 | UGAAGCUGCCAGCAUGAUCU | 20 | 5′ | 268 | 32.46 | 56.8 | 21.19 | 0.652 |
| EX041799.1 | ||||||||||
| BrMiR 167d | contig181922 | AT3G22886.1 | UGAAGCUGCCAGCAUGAUCU | 20 | 5′ | 116 | 37.93 | 43.1 | 37.16 | 0.979 |
| BrMiR 168a | DU984956 | AT5G45307.1 | UCGCUUGGUGCAGGUCGGGAA | 21 | 5′ | 111 | 57.66 | 31 | 27.93 | 0.484 |
| BrMiR 168b | contig164758 | AT4G19395.1 | UCGCUUGGUGCAGGUCGGGAA | 21 | 5′ | 136 | 50.74 | 53.6 | 39.41 | 0.776 |
| BrMiR 168c | contig127394 | AT5G45307.1 | UCGCUUGGUGCAGGUCGGGAA | 21 | 5′ | 167 | 52.12 | 58.3 | 34.91 | 0.677 |
| BrMiR 168d | contig043838 | AT4G19395.1 | UCGCUUGGUGCAGGUCGGGA | 20 | 3′ | 134 | 52.99 | 33.5 | 25 | 0.471 |
| BrMiR 169a | DX901870 | AT3G13405.1 | CAGCCAAGGAUGACUUGCCGA | 21 | 5′ | 186 | 33.87 | 51.4 | 27.63 | 0.815 |
| BrMiR 169b | DU982154 | AT1G53687.1 | CAGCCAAGGAUGACUUGCCGA | 21 | 5′ | 109 | 33.03 | 40.6 | 37.25 | 1.128 |
| BrMiR 169c | DU988169 | AT3G13405.1 | CAGCCAAGGAUGACUUGCCGA | 21 | 5′ | 112 | 33.04 | 41.1 | 36.7 | 1.111 |
| BrMiR 169d | ED535040 | AT3G26818.1 | AGCCAAGGAUGACUUGCC | 18 | 5′ | 139 | 36.69 | 27.2 | 19.57 | 0.533 |
| BrMiR 169e | KFRT-048E03 | AT5G12840.3 | AGCCAAGAAUGAUUUGCCGG | 20 | 5′ | 271 | 43.91 | 63.5 | 23.43 | 0.534 |
| BrMiR 169f | ED527257 | AT3G26812.1 | UAGCCAAGGAGACUGCCUA | 19 | 5′ | 161 | 36.02 | 43 | 26.71 | 0.741 |
| BrMiR 169g | contig153738 | AT3G13405.1 | CAGCCAAGGAUGACUUGCCGA | 21 | 3′ | 181 | 32.6 | 46 | 25.41 | 0.779 |
| BrMiR 169h | contig132829 | AT3G13405.1 | CAGCCAAGGAUGACUUGCCGA | 21 | 5′ | 134 | 36.57 | 47.9 | 35.75 | 0.977 |
| BrMiR 169i | contig122748 | AT3G13405.1 | CAGCCAAGGAUGACUUGCCGA | 21 | 3′ | 195 | 31.28 | 39.9 | 20.46 | 0.654 |
| BrMiR 169j | contig062782 | AT4G21595.1 | AGCCAAGGAUGACUUGCCGA | 20 | 5′ | 129 | 39.53 | 44.1 | 34.19 | 0.864 |
| BrMiR 169k | contig176484 | AT5G24825.1 | CAGCCAAGGAUGACUUGCCG | 20 | 5′ | 148 | 41.89 | 50.8 | 34.32 | 0.819 |
| BrMiR 169l | contig139483 | AT3G14385.1 | AGCCAAGGAUGACUUGCCGG | 20 | 3′ | 149 | 44.97 | 35.2 | 23.62 | 0.525 |
| BrMiR 169m | contig063427 | AT4G21595.1 | UGAGCCAAGGAUGACUUGCC | 20 | 5′ | 108 | 46.3 | 31 | 28.7 | 0.619 |
| BrMiR 169n | contig172893 | AT4G21595.1 | UGAGCCAAAGAUGACUUGCCG | 21 | 5′ | 122 | 40.98 | 43.1 | 35.33 | 0.862 |
| BrMiR 169o | contig131930 | AT1G53687.1 | UGAGCCAAAGAUGACUUGCCG | 21 | 5′ | 110 | 38.18 | 40.4 | 36.73 | 0.961 |
| BrMiR 169p | contig023436 | AT1G19371.1 | UAGCCAAGGAUGACUUGCCUG | 21 | 5′ | 131 | 47.33 | 46.4 | 35.42 | 0.748 |
| BrMiR 169q | contig155478 | AT1G19360.1 | UAGCCAAGGAUGACUUGCCUG | 21 | 5′ | 155 | 41.29 | 44.5 | 28.71 | 0.695 |
| BrMiR 169r | contig000299 | AT3G26813.1 | UAGCCAAGGAUGACUUGCCUG | 21 | 3′ | 193 | 38.86 | 51.4 | 26.63 | 0.685 |
| BrMiR 169s | contig000299 | AT3G26813.1 | UAGCCAAGGAUGACUUGCCU | 20 | 3′ | 162 | 34.57 | 38 | 23.46 | 0.678 |
| BrMiR 170a | contig152783 | AT5G66045.1 | UGAUUGAGCCGCGUCAAUAUC | 21 | 3′ | 108 | 47.22 | 26.3 | 24.35 | 0.515 |
| BrMiR 170b | contig043748 | UGAUUGAGCCGUGCCAAUAUC | 21 | 5′ | 111 | 36.94 | 24.2 | 21.8 | 0.59 | |
| BrMiR 171a | DX044654 | AT1G11735.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 3′ | 97 | 44.33 | 39 | 40.21 | 0.907 |
| BrMiR 171b | ED531830 | AT1G62035.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 3′ | 89 | 38.2 | 32.8 | 36.85 | 0.964 |
| BrMiR 171c | ED514994 | AT1G11720.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 3′ | 91 | 39.56 | 37.2 | 40.88 | 1.033 |
| BrMiR 171d | ED515731 | AT1G11735.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 3′ | 93 | 37.63 | 32.8 | 35.27 | 0.937 |
| BrMiR 171e | DU980843 | AT1G62035.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 3′ | 102 | 40.2 | 27.8 | 27.25 | 0.678 |
| BrMiR 171f | DU827625 | AT3G51375.1 | UGAUUGAGCCGCGCCAAUAUC | 21 | 3′ | 126 | 44.44 | 38.4 | 30.48 | 0.685 |
| BrMiR 171g | DY013547.1 | AT1G11735.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 3′ | 89 | 38.2 | 32.8 | 36.85 | 0.964 |
| BrMiR 171h | contig183747 | AT3G51375.1 | UGAUUGAGCCGCGCCAAUAUC | 21 | 3′ | 127 | 43.31 | 32.9 | 25.91 | 0.598 |
| BrMiR 171i | contig179438 | AT3G51375.1 | UGAUUGAGCCGCGCCAAUAUC | 21 | 5′ | 111 | 44.14 | 34.8 | 31.35 | 0.71 |
| BrMiR 171j | contig134903 | AT1G11735.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 5′ | 116 | 42.24 | 31.2 | 26.9 | 0.636 |
| BrMiR 171k | contig128488 | AT1G11735.1 | UUGAGCCGUGCCAAUAUCACG | 21 | 3′ | 86 | 39.53 | 25.9 | 30.12 | 0.761 |
| BrMiR 172a | AJ860723 | AT2G39730.3 | GAAUCUUGAUGAUGUUACA | 19 | 5′ | 111 | 48.65 | 29.7 | 26.76 | 0.55 |
| BrMiR 172b | DX011558 | AT5G59505.1 | GAAUCUUGAUGAUGCUGCAU | 20 | 3′ | 90 | 42.22 | 36 | 40 | 0.947 |
| BrMiR 172c | DX034851 | AT2G28056.1 | GAAUCUUGAUGAUGCUGCAU | 20 | 3′ | 99 | 41.41 | 35.6 | 35.96 | 0.868 |
| BrMiR 172d | DX078594 | AT5G59505.1 | GAAUCUUGAUGAUGCUGCAU | 20 | 3′ | 89 | 42.7 | 39.1 | 43.93 | 1.029 |
| miRNA family | Br-GSS/EST | At-gene | Mature miRNA | ML | MAS | Len(Pre) | GC% | MFE | AMFE | MFEI |
| BrMiR 172e | ED520082 | AT5G59505.1 | GAAUCUUGAUGAUGCUGCAU | 20 | 3′ | 86 | 41.86 | 36 | 41.86 | 1 |
| BrMiR 172f | ED525000 | AT5G59505.1 | GAAUCUUGAUGAUGCUGCAU | 20 | 116 | 44.83 | 46 | 39.66 | 0.884 | |
| BrMiR 172g | ED516998 | AT2G28056.1 | GAAUCUUGAUGAUGCUGCAU | 20 | 3′ | 109 | 45.87 | 30 | 27.52 | 0.6 |
| BrMiR 172h | ED526567 | AT5G59505.1 | GAAUCUUGAUGAUGCUGCAU | 20 | 3′ | 91 | 41.76 | 40.6 | 44.62 | 1.068 |
| BrMiR 172i | EX060396.1 | AT2G39730.3 | GAAUCUUGAUGAUGUUACA | 19 | 5′ | 111 | 48.65 | 29.7 | 26.76 | 0.55 |
| BrMiR 172j | EX059018.1 | AT2G39730.3 | GAAUCUUGAUGAUGUUACA | 19 | 5′ | 111 | 48.65 | 29.7 | 26.76 | 0.55 |
| BrMiR 172k | EX057674.1 | AT2G39730.3 | GAAUCUUGAUGAUGUUACA | 19 | 5′ | 111 | 48.65 | 29.7 | 26.76 | 0.55 |
| BrMiR 172l | EX019450.1 | AT2G39730.3 | GAAUCUUGAUGAUGUUACA | 19 | 5′ | 111 | 48.65 | 29.7 | 26.76 | 0.55 |
| BrMiR 172m | contig138929 | AT2G28056.1 | AGAAUCUUGAUGAUGCUGCAU | 21 | 5′ | 122 | 50 | 40.2 | 32.95 | 0.659 |
| BrMiR 172n | contig084904 | AT3G55512.1 | AGAAUCUUGAUGAUGCUGCA | 20 | 3′ | 126 | 35.71 | 41.7 | 33.1 | 0.926 |
| BrMiR 172o | contig164892 | AT3G11435.1 | AGAAUCUUGAUGAUGCUGCA | 20 | 5′ | 133 | 39.1 | 41.7 | 31.35 | 0.801 |
| BrMiR 319a | DX903478 | AT4G23713.1 | UUGGACUGAAGGGAACUCCCU | 21 | 5′ | 176 | 40.91 | 66.9 | 38.01 | 0.929 |
| BrMiR 319b | AJ856769 | AT2G40805.1 | UUGGACUGAAGGGAGCUC | 18 | 3′ | 202 | 40.1 | 68.7 | 34.01 | 0.848 |
| BrMiR 319c | contig173636 | AT5G41663.1 | UUGGACUGAAGGGAGCUCCUU | 21 | 3′ | 200 | 38.5 | 68.8 | 34.4 | 0.893 |
| BrMiR 390a | DU830650 | AT5G58465.1 | AAGCUCAGGAGGGAUAGCGCC | 21 | 5′ | 96 | 43.75 | 30.3 | 31.56 | 0.721 |
| BrMiR 390b | DX059137 | AT2G38325.1 | AAGCUCAGGAGGGAUAGCGCC | 21 | 5′ | 97 | 43.3 | 31.3 | 32.27 | 0.745 |
| BrMiR 390c | DX890076 | AT5G58465.1 | AAGCUCAGGAGGGAUAGCGCC | 21 | 5′ | 139 | 41.73 | 58.3 | 41.94 | 1.005 |
| BrMiR 390d | contig070593 | AT2G38325.1 | AAGCUCAGGAGGGAUAGCGCC | 21 | 5′ | 96 | 44.79 | 37 | 38.54 | 0.86 |
| BrMiR 390e | contig054389 | AT5G58465.1 | AAGCUCAGGAGGGAUAGCGCC | 21 | 3′ | 94 | 42.55 | 32.6 | 34.68 | 0.815 |
| BrMiR 390f | contig039203 | AAGCUCAGGAGGGAUAGCGCC | 21 | 3′ | 120 | 40 | 40.2 | 33.5 | 0.837 | |
| BrMiR 390g | contig110394 | AAGCUCAGGAGGGAUAGCGCC | 21 | 3′ | 119 | 44.54 | 34.1 | 28.66 | 0.643 | |
| BrMiR 391a | contig125684 | AT5G60408.1 | UUCGCAGGAGAGAUAGCGCCA | 21 | 5′ | 112 | 41.96 | 36.6 | 32.68 | 0.778 |
| BrMiR 391b | contig083747 | AT5G60408.1 | UUCGCAGGAGAGAUAGCGCCA | 21 | 5′ | 111 | 43.24 | 37.2 | 33.51 | 0.775 |
| BrMiR 393a | contig164532 | AT3G55734.1 | UCCAAAGGGAUCGCAUUGAUCC | 22 | 3′ | 156 | 35.9 | 37.7 | 24.17 | 0.673 |
| BrMiR 393b | contig009383 | AT3G55734.1 | UCCAAAGGGAUCGCAUUGAUCC | 22 | 5′ | 171 | 34.5 | 47.8 | 27.95 | 0.81 |
| BrMiR 393c | contig053732 | AT2G39885.1 | UCCAAAGGGAUCGCAUUGAUCC | 22 | 3′ | 129 | 33.33 | 33.2 | 25.74 | 0.772 |
| BrMiR 394a | DU120003 | AT1G20375.1 | UUGGCAUUCUGUCCACCUCC | 20 | 5′ | 100 | 39 | 26.7 | 26.7 | 0.684 |
| BrMiR 394b | contig137399 | AT1G20375.1 | UUGGCAUUCUGUCCACCUCC | 20 | 5′ | 107 | 40.19 | 31.4 | 29.35 | 0.73 |
| BrMiR 394c | contig133329 | AT1G20375.1 | UUGGCAUUCUGUCCACCUCC | 20 | 5′ | 131 | 38.17 | 40.1 | 30.61 | 0.801 |
| BrMiR 395a | DX020424 | AT1G26973.1 | CUGAAGUGUUUGGGGGAACUC | 21 | 3′ | 88 | 44.32 | 32.8 | 37.27 | 0.841 |
| BrMiR 395b | DU832403 | AT1G26975.1 | CUGAAGUGUUUGGGGGGACUC | 21 | 3′ | 115 | 40 | 39.8 | 34.61 | 0.865 |
| BrMiR 395c | contig143422 | AT1G69795.1 | CUGAAGUGUUUGGGGGAACUC | 21 | 3′ | 111 | 38.74 | 41.4 | 37.3 | 0.962 |
| BrMiR 395d | contig124372 | AT1G69797.1 | CUGAAGUGUUUGGGGGGACUC | 21 | 3′ | 130 | 43.08 | 50.1 | 38.54 | 0.894 |
| BrMiR 395e | contig153647 | CUGAAGUGUUUGGAGGAACUC | 21 | 5′ | 123 | 34.96 | 32.8 | 26.67 | 0.762 | |
| BrMiR 396a | DX897816 | AT3G52910.1 | UCCACAGGCUUUCUUGAAC | 19 | 5′ | 85 | 51.76 | 21 | 24.71 | 0.477 |
| BrMiR 396b | contig029367 | AT2G10606.1 | UUCCACAGCUUUCUUGAACUG | 21 | 5′ | 161 | 33.54 | 44 | 27.33 | 0.814 |
| BrMiR 396c | contig159446 | AT5G35407.1 | UUCCACAGCUUUCUUGAACU | 20 | 3′ | 164 | 31.1 | 50.9 | 31.04 | 0.997 |
| BrMiR 398a | AJ861920 | AT5G14565.1 | UGUGUUCUCAGGUCACCCCU | 20 | 3′ | 121 | 47.11 | 32.7 | 27.02 | 0.573 |
| BrMiR 398b | EX049578.1 | AT5G14545.1 | UGUGUUCUCAGGUCACCCCU | 20 | 3′ | 126 | 46.83 | 41.5 | 32.94 | 0.703 |
| BrMiR 398c | contig015561 | AT2G03445.1 | UGUGUUCUCAGGUCACCCCUU | 21 | 3′ | 96 | 39.58 | 31.8 | 33.13 | 0.836 |
| BrMiR 399a | contig182431 | AT1G29265.1 | UGCCAAAGGAGAUUUGCCCUG | 21 | 3′ | 179 | 35.2 | 53.2 | 29.72 | 0.844 |
| BrMiR 399b | contig182431 | AT1G29265.1 | UGCCAAAGGAGAUUUGCCCGG | 21 | 3′ | 160 | 38.75 | 67.34 | 42.09 | 1.086 |
| BrMiR 399c | contig152434 | AT5G62162.1 | UGCCAAAGGAGAGUUGCCCUG | 21 | 5′ | 128 | 46.88 | 40.5 | 31.64 | 0.674 |
| BrMiR 399d | contig153243 | AT2G34202.1 | UGCCAAAGGAGAUUUGCCCCG | 21 | 5′ | 135 | 33.33 | 48.2 | 35.7 | 1.071 |
| BrMiR 399e | contig135242 | AT2G34208.1 | UGCCAAAGGAGAUUUGCCCGG | 21 | 3′ | 111 | 39.64 | 42.1 | 37.93 | 0.956 |
| BrMiR 399f | contig122533 | AT1G29265.1 | UGCCAAAGGAGAUUUGCCCGG | 21 | 3′ | 105 | 41.9 | 46.6 | 44.38 | 1.059 |
| BrMiR 399g | contig176382 | AT2G34208.1 | UGCCAAAGGAGAUUUGUCCGG | 21 | 5′ | 113 | 42.48 | 32 | 28.32 | 0.666 |
| BrMiR 400a | ED535204 | AT1G32582.1 | UAUGAGAGUAUUAUAAGUCAC | 21 | 5′ | 134 | 32.84 | 46.6 | 34.78 | 1.059 |
| BrMiR 400b | DX912125 | AT1G64580.1 | UAUGAGAGUAUUAUACGUCAC | 21 | 3′ | 221 | 46.61 | 60.4 | 27.33 | 0.586 |
| BrMiR 400c | DU827877 | AT1G62680.1 | AUGAGAGUAUUAUAACUCAC | 20 | 3′ | 124 | 45.16 | 29.1 | 23.47 | 0.519 |
| miRNA family | Br-GSS/EST | At-gene | Mature miRNA | ML | MAS | Len(Pre) | GC% | MFE | AMFE | MFEI |
| BrMiR 403 | contig102674 | AT2G47275.1 | UUAGAUUCACGCACAAACUCG | 21 | 5′ | 91 | 35.16 | 27 | 29.67 | 0.843 |
| BrMiR 408a | DX037057 | AT2G47015.1 | AUGCACUGCCUCUUCCCUGGC | 21 | 3′ | 149 | 42.95 | 38.2 | 25.64 | 0.596 |
| BrMiR 408b | EX141550.1 | AT2G02850.1 | AUGCACUGCCUCUUCCCU | 18 | 3′ | 151 | 54.3 | 43.9 | 29.07 | 0.535 |
| BrMiR 472a | ED514754 | AT1G14850.1 | UCCCUACUCCACUCAUCCC | 19 | 5′ | 241 | 43.15 | 63.9 | 26.51 | 0.614 |
| BrMiR 472b | CX271339.1 | AT1G14850.1 | UCCCUACUCCACUCAUCCC | 19 | 5′ | 241 | 43.15 | 63.9 | 26.51 | 0.614 |
| BrMiR 473 | DU120348 | UCUCCCUCAAGGUUUCCA | 18 | 5′ | 179 | 34.64 | 29.6 | 16.54 | 0.477 | |
| BrMiR 776 | DX019504 | UCUAAGUCUUCUUUUGAU | 18 | 5′ | 251 | 22.31 | 29.2 | 11.63 | 0.521 | |
| BrMiR 824 | contig141281 | AT4G24415.2 | UAGACCAUUUGUGAGAAGGGA | 21 | 5′ | 66 | 40.91 | 11.1 | 16.82 | 0.411 |
| BrMiR 837 | DX902366 | AT4G16143.2 | AUCAGUUUCUUGUUCUUUUC | 20 | 5′ | 202 | 29.21 | 25.9 | 12.82 | 0.439 |
| BrMiR 838 | DU127124 | AT1G65590.1 | UUUUCUUAUACUUCUUGCA | 19 | 3′ | 280 | 43.57 | 59.4 | 21.21 | 0.486 |
| BrMiR 842 | DX012634 | AT2G14370.1 | AUGGUCAGAUCGGUCAUC | 18 | 5′ | 196 | 53.57 | 52.6 | 26.84 | 0.5 |
| BrMiR 845a | CT021891 | AT5G04290.1 | GGCUCUGAUACCAAUUGA | 18 | 5′ | 139 | 53.24 | 40.2 | 28.92 | 0.543 |
| BrMiR 845b | DX897026 | AT4G31200.1 | UGGCUCUGAUACCAACUGAUG | 21 | 3′ | 141 | 32.62 | 50.3 | 35.67 | 1.093 |
| BrMiR 845c | contig151543 | UGGCUCUGAUACCAACUGAUG | 21 | 3′ | 103 | 34.95 | 40.6 | 39.42 | 1.127 | |
| BrMiR 846 | ED536287 | AT1G52110.1 | UUGAACUGAAGUGCUUGAAU | 20 | 5′ | 195 | 32.82 | 42.9 | 22 | 0.67 |
| BrMiR 850 | DU832285 | AT1G32990.1 | GAUCCGGACUAAAACAAAG | 19 | 5′ | 83 | 43.37 | 17.3 | 20.84 | 0.48 |
| BrMiR 854a | DX017286 | AT2G44710.1 | UGAGGAGAGGGAGGAGGAG | 19 | 3′ | 305 | 40 | 62.9 | 20.62 | 0.515 |
| BrMiR 854b | DX899834 | AT5G58490.1 | GAGGAGAGGGAGGAGGAG | 18 | 3′ | 98 | 63.27 | 32 | 32.65 | 0.516 |
| BrMiR 857 | DX077868 | AT1G30400.2 | UUUUGCAUGUUGAAGGUGU | 19 | 5′ | 109 | 37.61 | 22.6 | 20.73 | 0.551 |
| BrMiR 1132a | DX047066 | CAUUAAGGAACGGAAGGAG | 19 | 3′ | 271 | 19.93 | 42.3 | 15.61 | 0.783 | |
| BrMiR 1132b | DX910272 | AT3G26570.2 | AUUAUGAAACGGAAGGAG | 18 | 3′ | 255 | 22.35 | 46.3 | 18.16 | 0.812 |
| BrMiR 1139 | AJ855061 | AGAGUAAAAUACACUAGUA | 19 | 3′ | 85 | 29.4 | 10.1 | 11.88 | 0.404 | |
| BrMiR 1140 | contig123251 | ACAGCCUAAACCAAUCGGAGC | 21 | 3′ | 151 | 37.09 | 61.4 | 40.66 | 1.096 | |
| BrMiR 1436 | DX082315 | AT5G49680.2 | UUAUGGGACGGAAGGAGU | 18 | 3′ | 214 | 22.43 | 30.7 | 14.35 | 0.639 |
| BrMiR 1439 | contig149192 | UUUUGGAACGGAGAGAGUAUU | 21 | 3′ | 271 | 23.62 | 43.9 | 16.2 | 0.685 | |
| BrMiR 1514 | DX899429 | AT2G45560.1 | UUCAUUUUUAUAAAUAGACAUU | 22 | 3′ | 183 | 18.03 | 19.8 | 10.82 | 0.6 |
| BrMiR 1520a | DX016618 | AGAACUUGACACGUGACAA | 19 | 5′ | 164 | 31.1 | 25.7 | 15.67 | 0.504 | |
| BrMiR 1520b | ED528582 | AGAACUUGACACGUGACAA | 19 | 5′ | 153 | 28.1 | 23.6 | 15.42 | 0.548 | |
| BrMiR 1521a | CW982899 | AT1G79580.3 | CUGUUAAUGGAAAAAGUUG | 19 | 5′ | 136 | 36.03 | 17.1 | 12.57 | 0.349 |
| BrMiR 1521b | CT022839 | AT5G39862.1 | CUGUUGAUGGAAAAUGUU | 18 | 5′ | 268 | 41.33 | 69.2 | 25.82 | 0.624 |
| BrMiR 1522 | AJ858836 | ATMG00110.1 | UUUAUUUCUUAAAAUGAAA | 19 | 3′ | 166 | 32.53 | 32 | 19.28 | 0.592 |
| BrMiR 1527 | DX023754 | AACUCAACCUUACAAAAC | 18 | 3′ | 167 | 34.73 | 34.5 | 20.66 | 0.605 | |
| BrMiR 1863 | DX895479 | AT2G36990.1 | AGCUCUGAUACCAUGUUAGAUU | 22 | 5′ | 236 | 30.08 | 40.5 | 17.16 | 0.57 |
| BrMiR 1886 | KBFL-140B03 | AT5G53120.1 | UGAGAGAAGUGAGAAGAAA | 19 | 5′ | 124 | 44.88 | 24.6 | 19.84 | 0.439 |
| BrMiR 2091 | DX017320 | AT5G65683.1 | AACCGAGCCGAAGAGGAG | 18 | 5′ | 96 | 52.08 | 33.1 | 34.48 | 0.662 |
| BrMiR 2111a | contig014252 | UAAUCUGCAUCCUGAGGUUUA | 21 | 3′ | 113 | 42.11 | 36.1 | 31.95 | 0.644 | |
| BrMiR 2111b | contig148483 | UAAUCUGCAUCCUGGGGUUUA | 21 | 3′ | 135 | 40.74 | 27.4 | 20.3 | 0.498 | |
| BrMiR 2673 | contig154626 | CCUCUUCCUCUUCCUCUUCC | 20 | 5′ | 119 | 47.06 | 32 | 26.89 | 0.571 | |
Br, Brassica rapa; At, Arabidopsis thaliana; ML, mature miRNA length; MAS, mature miRNA arm side in hairpin secondary structure; MFE, minimum folding free energy; AMFE, adjusted minimum folding free energy; MFEI, minimum folding free energy index.
Fig. 2. Number of Brassica rapa miRNAs against different lengths of mature miRNAs.
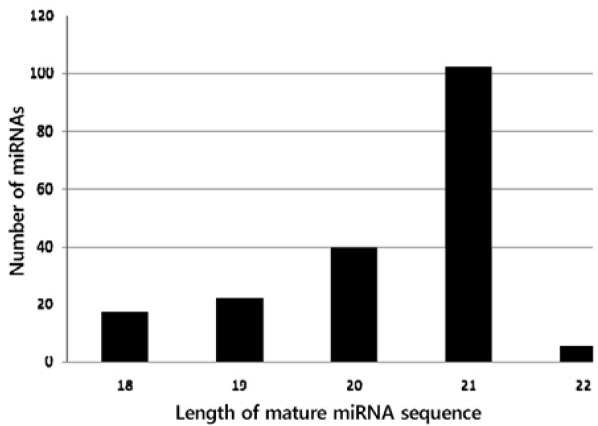
Fig. 3. Stem-loop hairpin structures of representative miRNA families. Red indicates matured miRNA sequences. The actual size of precursor sequences may be slightly shorter or longer than presented.

Precursor sequence characteristics
The 186 B. rapa miRNAs were derived from varying sizes of precursor sequences. The precursor miRNAs (pre-miRNA) ranged from 66 to 305 nucleotides with an average of 135.62 (47.79) nucleotides (Supplementary Table 1). The majority (77.69%) of the precursor miRNAs were 81-202 nt in length (Supplementary Table 1 and Fig. 4). The findings of pre-miRNA sequences ranging from 66 to 305 nt in the present study shows that B. rapa also contains miRNAs with varying premiRNA lengths as is the case in other plant species (Song et al., 2009; Zhang et al., 2007a; 2008). The distributions of A, U, G, and C nucleotides are different and uneven. A [28.89% (5.51%)] and U [30.23% (5.80%)] nucleotides were predominant compared to G [20.30% (4.39%)] and C [20.58% (4.43%)]. The percentage of GC content ranged from 18.03% to 63.27% with an average of 40.88% (6.86%) (Supplementary Table 1). Lower minimal folding free energy (MFE) is important for high thermodynamic stability of the sequences to form stable secondary loop structures (Zhang et al., 2008). In the present study, the MFE of 186 identified B. rapa miRNAs ranged from -10.1 to -75.4 kcal/mol with an average of -39.22 (12.52) kcal/mol. However, due to the strong positive influence of DNA/RNA length in the MFE calculations, a new method, adjusted minimal folding free energy (AMFE), was adopted to normalize the potential effects of nucleotide sequence length (Zhang et al., 2008). AMFE ranged from -10.82 to -52.28 kcal/mol with an average of -29.96 (7.84) kcal/mol (Table 2). The observation of high MFE and AMFE of pre-miRNAs in the present study is consistent with previous reports (Bonnet et al., 2004b; Zhang et al., 2006a; 2008). Recently, the minimum folding free energy index (MFEI), which is calculated on the basis of MFE, sequence length, and nucleotide GC content, was widely adopted for distinguishing miRNAs from other RNAs (Zhang et al., 2006a; 2006b). The MFEI in our study for precursor miRNA sequences ranged from 0.349 to 1.210 with an average of 0.737 (0.178). Although it is suggested that MFEI values ≥ 0.85 are strongly indicative of actual miRNAs, lower values cannot be ruled out as potential miRNAs (Zhang, 2007a; Zhang et al., 2006a; 2006b) (Table 2). In the present study, only 48 (25.81%) of 186 identified B. rapa miRNAs had MFEI values ≥ 0.85, in contrast to that of soybean miRNAs having values ≥ 0.85 (Zhang et al., 2008).
Identification of novel B. rapa and Brassica lineagespecific miRNAs
Of the 186 miRNAs belonging to 55 families identified in the present study 15 miRNAs belonging to 10 families were reported previously (He et al., 2008); thus, the remaining 40 miRNA families in our study are newly identified. Furthermore, we identified more members belonging to those previously reported B. rapa miRNA families. He et al. (2008) identified 1 member each for miRNA families 157, 159, 164, 167, 390, and 1885 and 2 members each for miRNA families 160, 172 and 398; only miRNA family 171 had 3 identified members (He et al., 2008). In the present study, we identified 15 members each for BrMiR 159 and 172; 6 for miRNA 164; 8 for miRNA 160; 4 for miRNA 167; 7 for miRNA 390; and 3 for miRNA 398 (Table 2). Further analysis of the miRNA families with previously identified miRNAs present in the miRBase revealed that of 186 miRNAs belonging to 55 families, 48 miRNAs belonging to 30 miRNA families were not identical and had 1-2 mismatches with the previously reported miRNAs from one or more plant species; this suggests that these miRNA nucleotide mismatches are specific to B. rapa. One such example is the BrMiR 162 family, which has a guanine in B. rapa instead of a conserved cytosine in other plant species at the 12th nucleotide position of the mature miRNA. The single nucleotide difference is also observed between B. rapa and other Brassica species, such as B. napus and B. oleracea, apart from Arabidopsis, although these 4 species belongs to same family of Brassicaceae (Fig. 5). The conserved and nonconserved regions of miRNA family 162 of different plant species such as B. napus, B. oleracea, Solanum lycopersicum, Vitis vinifera, O. sativa, A. thaliana, and Carica papaya are shown in Fig. 5. Further analyses could identify 5 Arabidopsis-Brassica lineage-specific miRNA families (BrMiR 158, 391, 400, 824, and 1140) in B. rapa. Sunkar and Jagdeeswaran (2008) identified miRNA family 158 in B. rapa and B. oleracea, and family 391 in B. oleracea. In the present study, through computational analysis, we identified BrMiR 391 in B. rapa. Of the remaining 3 newly identified Brassica lineagespecific miRNAs identified in B. rapa, BrMiR 400 is only reported in A. thaliana; BrMiR 824 in B. oleracea, B. napus, and A. thaliana; and BrMiR 1140 only in B. napus.
Fig. 5. Brassica rapa miRNA162 family showing a single nucleotide difference at the 12th nucleotide position of mature miRNA in comparison to the miRNA 162 family of other plants; BrMiR, Brassica rapa miRNA; Bnp, Brassica napus; Bol, Brassica oleracea; Sly, Solanum lycopersicum; Vvi, Vitis vinifera; Osa, Oryza sativa; Ath, Arabidopsis thaliana; Cpa, Carica papaya.

Detection of sense, antisense, and clusters of B. rapa miRNAs
Although many animals studies found that miRNAs are transcribed and processed from the sense and antisense strands of the same genomic locus, this was reported only recently in plants (Xie et al., 2010; Zhang et al., 2008). In our study, we found only a DNA sequence producing identical matured BrMiR 399 from both sense and antisense strands. Although sense and antisense miRNAs are transcribed from the same genomic locus, they are not identical with respect to the, 20th nucleotide position; U and G nucleotides were present in sense and antisense BrMiR 399, respectively. Previous studies also report the occurrence of miRNA clusters in plants, especially in cotton and soybean (Xie et al., 2010; Zhang et al., 2008). miRNAs in these clusters are produced from the same precursors or from different precursor sequences separated by a few nucleotides on the same genomic locus (Xie et al., 2010; Zhang et al., 2008). In this study, we identified only 1 miRNA cluster in B. rapa. This cluster includes 2 miRNAs belonging to the BrMiR family 169 (BrMiR 169r and 169s), which are 353 nucleotides apart. These 2 BrMiRs differ with respect to a single nucleotide difference of their mature miRNA sequences (Table 2 and Fig. 6).
Fig. 6. BrMiR 169r-BrMiR 169s cluster in B. rapa contig000299. (A) Brassica rapa genomic contig000299 showing 2 miRNA clusters. Shaded sequences indicate pre-miRNAs and red sequences indicate mature miRNA. (B) Predicted miRNA secondary structure of BrMir 169r. (C) Predicted miRNA secondary structure of BrMir 169s.

Expression analyses of identified B. rapa miRNAs
The expression of B. rapa miRNAs was identified in 2 ways. First, we examined the original tissues of the EST sequences obtained from public databases from which miRNAs were derived. A total of, 20 miRNAs belonging to 9 BrMiR families were found to be expressed in different tissues in B. rapa. The tissues where miRNAs were expressed include floral buds (BrMiR 159, 160, 167, and 398), roots (BrMiR 162), callus (BrMiR 160), seedlings (BrMiR 162), primary leaves (BrMiR 408), etiolated mature leaves (BrMiR 166), and salt-treated whole plant (BrMiR 172) (Table 3). This observation suggests that some of these identified B. rapa miRNAs exhibit tissue-specific expression whereas some are expressed throughout the plant.
Table 3.
Identified Brassica rapa expressed sequence tags (ESTs) producing miRNA and their expressing tissues
| Brassica rapa miRNA | Brassica rapa EST | Expressing tissue |
|---|---|---|
| BrMiR 156j | CV432746.1 | Root |
| BrMiR 159b | EX050542.1 | Floral buds |
| BrMiR 159c | EX048967.1 | Floral buds |
| BrMiR 159d | EX047628.1 | Floral buds |
| BrMiR 159g | EX039355.1 | Floral buds |
| BrMiR 160b | EX044968.1 | Floral buds |
| BrMiR 160h | EX025484.1 | Callus |
| BrMiR 162b | EX133401.1 | Root |
| BrMiR 162c | EX071919.1 | Seedling |
| BrMiR 162d | EX071254.1 | Seedling |
| BrMiR 162e | EX069816.1 | Seedling |
| BrMiR 166b | EX063968.1 | Etiolated mature leaf |
| BrMiR 166c | EX063242.1 | Etiolated mature leaf |
| BrMiR 167c | EX041799.1 | Floral buds |
| BrMiR 172i | EX060396.1 | Salt-treated whole plant |
| BrMiR 172j | EX059018.1 | Salt-treated whole plant |
| BrMiR 172k | EX057674.1 | Salt-treated whole plant |
| BrMiR 172l | EX019450.1 | Light-chilled whole plant |
| BrMiR 398b | EX049578.1 | Floral buds |
| BrMiR 408b | EX141550.1 | Primary leaf |
Second, the expression of identified miRNAs in various tissues was further examined using microarray data from the tissues of various stages of B. rapa development from another experiment. The 50K B. rapa DNA chip contains 12 identified miRNA-producing genes belonging to 8 miRNA families; these were confirmed by computational identification and secondary structure prediction by using the sequences found in microarray DNA chip (data not shown). Microarray analysis was performed to analyze gene expression at different stages of floral buds, stamens, carpels, siliques, and different parts of leaves (e.g., inner and outer parts). The expression of 12 miRNA genes was observed in microarray data taken at different stages of bud development from male sterile and fertile plants, photosynthetic (source) and mid-leaves (sink), and young shoots including the white and yellow portions of leaves. The microarray data showed very low to high expression of 12 miRNA genes in different tissues (data not shown).
To confirm the results of the microarray data analysis, we performed a subsequent Northern blot analysis of 5 miRNA families from 3 tissues, namely the leaves, stem, and roots (Fig. 7). Among these 5 miRNA families, BrMiR 159 was highly expressed in all analyzed tissues (Fig. 7). Furthermore, we observed different sizes of miRNAs in stem tissues from the northern blot analysis using these miRNAs; this convincingly supports the computational analysis identification that the matured miRNAs are 18-22 nt long (Table 2 and Fig. 7). BrMiR 160 exhibited a low level of expression in the 3 tissues; BrMiR 167 was highly expressed in leaves and roots, but very little expression was observed in the stem; BrMiR 398 was highly expressed in leaves, whereas expression was low in the stem and no expression was detected in the roots; BrMiR 408 exhibited medium expression in leaves but low expression in the stem and roots (Fig. 7).
Fig. 7. Northern blot analysis of leaf, stem, and root tissues of Brassica rapa seedlings. Low-molecular-weight RNA (10 μg) was used for Northern analysis. Antisense oligonucleotide probes were designed, radiolabeled, and used as probes for the detection of the B. rapa miRNAs. 5s rRNA was used as a loading control.

Further, we performed quantitative real time polymerase chain reaction (qRT-PCR) for 12 miRNAs (Table 1) in six different tissues of B. rapa cultivar Chiifu. The qRT-PCR analyses demonstrated that all miRNAs were expressed in all the six tissues tested. However, while analyzing the results from qRTPCR, we observed that the expression level of miRNAs differ from each other in the six B. rapa tissues tested (Fig. 8). qRTPCR result showed that BrMir158b expression levels was not significantly different in all tested tissues except for in midrib tissue where there was decreased expression level of this miRNA was detected. BrMir162b showed almost equal expression levels in all tested tissues except the increased expression in midrib tissues. BrMir164c, BrMir396b, BrMir396b and BrMir 1132b showed very high expression in root tissues, BrMir171f in shoot apex, BrMir172g and BrMir1521b in stem, respectively. BrMir170b, BrMir837 and BrMir842 did not show any significant expression differences in the six tissues of B. rapa.
Fig. 8. The characterization of Brassica rapa miRNA expression patterns by real-time PCR analysis. Relative expression levels of BrMiR 158b, BrMiR 162b, BrMiR 164c, BrMiR 170b, BrMiR 171f, BrMiR 172g, BrMiR 396b, BrMiR 400b, BrMiR 837, BrMiR 842, BrMiR 1132b, and BrMiR 1521b were analyzed in shoot apex, stem, old leaf, young leaf, midrib, and in root tissues of B. rapa plants. The expression levels of all miRNAs by real-time PCR were normalized to the expression of BrACT.

Identification of target genes
Due to the perfect or near-perfect complementarities of mature miRNAs and target mRNAs, it is possible to find target genes that are regulated by specific miRNA. Several studies utilized this fact and identified many target genes in different plant species while allowing 1-4 nucleotide mismatches between target mRNA and miRNAs (Song et al., 2009; Zhang et al., 2008). In this study, to identify miRNA targets, we searched for B. rapa mRNA sequences available in the laboratory and public databases showing complementarity with < 3 mismatches. Gaps, G:U pairs, and other noncanonical pairs were not allowed and considered mismatches (Xie et al., 2007). By using this criterion, the target genes of 33 out of 55 B. rapa miRNA families were identified (Table 4). We could not identify the target genes for the following 22 miRNA families: BrMiR 157, 161, 165, 166, 169, 170, 319, 391, 393, 396, 399, 403, 408, 824, 842, 850, 1140, 1436, 1439, 1514, 2111, and 2673. A total of 66 target genes that are involved in different biological functions were identified for 33 miRNA families. The number of potential target genes identified per miRNA family ranged from 1 to 10. All target genes share high homology with Arabidopsis orthologues. The majority of these potential target genes are transcription factor/regulator genes (Table 4). The BrMiR 156 family largely targets squamosa promoter-binding protein like (SPL) transcription factor gene families, which are important in leaf development and phase change. Targeting of the SBP genes by miRNA 156 is reported in other plant species (Song et al., 2009; Zhang et al., 2008). The other miRNA families that target transcription factors are as follows: BrMiR 160, 164, 167, 171, 172, 394, 473, 845, 1132, 1139, 1521, 1527, 1886, and, 2091 (Table 4). The conserved targets between Arabidopsis and B. rapa confirm the conserved function of miRNAs in both species. Apetala 2 is a transcription factor that establishes floral meristem identity and ovule and seed coat development (Chen, 2004); BrMiR 172 targets AP2.
Table 4.
Potential targets of identified Brassica rapa miRNAs and similar Arabidopsis thaliana genes
| miRNA family | Brassica rapa genes | Target protein | Target function | Arabidopsis thaliana gene |
|---|---|---|---|---|
| BrMiR 156 | AC155337.1, DX032440 | Squamosa promoter-binding protein-like 9 | Transcriptional factor (TF) | AT2G42200.1 |
| AC189445.2 | Squamosa promoter-binding protein | TF | AT3G57920.1 | |
| AC189413.1 | Squamosa promoter-binding protein-like 2 | TF | AT5G43270.1 | |
| AC189458.2, AC189298.1 | Squamosa promoter-binding protein-like 6 | TF | AT1G69170.1 | |
| AC189298.1 | Squamosa promoter-binding protein-like 3 | TF | AT2G33810.1 | |
| AC189649.2 | Squamosa promoter-binding protein-like 10 | TF | AT1G27370.4 | |
| AC232495.1 | TOC1 (TIMING OF CAB1 1) | Transcription regulator | AT5G61380.1 | |
| BrMiR 158 | AC189530.2 | Alpha-Dioxygenase | Oxidoreductase | AT1G73680.1 |
| BrMiR 159 | CW988048 | Unknown | ||
| BrMiR 160 | AC189370.2 | ARF17 (Auxin Response Factor 17) | TF | AT1G77850.1 |
| BrMiR 162 | DX038960 | Unknown | ||
| BrMiR 164 | AC232542.1 | NAC100 (NAC domain containing protein 100) | TF | AT5G61430.1 |
| AC189572.1 | VIM1 (Variant In Methylation 1) | TF | AT1G57820.1 | |
| BrMiR 167 | AC189484.2 | ARF8 (Auxin Response Factor 8) | TF | AT5G37020.2 |
| BrMiR 168 | AC189554.2 | Pyruvate kinase | Catalytic activity | AT5G56350.1 |
| BrMiR 171 | AC189445.2, DX039307, DX039875, DX050715, DX079200, DX080294 | Scarecrow transcription factor family protein | TF | AT2G45160.1 |
| BrMiR 172 | AC189306.2 | RCA (RUBISCO ACTIVASE) | ATP binding | AT2G39730.3 |
| AC234737.1 | TOE2 (Target Of Eat 2) | TF | AT5G60120.1 | |
| AC172870.1 | SMZ (SCHLAFMUTZE) | TF | AT3G54990.1 | |
| AC189433.2 | CDT1A | Protein binding | AT2G31270.1 | |
| AC232512.1 | AP2 (APETALA 2) | TF | AT4G36920.1 | |
| AC189432.2 | TOE1 (Target Of Eat 1) | TF | AT2G28550.2 | |
| BrMiR 390 | DX043165 | Unknown | ||
| BrMiR 394 | AC189649.2 | F-box family protein | TF | AT1G27340.1 |
| BrMiR 395 | AC189402.2, DU984698 | APS4 | ATP activity | AT5G43780.1 |
| DX024391, DX075472 | APS1 (ATP sulfurylase 3) | AT3G22890.1 | ||
| BrMiR 398 | AC189227.2 | Unknown protein | AT5G14550.1 | |
| BrMiR 400 | AC189437.2 | Unknown protein | AT1G63300.1 | |
| BrMiR 472 | DX049611, DX038688 | Unknown | ||
| BrMiR 473 | AC189542.2 | Agenet domain-containing protein/bromoadjacent homology (BAH) domaincontaining protein | DNA binding | AT5G55600.1 |
| AC189298.1 | Zinc finger (C3HC4-type RING finger) family protein | Protein binding, zinc ion binding | AT1G69330.1 | |
| BrMiR 776 | AC189308.2 | CUT1 (CUTICULAR 1) | Acyltransferase | AT1G68530.2 |
| BrMiR 837 | L31937.1 | LCR69/PDF2.2 (Low-molecular-weight cysteine-rich 69) | Peptidase inhibitor activity | AT2G02100.1 |
| BrMiR 838 | EU117118.1 | SPT1 | Serine C-palmitoyltransferase | AT3G48780.1 |
| BrMiR 845 | AC232527.1 | KOW domain-containing transcription factor family protein | TF | AT5G04290.1 |
| AC232513.1, AC189578.2, EU180578.1 | Unknown | |||
| BrMiR 846 | AC189248.2 | Jacalin lectin family protein | AT5G38550.1 | |
| AC189524.2 | Jacalin lectin family protein | AT1G57570.1 | ||
| BrMiR 854 | AC189355.2 | Glycosyl hydrolase family 17 protein | Cation binding | AT5G58480.1 |
| AC189591.2, AC189402.2 | Beta-ketoacyl-CoA synthase | Fatty acid elongase activity | AT5G43760.1 | |
| BrMiR 857 | CW981541 | Unknown | ||
| BrMiR 1132 | AC189577.2 | Unknown protein | AT5G48610.1 | |
| miRNA family | Brassica rapa genes | Target protein | Target function | Arabidopsis thaliana gene |
| BrMiR 1132 | AC189568.2 | Nucleoporin interacting component family protein | Protein binding | AT2G41620.1 |
| BrMiR 1139 | AC189599.2 | Zinc finger (CCCH-type) family protein | Zinc-ion binding | AT5G12440.1 |
| BrMiR 1520 | AC189550.1 | UDP-glucose:sterol glucosyltransferase (UGT80A2) | Transferase activity | AT3G07020.2 |
| BrMiR 1521 | AC189564.2 | Transducin family protein/WD-40 repeat family protein | Nucleotide binding | AT5G14050.1 |
| BrMiR 1522 | AC232493.1 | UBX domain-containing protein | Unknown | AT4G10790.1 |
| BrMiR 1527 | AC189385.2 | Cullin 3A | Protein binding | AT1G26830.1 |
| BrMiR 1863 | AC189561.3 | ACO1 (ACC OXIDASE 1) | Unknown | AT2G19590.1 |
| AC189318.1, AC189464.2 | ||||
| BrMiR 1886 | AC189592.2 | UB2 (HISTONE MONO-UBIQUITINATION 2) | Protein binding | AT1G55255.1 |
| BrMiR 2091 | AC189359.2 | Zinc finger (C3HC4-type RING finger) family protein | Zinc ion binding | AT3G19950.1 |
Many studies report that miRNAs also target genes that are involved in signaling pathways. In the present study, BrMiR 167 and 160 were found to target Auxin response factor (ARF) 8 and 17, respectively; these mediate auxin response via the expression of auxin-regulated genes, and control stamen and flower maturation. B. rapa miRNAs were also found to target genes that control intracellular processes such as enzymes involved in metabolism, protein degradation, and transporters genes. BrMiR 168 targets the pyruvate kinase gene involved in glycolysis. BrMiR 854 targets glycosyl hydrolase family proteins, which have functions such as cation binding and hydrolase activity, including hydrolyzing O-glycosyl compounds mainly in carbohydrate metabolism. BrMiR 1520 targets the UDPglucose: sterol glucosyltransferase (UGT80A2) genes, which are involved in the transfer of glycosyl groups in carbohydrate and lipid metabolism. BrMiR 838 targets the SPT1 gene, which is involved in sphingolipid biosynthesis. BrMiR 776 and 854 target the cuticular 1 (CUT1) and beta-ketoacyl-CoA synthase genes, respectively, which are both involved in the biosynthesis of very-long-chain fatty acids (VLCFA). BrMiR 395 targets APS1 and APS4, which are involved in sulfate assimilation in Arabidopsis; this shows that the functions of different miRNA families are conserved in plant species. BrMiR 1132 targets nucleoporin-interacting component family proteins, which are involved in protein binding/transport. Many studies report that miRNAs also target genes that are involved in biotic and abiotic stress. In the present study, BrMiR 837 was found to target LCR69/PDF2.2 (low molecular weight cysteine-rich 69), which is a family of pathogenesis-related and plant defensin proteins that have peptidase activity.
DISCUSSION
Since the discovery of plant miRNAs in, 2002 (Llave et al., 2002b), miRNAs have become an important and integral topic in functional genomic research; consequently, several studies show that miRNAs are important regulatory elements involved in plant development, metabolism, biotic and abiotic stresses, and many other functions (Lu et al., 2005; Pulido and Laufs, 2010; Ruiz-Ferrer and Voinnet, 2009; Zhang et al., 2006a; 2007c). B. rapa is an important crop with varying leaf and plant morphophytes. It has a comparatively smaller genome size compared to other Brassica species, and is recently considered a model plant for Brassica A genome sequencing (http://www.brassica-rapa.org, http://www.brassica.info). Although a previous study reports the identification of few miRNAs in B. rapa (He et al., 2008), their presence remains largely unknown in this species. An increasing number of genomic/DNA sequences in the form of GSS, EST, mRNA, and cDNA are being deposited into the NCBI database every day. Comparative genome-based in silico screening of B. rapa EST, mRNA, cDNA, and GSS sequences, using previously reported miRNAs from different plant species, would allow us to systematically and comprehensively identify 186 miRNAs belonging to 55 families in the present study. Although earlier studies identified a few miRNAs in B. rapa, they could not find all of the miRNAs reported in the present study. Of the 55 miRNA families, 38 are reported for the first time; in addition, more members of previously identified B. rapa miRNA families were found. Furthermore, we identified 5 Arabidopsis-Brassica lineage-specific miRNA families in B. rapa by using computational analysis; of these, BrMiR 391, 400, 824, and 1140 are newly identified, and BrMiR 158 was previously reported (Sunkar and Jagdeeswaran, 2008). These findings suggest that with the continuous availability of genome sequences, we could identify many more miRNAs that regulate important genes in B. rapa.
The transcription of miRNAs from sense and antisense strands is recently reported in insects, human, and plants (Bender, 2008; Stark et al., 2008; Tyler et al., 2008; Zhang et al., 2008). In the present study, we found only 1 DNA sequence from which both sense and antisense mature miRNAs were produced compared to 5 miRNAs belonging to 3 families identified in soybean (Zhang et al., 2008); the sense and antisense miRNAs differ by 1-3 nt in that study. We also observed a single- nucleotide difference between the sense and antisense miRNAs belonging to the BrMiR 399 family. The identification of only 1 DNA sequence producing both sense and antisense miRNAs may be due to the limited availability of whole-genome sequence information for B. rapa. Several miRNA clusters, including miRNA family 166, 169, 171, and 2118, have been identified in plants (Xie et al., 2010; Zhang et al., 2007b; 2008). However, we only observed one miRNA cluster for BrMiR 169, which suggests that miRNA clusters are conserved among plant species. We believe that increased availability of B. rapa EST and complete genome sequences will help identify more miRNAs that are produced from either sense or antisense, or in clusters from the same genomic locus.
Computationally identified miRNAs need to be validated through expression analysis. First, we analyzed B. rapa ESTs derived from different tissues obtained from public databases and determined the expression of 9 miRNA families in different tissues such as floral buds, roots, callus, seedlings, primary and etiolated mature leaves, and in the entire plant (Table 3). Second we did Northern blot analyses of 5 miRNA families and qRT-PCR analyses of 12 different miRNA families which revealed both differential and tissue-specific expression in leaf, stem, shoot apex, midrib and root tissues. Northern blot analyses revealed that BrMiR 159 was highly expressed in all the analyzed tissues, BrMiR 167 in leaves and roots, and BrMiR 398 in leaves. On the other hand, BrMiR 408 was moderately expressed in leaves, but showed low expression in the other 2 tissues (Fig. 4). qRT-PCR analyses revealed that BrMir164c, BrMir 396b, BrMir 400b and BrMir 1132b showed significantly high expression in root tissues, BrMir172g and 1521b showed stem specific expression, BrMir171f showed shoot specific expression while BrMir162b showed increased expression in midrib tissues, respectively. This observation suggests that some of the identified miRNAs are expressed in tissue-specific manner, whereas some are expressed in whole plants as indicated by previous findings in other plant species (Song et al., 2009; Zhang et al., 2008). The comprehensive characterization of all the remaining identified B. rapa miRNAs in the different tissues would be helpful for understanding the tissue-specific expression of all the miRNAs as well as their regulatory roles with respect to different tissues, organs, and conditions.
Fig. 4. Number of matured miRNAs against different lengths of precursor miRNAs.

The identification of target genes for miRNAs is an important step in understanding the regulation of miRNA by structural genes. It is well understood that miRNAs and their counterpart target genes have perfect or near-perfect complementarities; computational identification coupled with experimental results have been successful in proving this in plants (Llave et al., 2002a; Park et al., 2002; Reinhart et al., 2002; Rhoades et al., 2002; Song et al., 2009). BLAST search analysis allowing for 1- 4 nt mismatches without gaps for 186 miRNA could identify a total of 66 candidate target genes in B. rapa for 33 miRNA families. Since Brassica and Arabidopsis are the closest relatives belonging to the same family and have nucleotide sequence identities of 80-90%, the target genes can be confirmed by aligning them with their orthologues in Arabidopsis. Our results show that most of the predicted targets have conserved functions with those of the miRNA targets in Arabidopsis. The highly conserved nature of miRNA target sequences among a wide variety of plant species has been reported by many researchers (Floyd and Bowman, 2004). In the present study, the majority of target genes were transcription factors such as SBPs, Apetala 2, and many other DNA and RNA binding proteins. The targets of BrMiR 156 are mainly SBP proteins, as reported by many studies in different plant species, suggesting the conserved function of this miRNA family (Zhang et al., 2006b; 2007a; 2008). The other notable transcription factor genes that were found to be miRNA targets were NAC domain-containing protein 100, scarecrow transcription factor family protein, and zinc finger protein genes in addition to above-mentioned genes. The genes of transcription factors that are involved in hormone signaling, such as ARF8 and ARF17, were also found to be targets of BrMiR 167 and 160, respectively. Several studies also report these genes as targets of miRNAs in different plant species including A. thaliana (Chen, 2004; Jones-Rhoades et al., 2004; Mallory et al., 2004; Song et al., 2009; Zhang et al., 2006b; 2007a; 2008). Apart from transcription factors, we observed genes involved in various functions, such as fatty acid metabolism, glycolysis, and other cellular functions, as targets of miRNAs in B. rapa. The targets of BrMiR 837 were pathogenesis- related protein genes such as low-molecular-weight cysteine-rich 69 (LCR69). Similar observations were made in several studies of other plants (Bonnet et al., 2004a; Rhoades et al., 2002; Zhang et al., 2006b). Our result supports the previous reports of extensive evolutionary and functional conservation of miRNAs in plant species.
This is the first comprehensive study that identifies and characterizes miRNAs in a diploid Brassica species, B. rapa. However, the number of identified miRNAs is substantially less compared to the, 199 miRNAs belonging to 121 miRNA families identified in Arabidopsis deposited in the miRNA database (http://www.mirbase.org). Many comparative mapping studies of A. thaliana show the presence of an average of 3 copies of an Arabidopsis chromosome segment in the Brassica genome (Panjabi et al., 2008; Parkin et al., 2005; Teutonica and Osborn, 1994; Truco et al., 1996), although this might not hold true for miRNA genes. Therefore, with the availability of complete genome sequences in the near future, the identification of more members of miRNA belonging to previously identified or new miRNA families is expected. Furthermore, direct cloning and sequencing of miRNAs from different tissues, organs, and developmental timing would speed up the identification of more unidentified miRNAs. The identification of 186 B. rapa miRNAs belonging to 48 families in the present study will aid in the study of the involvement of miRNAs in various plant development, cellular, metabolic, and other functions including biotic and abiotic stresses, which might be a part of the driving force behind expression variation, reflecting the phenotypic plasticity and numerous subspecies of B. rapa with diverse morphophytes.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
This work was supported by research grants from the Technology Development Program for Agriculture and Forestry (Grant no. 607003-05), the Ministry of Agriculture, Forestry, and Fisheries, Republic of Korea. NR was partially supported by the National Research Foundation, Republic of Korea.
References
- 1.Ambros V., Chen X.M. The regulation of genes and genomes by small RNAs. Development. (2007);134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 2.Alves-Junior L., Niemeier S., Hauenschild A., Rehmsmeier M., Merkle T. Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res. (2009);37:4010–4021. doi: 10.1093/nar/gkp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004);116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Bazzini A.A., Hopp H.E., Beachy R.N., Asurmendi S. Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc. Natl. Acad. Sci. USA. (2007);104:12157–12162. doi: 10.1073/pnas.0705114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. (2008);22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollman K.M., Aukerman M.J., Park M.Y., Hunter C., Berardini T.Z., Poethig R.S. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. (2003);130:1493–1504. doi: 10.1242/dev.00362. [DOI] [PubMed] [Google Scholar]
- 7.Bonnet E., Wuyts J., Rouzé P., Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA. (2004a);101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet E., Wuyts J., Rouze P., Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. (2004b);20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 9.Buhtz A., Springer F., Chappell L., Baulcombe D.C., Kehr J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. (2008);53:739–749. doi: 10.1111/j.1365-313X.2007.03368.x. [DOI] [PubMed] [Google Scholar]
- 10.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architectureand applications. BMC Bioinformatics. (2009);421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. (2003);301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 12.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. (2009);136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. (2004);303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell. (2006);18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depicker A., Montagu M.V. Post-transcriptional gene silencing in plants. Curr. Opin. Cell Biol. (1997);9:373–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- 16.Dugas D.V., Bartel B. MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. (2004);7:512–520. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Floyd S.K., Bowman J.L. Gene regulation: ancient microRNA target sequences in plants. Nature. (2004);428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 18.Fujii H., Chiou T.J., Lin S.I., Aung K., Zhu J.K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. (2005);15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S. MiRBase: the microRNA sequence database. Methods Mol. Biol. (2006);342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S., Saini H.K., Van D.S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. (2008);36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo H.S., Xie Q., Fei J.F., Chua N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. (2005);17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X.F., Fang Y.Y., Feng L., Guo H.S. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. (2008);582:2445–2452. doi: 10.1016/j.febslet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Jagadeeswaran G., Zheng Y., Li Y.F., Shukla L.I., Matts J., Hoyt P., Macmil S.L., Wiley G.B., Roe B.A., Zhang W., et al. Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. (2009);184:85–98. doi: 10.1111/j.1469-8137.2009.02915.x. [DOI] [PubMed] [Google Scholar]
- 24.Jin W., Li N., Zhang B., Wu F., Li W., Guo A., Deng Z. Identification and verification of microRNA in wheat (Triticum aestivum). J. Plant Res. (2008);121:351–355. doi: 10.1007/s10265-007-0139-3. [DOI] [PubMed] [Google Scholar]
- 25.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress induced miRNA. Mol. Cell. (2004);14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 26.Kim J., Jung J.-H., Reyes J.L., Kim Y.-S., Kim S.Y., Chung K.-S., Kim J.A., Lee M., Lee Y., Kim V.N., et al. microRNAdirected cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. (2005);42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klevebring D., Street N.R., Fahlgren N., Kasschau K.D., Carrington J.C., Lundeberg J., Jansson S. Genome-wide profiling of Populus small RNAs. BMC Genomics. (2009);10:620. doi: 10.1186/1471-2164-10-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauter N., Kampani A., Carlson S., Goebel M., Moose S.P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA. (2005);102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. (2001);25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Llave C., Xie Z.X., Kasschau K.D., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. (2002a);297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 31.Llave C., Kasschau K.D., Rector M., Carrington J.C. Endogeneous and silencing-associated small RNAs in plants. Plant Cell. (2002b);14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu S., Sun Y.H., Shi R., Clark C., Li L., Chiang V.L. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. (2005);17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S., Sun Y.H., Amerson H., Chiang V.L. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. (2007);51:1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- 34.Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. MicroRNA regulation of NAC domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. (2004);14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Meyers B.C., Axtell M.J., Bartel B., Bartel D.P., Baulcombe D., Bowman J.L., Cao X.F., Carrington J.C., Chen X., Green P.J., et al. Criteria for annotation of plant MicroRNAs. Plant Cell. (2008);20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li. S., Zhou H., Long C., et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. (2008);133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. (2008);133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 38.Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. (2006);21:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 39.Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. Control of leaf morphogenesis by microRNAs. Nature. (2003);425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 40.Panjabi P., Jagannath A., Bisht N.C., Padmaja K.L., Sharma S., Gupta V., Pradhan A.K., Pental D. Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics. (2008);9:113. doi: 10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park W., Li J.J., Song R.T., Messing J., Chen X.M. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. (2002);12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2005);102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkin I.A.P., Gulden S.M., Sharpe A.G., Lukens L., Trick M., Osborn T.C., Lydiate D.J. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. (2005);171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pulido A., Laufs P. Co-ordination of developmental processes by small RNAs during leaf development. J. Exp. Bot. (2010);61:1277–1291. doi: 10.1093/jxb/erp397. [DOI] [PubMed] [Google Scholar]
- 45.Qiu C.X., Xie F.L., Zhu Y.Y., Guo K., Huang S.Q., Nie L., Yang Z.M. Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene. (2007);395:49–61. doi: 10.1016/j.gene.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 46.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. Micro-RNAs in plants. Genes Dev. (2002);16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. Prediction of plant microRNA targets. Cell. (2002);110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Ferrer V., Voinnet O. Roles of plant small RNAs in biotic stress responses. Ann. Rev. Plant Biol. (2009);60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 49.Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. (2005);8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 50.Shi R., Chiang V.L. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques. (2005);39:519–524. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 51.Song C., Fang J., Li X., Liu H., Chao C.T. Identification and characterization of 27 conserved microRNAs in citrus. Planta. (2009);230:671–685. doi: 10.1007/s00425-009-0971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stark A., Bushati N., Jan C.H., Kheradpour P., Hodges E., Brennecke J., Bartel D.P., Cohen S.M., Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. (2008);22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sunkar R., Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. (2008);8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunkar R., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabdopsis. Plant Cell. (2004);16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sunkar R., Girke T., Jain P.K., Zhu J.K. Cloning and characterization of microRNAs from rice. Plant Cell. (2005);17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunkar R., Kapoor A., Zhu J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. (2006);18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunkar R., Chinnusamy V., Zhu J., Zhu J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. (2007);12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Sunkar R., Zhou X., Zheng Y., Zhang W., Zhu J.K. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. (2008);8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szittya G., Moxon S., Santos D.M., Jing R., Fevereiro M.P., Moulton V., Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. (2008);9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. (2008);49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 61.Teutonico R.A., Osborn T.C. Mapping of RFLP and quantitative trait loci in Brassica rapa and comparison to the linkage maps of B. napus, B. oleracea, and Arabidopsis thaliana. Theor. Appl. Genet. (1994);89:885–894. doi: 10.1007/BF00224514. [DOI] [PubMed] [Google Scholar]
- 62.Truco M.J., Hu J., Sadowski J., Quiros C.F. Interand intra-genomic homology of the Brassica genomes: implications for their origin and evolution. Theor. Appl. Genet. (1996);93:1225–1233. doi: 10.1007/BF00223454. [DOI] [PubMed] [Google Scholar]
- 63.Tyler D.M., Okamura K., Chung W.J., Hagen J.W., Berezikov E., Hannon G.J., Lai E.C. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. (2008);22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaucheret H., Fagard M. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. (2001);17:29–35. doi: 10.1016/s0168-9525(00)02166-1. [DOI] [PubMed] [Google Scholar]
- 65.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. (2009);136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 66.Wang J.F., Zhou H., Chen Y.Q., Luo Q.J., Qu L.H. Identification of, 20 microRNAs from Oryza sativa. Nucleic Acids Res. (2004a);32:1688–1695. doi: 10.1093/nar/gkh332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X.L., Reyes J.L., Chua N.H., Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. (2004b);5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu G., Poethig R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. (2006);133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie F.L., Huang S.Q., Guo K., Xiang A.L., Zhu Y.Y., Nie L., Yang Z.M. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett. (2007);581:1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- 70.Xie F., Frazier T.P., Zhang B. Identification and characterization of microRNAs and their targets in the bioenergy plant switchgrass (Panicum virgatum). Planta. (2010);232:417–434. doi: 10.1007/s00425-010-1182-1. [DOI] [PubMed] [Google Scholar]
- 71.Yang T.J., Kim J.S., Lim K.B., Kwon S.J., Kim J.A., Jin M., Park J.Y., Lim M.H., Kim H.I., Kim S.H., et al. The Korea Brassica Genome Project: A glimpse of the Brassica genome based on comparative genome analysis with Arabidopsis. Comp. Funct. Genom. (2005);6:138–146. doi: 10.1002/cfg.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. (2005);33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B.H., Pan X.P., Cobb G.P., Anderson T.A. Plant microRNA: a small regulatory molecule with big impact. Dev. Biol. (2006a);289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 74.Zhang B.H., Pan X.P., Anderson T.A. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett. (2006b);580:3753–3762. doi: 10.1016/j.febslet.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 75.Zhang B.H., Pan X.P., Cox S.B., Cobb G.P., Anderson T.A. Evidence that miRNAs are different from other RNAs. Cell. Mol. Life Sci. (2006c);63:246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang B.H., Wang Q., Wang K., Pan X., Liu F., Guo T., Cobb G.P., Anderson T.A. Identification of cotton microRNAs and their targets. Gene. (2007a);397:26–37. doi: 10.1016/j.gene.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 77.Zhang B.H., Pan X.P., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. (2007b);302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 78.Zhang B.H., Wang Q.L., Pan X.P. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol. (2007c);210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 79.Zhang B.H., Pan X.P., Stellwag E.J. Identification of soybean microRNAs and their targets. Planta. (2008);229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- 80.Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., Ruan K., Jin Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. (2007);354:585–590. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 81.Zhou X., Wang G., Zhang W. UV-B responsive micro-RNA genes in Arabidopsis thaliana. Mol. Syst. Biol. (2007);3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. (2003);31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuker M., Mathews D.H., Turner D.H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In RNA Biochemistry and Biotechnology, J. Barciszewski, and C.C. Brian Frederic, eds. (NATO Science series), Vol. 70. (1998):11–43.


