Abstract
Reversible conjugation of the small ubiquitin modifier (SUMO) peptide to proteins (SUMOylation) plays important roles in cellular processes in animals and yeasts. However, little is known about plant SUMO targets. To identify SUMO substrates in Arabidopsis and to probe for biological functions of SUMO proteins, we constructed 6xHis-3xFLAG fused AtSUMO1 (HFAtSUMO1) controlled by the CaMV35S promoter for transformation into Arabidopsis Col-0. After heat treatment, an increased sumoylation pattern was detected in the transgenic plants. SUMO1-modified proteins were selected after two-dimensional gel electrophoresis (2-DE) image analysis and identified using matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). We identified 27 proteins involved in a variety of processes such as nucleic acid metabolism, signaling, metabolism, and including proteins of unknown functions. Binding and sumoylation patterns were confirmed independently. Surprisingly, MCM3 (At5G46280), a DNA replication licensing factor, only interacted with and became sumoylated by AtSUMO1, but not by SUMO1ΔGG or AtSUMO3. The results suggest specific interactions between sumoylation targets and particular sumoylation enzymes.
Keywords: Arabidopsis, mass spectrometry, proteomics, SUMO binding proteins, Sumoylation
INTRODUCTION
Eukaryotic proteins are regulated by many types of post translational modifications, and in some case the modifying group is itself a protein. Ubiquitin, a small, highly conserved polypeptide, provides the prototypical example. It is reversibly linked to target proteins in the cell in a process termed ubiquitination (Hochstrasser, 2009). As another example, the small ubiquitin-like modifier (SUMO) resembles ubiquitin in its structure and mechanism. Activation and conjugation reactions involving SUMO have much in common with what we know about ubiqutin (Johnson, 2004; Melchior, 2000; Miura and Hasegawa, 2010). SUMO attachment (sumoylation) to substrates depends on three components: heterodimeric SUMO-activating enzyme (E1), SUMO-conjugating enzyme (E2) and the substrate recognition factor (E3) that stimulates SUMO transfer from E2 to substrates (Johnson, 2004; Seeler and Dejean, 2003). Finally, sumoylated proteins can be removed from conjugates by SUMO proteases for SUMO recycling.
Although the sumoylation process resembles ubiquitination, unlike ubiquitin ligation, target proteins typically include several sumoylation sites consisting of a short consensus sequence, Ψ- K-X-EED (Ψ, large hydrophobic Ψ amino acid; K, the acceptor lysine; X, any amino acid; EED, glutamate or aspartate) (Schmidt and Müller, 2003). SUMO modification of substrate proteins in animals and yeasts participate in critical and diverse cellular processes, such as innate immunity, chromatin stability and cell division, DNA repair, nucleocytoplasmic trafficking, transcriptional regulation, and ubiquitination antagonism (Gill, 2005; Hay, 2005). Vertebrate RanGAP1, in which the modification by SUMO is required for its localization to the nuclear pore complex, has been identified as the first target of SUMO conjugation. To date, more than 50 mammalian proteins are known as sumoylation targets (Hannich et al., 2005; Matunis et al., 1996). Global analyses in yeast and metazoans indicated >100 target proteins (Denison et al., 2005; Panse et al., 2004; Wykoff and O’Shea, 2005; Zhao et al., 2004; Zhou et al., 2004).
In Arabidopsis, the SUMO gene family amounts to eight genes of which only four appear to be expressed (SUM1-3, and 5) (Budhiraja et al., 2009; Colby et al., 2006). Levels of SUMO1E2 conjugates are dramatically but transiently increased by various types of cellular stress, including exposure of the plants to heat shock, H2O2, ethanol, and canavanine, suggesting sumoylation as an important post-translational feature of the plant stress response. Indicating some specificity is the fact that levels and distribution of SUMO3 conjugates appear to be unaffected by any stress (Kurepa et al., 2003). Although our understanding of sumoylation remains limited in plants, the system appears to play an exceptionally important role in various plant environmental responses, including heat shock, pathogen defense, drought and cold tolerance, ABA signaling, flowering, and phosphate deficiency (Catala et al., 2007; Jin et al., 2008; Lee et al., 2007; Miura et al., 2005; 2007; 2009; Saracco et al., 2007; Yoo et al., 2006). Recently, van den Burg et al. (2010) reported that AtSUMO3 acted downstream of SA accumulation, while involvement of AtSUMO1 and AtSUMO2 has been reported in suppressing SA accumulation. Therefore, it implies that the Arabidopsis SUMO paralogs form a regulatory network that differentially modulates target modifications in a response to various environmental conditions. However, many aspects of biological function and regulation in plants, including the nature of SUMO target proteins, remain poorly defined. To date, a small number of SUMO target proteins have been uncovered in the form of seven genuine Arabidopsis proteins, MYB30, GTE3/5, PHR1, ICE1, ABI5 and FLD, that harbor sumoylation consensus motifs (Garcia-Dominguez et al., 2008; Jin et al., 2008; Miura et al., 2005; 2007; 2009; Okada et al., 2009). In addition, very recent are reports about systematic approaches using yeast two-hybrid assays and affinity enrichment methods with mass spectrometry for mapping SUMO target proteins (Elrouby and Coupland, 2010; Miller et al., 2010).
To extend our knowledge about biological implications of putative SUMO targets and SUMO binding proteins in plants, we have begun to screen in-planta for SUMO-binding proteins using by a mass spectrometry-based proteomics approach using a transgenic Arabidopsis line overexpressing AtSUMO1. Here we report the isolation of 27 proteins that fit the criteria. They include proteins with a variety of putative functions, in DNA or RNA-related metabolism, signaling pathway, general metabolism, and several functional unknown proteins. Specifically, based on a yeast split ubiquitin assay and E. coli sumoylation assay using SUMO1, SUMO1ΔGG, or SUMO3 proteins in combination with MCM3 (At5G46280) protein, the functional properties of SUMO proteins detected in vivo might reflect a capability to conditionally differentiate sumoylation activities in plants.
MATERIALS AND METHODS
Plant materials and ABA treatment
Wild-type, Arabidopsis thaliana Columbia-0 (Col-0) was used in these experiments. Seeds were surface sterilized, stratified for three days at 4℃ and then sown to a medium in petri plates containing 1× Murashige and Skoog basal salt mixture, 3% sucrose, 2.5 mM MES, pH 5.7, and 0.8% agar or soil. Plants were grown under a 16/8 h light/dark at 22℃, light intensity of 100 μmol m-2s-1 and relative humidity of 57-80%. To investigate the inhibition of root growth by ABA, 4-day-old seedlings were transferred onto plates supplemented with ABA. Root growth was measured as the difference in root length between the beginning and the end of the growth evaluation period. 14-dayold seedlings were photographed.
Plasmid constructs and plant transformation
To generate Arabidopsis transgenic plants over-expressing AtSUMO1, the protein coding region was amplified by RT (Reverse transcription)-PCR using Arabidopsis thaliana cDNAs. The 6xHis-3xFlag fused AtSUMO1 (HFAtSUMO1) was cloned into the pGEM-T Easy vector (Promega, USA), sequenced to verify the correct DNA sequence and then sub-cloned into the pCAMBIA1300PT-derived vector using the appropriate restriction enzymes. Agrobacterium tumefaciens GV3101 strains harbored the construct over-expressing HFAtSUMO1 were grown in LB liquid culture with 50 mg/L gentamycin, 50 mg/L rifampicin, and 50 mg/L kanamycin at 30℃. The Col-0 plants were transformed by the floral deep method as previously described (Clough and Bent, 1998; Park et al., 2009). Hygromycinresistant transgenic plants were selected on 1× MS medium containing 30 mg/L hygromycin.
Immunoblot analysis
Plant tissues were frozen and ground with mortar and pestle in liquid nitrogen. Protein extracts were prepared in protein extraction buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP40, 1 mM EDTA, 3 mM DTT, 1 mM phenylmethysulfonyl fluoride containing 1× Complete Protease Inhibitor (Roche)]. After centrifugation at 14,000 rpm for 20 min twice, the supernatant was used immediately or stored at -80℃. Protein concentration was determined using a protein assay kit (Bio-Rad), and 40 μg of total protein was separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, probed with an anti-His antibody, and detected using the ECL Western blot detection system (Amersham Biosciences).
2-DE analyses and MALDI-TOF MS
SUMO conjugates were affinity purified using nickel-nitrilotriacetic acid agarose (Ni-NTA) resin according to the manufacturer’s instruction (Qiagen, USA). Purified proteins were examined by immunoblot analysis using an anti HA-antibody. Quantified proteins (170 μg) were mixed in sample buffer and then loaded onto IEF gel (18 cm tube gel) (O’Farrell, 1975). In the second dimension, proteins were separated in 12% SDSpolyacrylamide gels and visualized by silver staining with no glutaraldehyde (Blum et al., 1987). Gel images were scanned using a GS-800 Imaging Densitometer (Bio-Rad) and analyzed with the software PDQuest version 7.2.0 (Bio-Rad). For each sample, quantitation was performed with three analytical gels originating from three independent biological replicas. The silver- stained protein spots were excised from the gel, subjected to in-gel tryptic digestion (Promega, USA), and extracted as previously described (Kim et al., 2004). Peptide mass fingerprinting was carried out on a Voyager-DE STR MALDI-TOF mass spectrometer (PerSeptive Biosystems, USA) according to previously reported methods (Kim et al., 2004). For data processing, the software package PerSeptive-Grams was used.
Yeast split ubiquitin assay
The yeast split ubiquitin assay was performed as described previously (Laser et al., 2000; Yoo et al., 2005). Saccharomyces cerevisiae strain JD53 was used for all experiments. The putative SUMO1-binding cDNAs were cloned into pMET-Ste14-Cub-RUra3, replacing yeast Ste14. AtSUMO1, AtSUMO1ΔGG, and AtSUMO3 cDNAs were cloned into modified versions of the pCup-Nub-Sec62 vector, replacing yeast Sec62, respectively (Stagljar et al., 1998). Interactions between each pair of proteins were tested on selective medium containing 2 mg/ml 5-FOA and selective medium lacking uracil. Plates were incubated at 30℃ for 3-5 days, unless specified otherwise.
Agrobacterium-mediated transient expression assay in tobacco plants
Seven-week-old N. benthamiana plants were used for Agrobacterium- mediated transient expression. AtSUMO1 was cloned into pCAMBIA 1300-cLUC and genes encoding putative SUMO binding proteins were into pCAMBIA1300-nLUC (Chen et al., 2008). The DNA constructs were introduced into Agrobacterium tumefaciens strain GV3101. The bacteria grown in Luria- Bertani medium supplemented with 10 mM MES buffer, 20 μM acetosyringone, and appropriate antibiotics at 28℃ were washed with infiltration solution (10 mM MgCl2, 10 mM MES buffer, and 100 μM acetosyringone) twice to avoid the toxicity of antibiotics. In addition, Agrobacterium transformed with p19, a suppressor of gene silencing, was cultured and prepared (Lakatos et al., 2004). For co-infiltration, each of Agrobacterium cultures was OD600 = 0.5 in final infiltration solution and mixed in equal volumes. Bacterial suspensions were infiltrated into tobacco leaves using a needleless syringe. After infiltration, plants were immediately covered with plastic bags and placed at 23℃ for 36 h. The tobacco leaves were sprayed three times with 1 mM luciferin in solution, and imaged by an EM CCD camera (iXon, Andor Technology plc, Ireland). Bioluminescence was recorded after quenching for 5 min in the dark.
Sumoylation assay in E. coli
The sumoylation assay in E. coli was carried out as described previously (Okada et al., 2009). Escherichia coli BL21(DE3) cells were transformed with pACYCDuet-AtSAE1b-AtSAE2, and pCDFDuet-AtSUMO1/3 (AA or GG)-AtSCE1a. The transformed cells were used for the preparation of competent cells. For in vivo sumoylation reactions, the competent cells were transformed with pET28a-AtMYB30 or pET28a-At5g46280, respectively. Transformed cells were cultured in 5 ml of LB medium at 37℃ until the OD600 was 1.0, followed by the addition of 0.4 mM IPTG. After an approximately 12 h induction at 25℃, 2 ml of the culture cells was harvested, washed with 1× PBS buffer and suspended with 200 μl of 1× PBS buffer. The samples were lysed by sonication, and boiled at 95℃ for 5 min. 20 μl of total protein was loaded to 7.5% continuous gradient gel, transferred to a polyvinylidene difluoride membrane, probed with an anti-T7 antibody, and detected using the ECL Western blot detection system (Amersham Biosciences).
RESULTS AND DISCUSSION
Construction of AtSUMO1 transgenic Arabidopsis plants
To identify SUMO-modified proteins in Arabidopsis, we constructed AtSUMO1 fused to His6-FLAG3 (HFAtSUMO1) and generated transgenic Arabidopsis lines. The His6-FLAG3 was added to the N-terminal end, and expression of the construct was under control of the CaMV35S promoter (Fig. 1A). A free His6-FLAG3 construct was compared as the control vector. Twenty-five individual transgenic Arabidopsis lines were selected using hygromycin resistance and then further selected by protein gel blot analysis using anti-His tag Antibody. Figure 1B shows protein expression levels of five representative HFAtSUMO1 lines from transgenic plants expressing fusion protein. Western blot analysis using anti-His tag antiserum showed two different bands in HFAtSUMO1 transgenic Arabidopsis plants. These two bands represent precursor and mature forms with SUMOs covalently attached to lysine residues in target proteins via cleavage of the C-terminal GG motif (Meulmeester and Melchior, 2008).
Fig. 1. Expression of AtSUMO1 in Arabidopsis. (A) Structure of the binary vector used for expression of AtSUMO1 in transgenic Arabidopsis. The 6xHis-3xFlag fused AtSUMO (HFAtSUMO1) were placed between the CaMV35S promoter and the OCS terminator of the pCAMBIA1300PT-derived binary vector. (B) Expression of HFAtSUMO1 in homozygous T3 over-expressing transgenic plants and control plants (Col-0 and HF inserted vector). The resulting crude extracts (40 μg) were subjected to SDS-PAGE, and immunoblot analysis was carried out with anti-HA antibody. (C) Identification of AtSUMO1 protein by MALDI-TOP MS. MALDI-TOP mass spectrometry-based analysis identified the peptide sequence of the expressed protein as AtSUMO1.
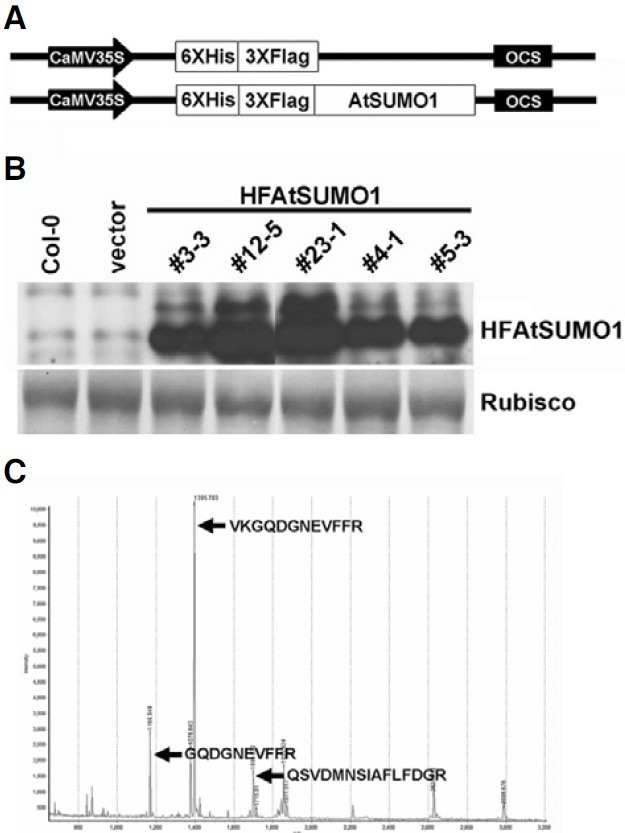
To confirm whether HFAtSUMO1 protein was over-expressed in the transgenic plants, we performed peptide mass fingerprinting (PMF) using MALDI-TOF mass spectrometry. After purification of plant crude extracts by histidine affinity chromatography, the purified sample was applied to SDS-PAGE analysis and a band containing the putative HFAtSUMO1 was excised from the gels, digested with trypsin, and identified by MALDI-TOF mass spectrometry. Peptide mass fingerprinting of the purified protein showed a match with the SUMO1 peptide sequence, indicating that the protein was derived from the HFAtSUMO1 DNA sequence (Fig. 1C).
Effect of ABA and accumulation of SUMO1 conjugates by heat stress in AtSUMO1 transgenic plants
Overexpression of SUMO1/2 had been reported to suppress abiotic stress responses mediated by abscisic acid (Lois et al., 2003). We also analyzed whether abscisic acid affected root growth in HFAtSUMO1 transgenic Arabidopsis plants. Fig. 2A showed that ABA-mediated root growth inhibition was reduced significantly in plants that overexpressed AtSUMO1 in comparison to Col-0 and empty vector control plants. In addition, the transgenic plants that exhibited high transgene protein levels (#3-3, #12-5, and #23-1 lines) demonstrated approximately 33% reduction of ABA-mediated root growth inhibition, compared to those of Col-0 and empty vector control plants (Fig. 2B).
Fig. 2. Phenotype of transgenic plants expressing HFSUMO1 and heat shock-induced SUMO conjugation. (A) Root growth inhibition by ABA is decreased in HFSUMO1 over-expressing transgenic plants. Three-day-old seedlings were transferred to MS medium without or supplemented with 10 μM ABA and maintained under a 16-h-light/8-h-dark daily photoperiod at 24℃. Photographs were taken after 24 days. (B) Quantitative representation of the data in (A). Error bars represent standard deviations (n = 15). Mean values of over-expressing transgenic plants are significantly different form control plants at *P < 0.01. (C) SUMO conjugation patterns after heat treatment in HFSUMO1 over-expressing transgenic plants. Ten-day-old seedling of vector, and each of the SUMO overexpressing lines (#3-3, #12-5, #23-1, #4-1, and #5-3) were exposed to a 30 min-heat shock at 39℃ in the dark. Total proteins were extracted from untreated (t = 0) or heat shock-treated (t = 30) seedlings, and 30 μg of proteins was subjected to SDS-PAGE. The immunoblot was probed with anti-His-tag antiserum.
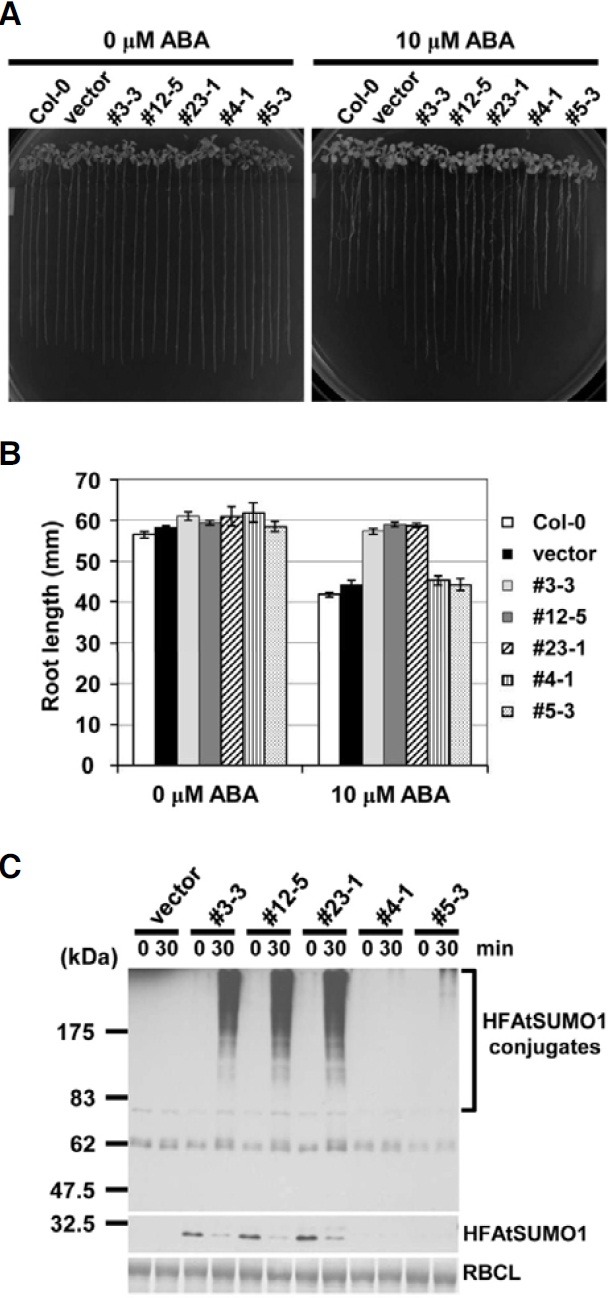
Sumoylation in animals is activated by various environmental stresses and some SUMO targets are part of the stress response (Goodson et al., 2001; Hong et al., 2001; Mao et al., 2000; Melchior, 2000; Saitoh and Hinchey, 2000). In Arabidopsis, the levels of SUMO1 and -2 conjugates but not SUMO3 conjugates increased after exposing seedlings to stress conditions, such as heat shock, H2O2, ethanol, and the amino acid analog canavanine. In addition, overexpression of SUMO2 enhanced both the steady state levels of SUMO2 conjugates under normal growth conditions and showed subsequent heat shock-induced accumulation in Arabidopsis plants (Kurepa et al., 2003). To examine endogenous SUMO conjugates in independent HFAtSUMO1 transgenic Arabidopsis, we performed gel blot analyses using anti-His tag antiserum after heat shock treatment (Fig. 2C). In His6-FLAG3 fused AtSUMO1 (HFAtSUMO1) controlled by the CaMV35S promoter, SUMO conjugates were not detected under no-stress conditions. This result is in agreement with SUMO conjugation data obtained in yeast, possibly because placement of the His6-FLAG3 tag at the Nterminus of AtSUMO1 reduced the efficiency of SUMO conjugations (Hannich et al., 2005). However, the conjugation pattern increased drastically over time and in amount in a protein expression-dependent manner (Fig. 2C). Thus, the #12-5 overexpression line was selected as a suitable candidate for screening of SUMO1-modified proteins by 2-DE analysis.
Identification of SUMO1-modified proteins by 2-DE analysis
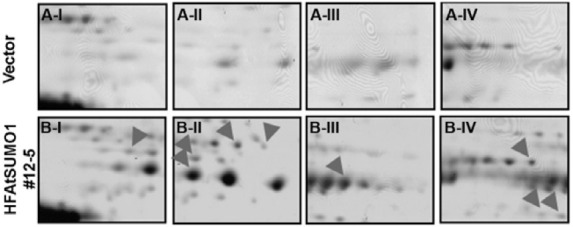
In order to isolate in vivo SUMO-modified proteins using His6-FLAG3 fused AtSUMO1 (HFAtSUMO1) overexpressing plant (#12-5 line), we established mass spectrometry-based screening combined with 2-DE. We first analyzed by performing Ni- NTA chromatography to purify the SUMO-modified proteins of AtSUMO1 overexpressing transgenic plants compared to the vector control plants after treatment with heat stress at 39℃ for 30 min. Using 2-DE analysis, differences in the protein spots between the vector control and AtSUMO1 transgenic plants were compared after heat-shock treatment. Fig. 3 shows representative images for vector control (Fig. 3A) and HFAtSUMO1 transgenic plants (Fig. 3B). New protein spots were detected in HFAtSUMO1 transgenic plant, suggesting appearance of SUMO-modified proteins by HFAtSUMO1 (arrow heads in Fig. 3). In total, 27 line-specific protein spots were excised from the gels and further analyzed by MALDI-TOF MS.
Fig. 3. Representative 2-DE gel images of vector control and AtSUMO1 (#12-5) overexpressing transgenic seedlings. The vector (A-I, II, III, and IV) and AtSUMO1 (B-I, II, III, and IV) over-expressing transgenic seedlings were grown for 7 days in liquid culture media under continous light at 24℃, and exposed to a 30 min-heat shock at 39℃. For isoelectric focusing, a total of 170 μg proteins were loaded onto pH 4-7L IPG strips (18 cm) followed by SDS-PAGE on 12% gel and silver staining. Arrow heads indicate the various putative SUMO modified proteins.
Excised protein spots were digested by trypsin, and the resulting peptides were extracted and identified by peptide mass fingerprinting (PMF) using MALDI-TOF MS. The list of SUMO1- modified proteins is compiled in Table 1. It shows that the putative SUMO targets are involved in DNA and RNA-related processes, signaling pathways, and in general metabolism. Generally, the SUMO proteins are predominantly nuclear proteins, but sumoylation also occurs in the cytoplasm, and the set of putative SUMO-modified proteins identified in this work encompasses both nuclear and cytoplasmic proteins. Recently, Budhiraja et al. (2009) reported that SUMO target proteins are involved in DNA-related or in RNA-dependent processes, such as the regulation of chromatin structure, splicing, or translation. We have as well obtained proteins involved in DNA and RNA metabolism, in particular MCM3 (At5G46280), UBA2A (At3G56860), an SMC-like protein (At3G54670), a zinc finger family protein (At3G08505), and RAD54 (At3G19210). Intriguingly, a disease resistance related protein (At5G46500) was also found. Together, these examples of SUMO-modified proteins indicated that SUMO modification is not limited to soluble proteins, and, like ubiquitin, might potentially have a membrane-based signal function. Indicating specificity in the putative target proteins, all SUMO1-modified candidate proteins contained predicted sumoylation sites with motifs of high probability (Table 1).
Table 1.
Identification of SUMO-modified proteins in HFAtSUMO1 Arabidopsis transgenic seedlings under heat stress
| Gene name | AGI code | Predicted sumoylation sitea (Motifs with high probability) | Description |
|---|---|---|---|
| DNA, RNA-related processes | |||
| MCM3 | At5G46280 | 3 | Replication licensing factor |
| UBA2A | At3G56860 | 1 | AU-rich element binding / RNA binding |
| SMC like protein | At3G54670 | 7 | Structure maintenance of chromosomes |
| Zinc finger family protein | At3G08505 | 1 | Zinc finger (CCCH-type/C3HC4-type RING finger) family protein |
| AtRAD54 | At3G19210 | 3 | ATP binding, DNA binding, helicase 1 |
| Signaling pathway | |||
| Transducin family protein | At2G37160 | 1 | WD-40 repeat family protein |
| Transducin family protein | At3G53390 | 1 | WD-40 repeat family protein |
| EIF4G | At3G60240 | 7 | Eukaryotic Translation Initiation Factor 4G |
| Disease resistance protein | At5G46500 | 4 | TIR-NBS-LRR class |
| Phospholipase C | At3G47290 | 3 | Phosphoinositide-specific phospholipase C family protein |
| EF-Tu | At4G02930 | 3 | Putative elongation factor Tu |
| RPT4a-like protein | At1G45000 | 3 | 26S proteasome regulatory complex subunit p42D |
| Metabolism | |||
| MLS | At5G03860 | 2 | Malate synthase |
| PYK10 | At3G09260 | 2 | Phosphate stravation response 3.1, hydrolase |
| Glucose-inhibited division family A | At2G13440 | 6 | Glucose-inhibited division family A protein |
| SHM1 | At4G37930 | 1 | Serine hydroxymethyltransferase 1 |
| Aminomethyltransferase | At1G11860 | 2 | Putative aminomethyltransferase |
| BXL1 | At5G49360 | 5 | Beta-xylosidase 1 |
| GAPA | At3G26650 | 2 | Glyceraldehyde 3-phosphate dehydrogenase A subunit |
| GAPC-2 | At1G13440 | 4 | Glyceraldehyde 3-phosphate dehydrogenase C subunit |
| KPHMT1 | At2G46110 | 3 | Ketopantoate hydroxymethyltransferase 1 |
| Aspartyl aminopeptidase | At5G60160 | 3 | Putative aspartyl aminopeptidase |
| ASP5 | At4G31990 | 3 | Aspartate aminotransferase 5 |
| LPD2 | At1G48030 | 2 | Lipoamide dehydrogenase 2 |
| Unknown protein | |||
| Unknown protein | At3G19870 | 4 | Unknown function |
| Unknown protein | At2G16760 | 2 | Unknown function |
| Unknown protein | At4G26920 | 2 | Putative homeodomain protein |
aPredicted sumoylation sites were analyzed by “http://www.abgent.com/tools/sumoplot_login”.
Interaction between SUMO1 and SUMO1-modified proteins in vivo
To verify that the SUMO1-modified proteins isolated were indeed SUMO-protein conjugates, we determined whether AtSUMO1 directly interacted with five representative SUMO1- modified proteins by using a yeast split-ubiquitin assay system, which is based on the reassembly of the N- and C- terminal halves (Nub and Cub) of ubiquitin (Ub). AtSUMO1 and SUMO1- modified proteins were fused to the C-terminus of Nub and the N-terminus of Cub, respectively (Fig. 4A). If AtSUMO1 directly interacted with SUMO1-modified proteins, Nub and Cub should reassemble into a native-like Ub (Varshavsky, 1996). Finally, the cells containing SUMO1-modified proteins-Cub-RUra3p and Nub-AtSUMO1 will be unable to grow on plates lacking uracil but will be able to grow on plates containing 5-FOA, which is converted into toxic 5-fluorouracil by RUra3p. In the opposite case, yeast cells are uracil prototrophs and 5-FOA sensitive (Fig. 4A). As shown in Fig. 4B, cells co-expressing SUMO1- modified proteins-Cub-RUra3p and Nub-AtSUMO1 were unable to grow on plates lacking uracil, but grew on plates containing 5-FOA, indicating that AtSUMO1 effectively forms stable complexes with the SUMO1-modified proteins in vivo. In negative controls, no interaction was observed between Nub-AtSUMO1 and Cub-RUra3p.
Fig. 4. Confirmation of the interaction between SUMO1 and putative SUMO1 binding proteins in yeast and tobacco expression systems. (A) Schematic diagram of the split ubiquitin system and its application to the interaction of SUMO1 with putative SUMO1 binding proteins. (B) Interaction between SUMO1 and putative SUMO1 binding proteins in the yeast split ubiquitin system. (C) Interaction between SUMO1 and SUMO1 binding proteins in N. benthamiana leaves. LUC image of N. benthamiana leaves co-infiltrated with the Agrobacterium strain containing the putative SUMO1 binding protein-NLuc and CLuc-SUMO1. (I, vector control; II, SGT1a-NLuc and CLuc-RAR1 Chen et al., 2007; III, Nluc; IV, At3G08505-Nluc; V, At3G08505K17R-Nluc; VI, At3G53390-Nluc; VII, At3G- 09260-NLuc and VIII, At5G46280-nLuc). (B) Quantification of LUC activity in leaves expressing the putative SUMO1 binding protein-NLuc and CLuc-SUMO1. (N; vector control, P; SGT1a-NLuc and CLuc-RAR1; see: Chen et al., 2007, Vector; NLuc). Data were collected 36 h after infiltration. The data are presented as mean ± SE of three independent samples.
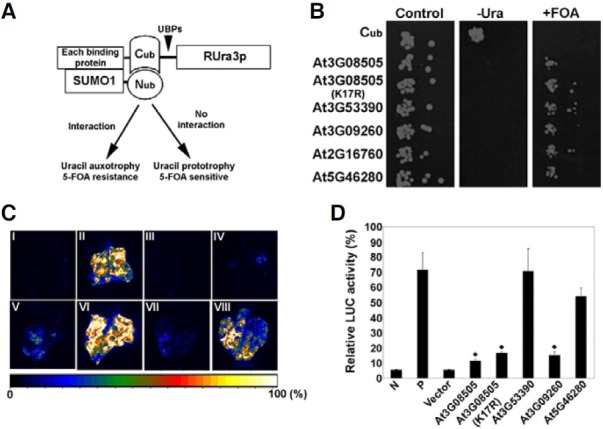
In addition, we constructed a zinc finger family protein (At3G08505)K17R carrying an amino acid substitution (lysine to arginine) at the indicated lysine residue of the putative sumoylation site. Interestingly, co-expression of AtSUMO1 and the zinc finger family protein (At3G08505)K17R mutant, which lacks Lys residue required to form a covalent bond with Gly residues of SUMO protein did not influence 5-FOA resistance, suggesting that AtSUMO1 still interacted with the zinc finger family protein (At3G08505)K17R mutant. It is speculated that the zinc finger family protein (At3G08505) may participate in non-covalent interaction with AtSUMO1 or that it may recognize and switch to other sumoylation sites. This may be similar to all finding in animals by Minty et al. (2000) who defined a Ser-Xaa-Ser motif surrounded by hydrophobic and acidic amino acids as a SUMO-interacting motif (SIM). Among SUMO-interacting proteins, several zinc finger-containing proteins have been determined to be involved in processes like DNA repair or transcriptional repression (Hecker et al., 2006).
Replacing the yeast assay, we next investigated whether these AtSUMO1-modified proteins interacted with AtSUMO1 under physiological conditions in-planta. Using an Agrobacterium-based tobacco LUC transient expression system, we tested the interactions between AtSUMO1 and AtSUMO1- modified proteins after the addition of luciferin, the substrate for firefly LUC. As shown in Fig. 4C, co-expression of CLuc-RAR1 and SGT1a-NLuc led to strong LUC activity in the tobacco leaf as positive control (Fig. 4C-II; Chen et al., 2008). In contrast, Nconjugated Luc coexpressed with C-attached Luc as vector control did not show LUC activity (Fig. 4C-III). Although LUC activity levels varied with respect to different targets (Fig. 4D), co-expression of CLuc-AtSUMO1 and AtSUMO1-modified proteins-NLuc showed LUC activity of higher levels compared to that of vector control. This result is in agreement with data obtained from the yeast split ubiquitin assays. Thus, the results imply that the SUMO-modified proteins isolated in this work appear to be bona fide SUMO binding proteins.
Specific interaction and sumoylation pattern between SUMO1 and MCM3 (At5G46280) protein
Arabidopsis contains eight SUMO genes (Kurepa et al., 2003; Novatchkova et al., 2004). Only four of these (SUMO1, SUMO2, SUMO3, and SUMO5) are highly expressed, functional and probably of major importance, although the other SUMOs could be expressed in very specific cells or under specific conditions only. SUMO1 and SUMO2 are most closely related, sharing 89% protein sequence identity, whereas SUMO3 shows 48% identity and SUMO5 only 35% identity to SUMO1. This suggests that individual SUMOs may have distinct targets and that functional diversification exists. Therefore, we investigated whether these SUMO-modified proteins bind preferentially to SUMO1, SUMO1ΔGG, or SUMO3 by using MCM3, (Mini Chromosome Maintenance3; DNA replication licensing factor (At5G46280) protein, which has an important role in the initiation of replication (Stevens et al., 2002). As shown in Fig. 5A, SUMO1 specifically bound with MCM3 protein, whereas both SUMO1ΔGG, or SUMO3 did not modify this protein as demonstrated by the yeast split ubiquitin assay. These results provide a first evidence that the MCM3 protein is covalently interacting with SUMO1 alone in sumoylation.
Fig. 5. Specific interaction and sumoylation pattern between SUMO1 and MCM3 (At5G46280) protein. (A) AtSUMO1 specifically interacts with MCM3 (At5G46280) protein in the yeast split ubiquitin system. Shown are interactions with MCM3 (At5G46280) protein and SUMOs (SUMO1, SUMO1ΔGG, or SUMO3) by serial dilutions of cells co-expressing Cub or Cub-SUMOs (SUMO1, SUMO1ΔGG, and SUMO3) fusions, respectively, on plates lacking tryptophan and histidine (control) additionally lacking uracil (-Ura) or containing 5-FOA (+FOA). Proteins were expressed from single copy vectors. (B) Specific sumoylation pattern of MCM3 (At5G46280) protein by AtSUMO1 using AtSAE1b as a subunit of the E1 heterodimer. Escherichia coli BL21(DE3) was transformed with each construct. Transformed E. coli cells were incubated at 37℃ until the OD600 reached 1.0, and the expression of tagged proteins and sumoylation was then induced overnight with 0.4 mM IPTG at 25℃. Cell lysates were prepared and Western blotting was carried out using an anti-T7-tag antibody. Sumoylated MCM3 (At5G46280) protein was observed as bands shifted to higher molecular weight in lanes using mature AtSUMO (GG) (indicated as GG), but not in lanes using mutated AtSUMO (AA) (indicated as AA). AtMYB30 was used as positive control for the sumoylation assay in an E. coli system. Arrow heads indicate sumoylated proteins.
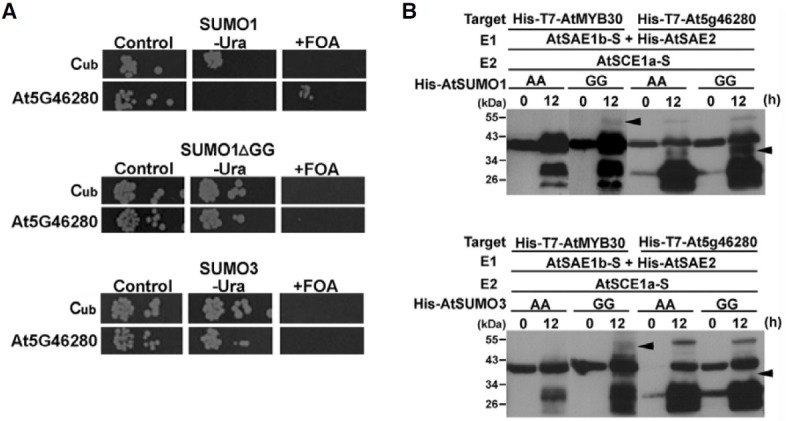
For a detailed examination of MCM3 (At5G46280) covalent modification by SUMO1, we investigated sumoylation patterns of AtSUMO1 and AtSUMO3 using a sumoylation assay system in E. coli. Recently, AtMYB30 has been used as a model SUMO target protein of AtSUMO1/2/3 and 5 for sumoylation assays in E. coli (Okada et al., 2009). AtSUMO1 and AtSUMO3 were modified to expose the C-terminal Gly-Gly sequence, AtSUMO1 (GG) or AtSUMO3 (GG), which is necessary for covalent attachment to a target protein. As negative controls, AtSUMO1 or AtSUMO3 with the C-terminal Gly-Gly mutated to Ala-Ala, AtSUMO1 (AA) or AtSUMO3 (AA) were constructed, respectively. As shown in Fig. 5B, sumoylation reactions were performed in E. coli using AtSUMO1 or AtSUMO3 in combination with MCM3 protein (At5G46280) as a substrate. As a positive control, AtMYB30 was sumoylated by AtSUMO1 (GG) or AtSUMO3 (GG), respectively. Whereas the MCM3 protein (At5G46280) as a substrate was strongly sumoylated by AtSUMO1 (GG), AtSUMO3 (GG) was not recognized and modified. In addition, we also investigated the sumoylation pattern of UBA2A (At3G56860) with AtSUMO1 (GG) using the sumoylation assay system in E. coli. UBA2A (At3G56860) was sumoylated by AtSUMO1 (GG) as well (Supplementary Fig. S1). Thus, the results imply that proteins sumoylated in plants are specifically sumoylated by different SUMO proteins.
CONCLUSION
Sumoylation regulates various biological functions. Examples, mostly obtained with animals and yeasts, include cell division, and DNA repair and transcription. In contrast, only few plant SUMO targets have to date been identified and their functions analyzed at the molecular level. In a very general sense, plant SUMO proteins are involved in the regulation of flowering time, and in orchestrating biotic and abiotic stress responses. To better understand the significance of sumoylation in plants, we have screened SUMO-binding proteins using by a mass spectrometry- based proteomics approach for identifying such target proteins in transgenic Arabidopsis overexpressing AtSUMO1. Based on the results, SUMO-modified proteins are ubiquitous in plants, indicating that sumoylation represents an important factor in the modulation of protein location and activity in different compartments of the cell. In addition, the MCM3 protein (At5G46280) represents a specifically sumoylated target by SUMO1, but not SUMO3, suggesting that individual SUMO proteins recognize distinct targets. Our study extends the knowledge on the SUMO pathway complexity and provides further insights into the function of sumoylation in plants. Determining the exact mechanism(s) leading to sumoylation of the many proteins that are apparently condition-specific targets of SUMO modification will be an important area for further study.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
We thank Dr. Jian-Min Zhou for providing the NLuc and CLuc plasmids and Dr. Katsunori Tanaka for providing the pCDFDuet- AtSUMO-AtSCE1, pACYCDuet-AtSAEb-AtSAE2, and pET28a- AtMYB30 plasmids. This work was supported by grants from the Biogreen 21 Program (grant No. PJ006654) of the Rural Development Administration, World Class University Program (R32-10148) funded by the Ministry of Education, Science and Technology in Korea, and the National Research Foundation of Korea Grant funded by the Korean Government (Ministry of Education, Science and Technology) [NRF-2010-359-F00006]. GS was supported by scholarship from the Brain Korea 21 program of the Ministry of Education, Science and Technology in Korea.
References
- 1.Blum H., Beier H., Gross H.J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. (1987);8:93–99. [Google Scholar]
- 2.Budhiraja R., Hermkes R., Müller S., Schmidt J., Colby T., Panigrahi K., Coupland G., Bachmair A. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. (2009);149:1529–1540. doi: 10.1104/pp.108.135053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catala R., Ouyang J., Abreu I.A., Hu Y., Seo H., Zhang X., Chua N.H. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. (2007);19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.-M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. (2008);146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clough S.J., Bent A.F. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. (1998);16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 6.Colby T., Matthai A., Boeckelmann A., Stuible H.P. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. (2006);142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denison C., Rudner A.D., Gerber S.A., Bakalarski C.E., Moazed D., Gygi S.P. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics. (2005);4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Elrouby N., Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA. (2010);107:17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Dominguez M., March-Diaz R., Reyes J.C. The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J. Biol. Chem. (2008);283:21469–21477. doi: 10.1074/jbc.M708176200. [DOI] [PubMed] [Google Scholar]
- 10.Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. (2005);15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Goodson M.L., Hong Y., Rogers R., Matunis M.J., Park-Sarge O.-K., Sarge K.D. SUMO-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. (2001);276:18513–18518. doi: 10.1074/jbc.M008066200. [DOI] [PubMed] [Google Scholar]
- 12.Hannich J.T., Lewis A., Kroetz M.B., Li S.-J., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. (2005);280:18513–18518. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 13.Hay R.T. Sumo: a history of modification. Mol. Cell. (2005);18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Hecker C.-M., Rabiller M., Haglund K., Bayer P., Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. (2006);281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. (2009);458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong Y., Rogers R., Matunis M.J., Mayhew C.N., Goodson M., Park-Sarge O.-K., Sarge K.D. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. (2001);276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 17.Jin J.B., Jin Y.H., Lee J., Miura K., Yoo C.Y., Kim W.-Y., Van Oosten M., Hyun Y., Somers D.E., Lee I., et al. The SUMO E3 ligase, AtSIZ1, regulates flo-wering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. (2008);53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. (2004);73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 19.Kim S.T., Kim S.G., Hwang D.H., Kang S.Y., Kim H.J., Lee B.H., Lee J.J., Kang K.Y. Proteomic analysis of pathogen- responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics. (2004);4:3569–3578. doi: 10.1002/pmic.200400999. [DOI] [PubMed] [Google Scholar]
- 20.Kurepa J., Walker J.M., Smalle J., Gosink M.M., Davis S.J., Durham T.L., Sung D.-Y., Vierstra R.D. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. (2003);278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 21.Lakatos L., Szittya G., Silhavy D., Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO. (2004);23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laser H., Bongards C., Schuller J., Heck S., Johnsson N., Lehming N. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc. Natl. Acad. Sci. USA. (2000);97:13732–13737. doi: 10.1073/pnas.250400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J., Nam J., Park H.C., Na G., Miura K., Jin J.B., Yoo C.Y., Baek D., Kim D.H., Jeong J.C., et al. Salicylic acidmediated innate immunity in Arabidopsis is regulated by SIZ SUMO E3 ligase. Plant J. (2007);49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 24.Lois L.M., Lima C.D., Chua N.H. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell. (2003);15:1347–1359. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao Y., Desai S.D., Liu L.F. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem. (2000);275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 26.Matunis M.J., Coutavas E., Blobel G. A novel ubiquitin- like modification modulates the partitioning of the Ran- GAPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. (1996);135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melchior F. SUMO-nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. (2000);16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 28.Meulmeester E., Melchior F. Cell biology: SUMO. Nature. (2008);452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 29.Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2010);107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minty A., Dumont X., Kaghad M., Caput D. Covalent modification of p73α by SUMO-1. J. Biol. Chem. (2000);275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 31.Miura K., Hasegawa P.M. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trend Cell Biol. (2010);20:223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A.S., Raghothama K.G., Baek D., Koo Y.D., Jin J.B., Bressan R.A., et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA. (2005);102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.-J., Hasegawa P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. (2007);19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA. (2009);106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novatchkova M., Budhiraja R., Coupland G., Eisenhaber F., Bachmair A. SUMO conjugation in plants. Planta. (2004);220:1–8. doi: 10.1007/s00425-004-1370-y. [DOI] [PubMed] [Google Scholar]
- 36.O’Farrell P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. (1975);250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 37.Okada S., Nagabuchi M., Takamura Y., Nakagawa T., Shinmyozu K., Nakayama J.-I., Tanaka K. Reconstitution of Arabidopsis thaliana SUMO pathway in E. coli: functional evaluation of SUMO machinery proteins and mapping of sumoylation sites by mass spectrometry. Plant Cell Physiol. (2009);50:1049–1061. doi: 10.1093/pcp/pcp056. [DOI] [PubMed] [Google Scholar]
- 38.Panse V.G., Hardeland U., Werner T., Kuster B., Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. (2004);279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 39.Park H.C., Kim M.L., Kang Y.H., Jeong J.C., Cheong M.S., Choi W., Lee S.Y., Cho M.J., Kim M.C., Chung W.S., et al. Functional analysis of the stress-inducible soybean calmodulin isoform-4 (GmCaM-4) promoter in transgenic tobacco plants. Mol. Cells. (2009);27:475–480. doi: 10.1007/s10059-009-0063-6. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO- 2/3. J. Biol. Chem. (2000);275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 41.Saracco S.A., Miller M.J., Kurepa J., Vierstra R.D. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. (2007);145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt D., Müller S. PIAS/SUMO: new partners in transcriptional regulation. Cell. Mol. Life Sci. (2003);60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seeler J.S., Dejean A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. (2003);4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 44.Stagljar I., Korostensky C., Johnsson N., te Heesen S. A new genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA. (1998);95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens R., Mariconti L., Rossignol P., Perennes C., Cella R., Bergounioux C. Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J. Biol. Chem. (2002);277:32978–32984. doi: 10.1074/jbc.M205125200. [DOI] [PubMed] [Google Scholar]
- 46.van den Burg H.A., Kini R.K., Schuurink R.C., Takken F.L.W. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. (2010);22:1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA. (1996);93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wykoff D.D., O’Shea E.K. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics. (2005);4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Yoo J.H., Park C.Y., Kim J.C., Heo W.D., Cheong M.S., Park H.C., Kim M.C., Moon B.C., Choi M.S., Kang Y.H., et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. (2005);280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]
- 50.Yoo C.Y., Miura K., Jin J.B., Lee J., Park H.C., Salt D.E., Yun D.-J., Bressan R.A., Hasegawa P.M. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. (2006);142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y., Kwon S.W., Anselmo A., Kaur K., White M.A. Broad spectrum identification of cellular small ubiquitinrelated modifier (SUMO) substrate proteins. J. Biol. Chem. (2004);279:20999–21002. doi: 10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 52.Zhou W., Ryan J.J., Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae: induction of protein sumoylation by cellular stresses. J. Biol. Chem. (2004);279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]


