Abstract
Since the identification of BRI1-Associated receptor Kinase 1 (BAK1), a member of the Somatic Embryogenesis Receptor Kinase (SERK) family, the dual functions of BAK1 in BR signaling and innate immunity in Arabidopsis have attracted considerable attention as clues for understanding developmental processes that must be balanced between growth and defense over the life of plants. Here, we extended our research to study cellular functions of OsSERKs in rice. As it was difficult to identify an authentic ortholog of AtBAK1 in rice, we generated transgenic rice in which the expression of multiple OsSERK genes, including OsBAK1, was reduced by OsBAK1 RNA interference. Resulting transgenic rice showed reduced levels of OsBAK1 and decreased sensitivity to BL, leading to semidwarfism in overall growth. Moreover, they resulted in abnormal growth patterns, especially in leaf development. Most of the OsBAK1RNAi transgenic rice plants were defective in the development of bulliform cells in the leaf epidermal layer. They also showed increased expression level of pathogenesis-related gene and enhanced susceptibility to a rice blast-causing fungal pathogen, Magnaporthe oryzae. These results indicate that OsSERK genes, such as OsBAK1, play versatile roles in rice growth and development.
Keywords: BR signaling, bulliform cell, Magnaporthe Oryzae, OsBAK1, OsSERKs, RNA interference
INTRODUCTION
SERK proteins, which are members of one of the subfamilies of leucine rich-repeat receptor-like kinases (LRR-RLKs), are involved in somatic embryogenesis, an artificial stimulation of natural embryogenesis (Toonen et al., 1993). Arabidopsis SERK proteins are encoded by a gene family consisting of five close homologs (AtSERK1-5) and are important not only as molecular markers in somatic embryogenesis but also because of their pleiotropic roles in plant development (Albrecht et al., 2008; Hecht et al., 2001). In these respects, BAK1 has received the most attention since identified as a co-receptor of BRI1, part of the plasma membrane localized brassinosteoroid (BR) receptor that mediates BR signaling independent of somatic embryogenesis (Li et al., 2002; Nam and Li, 2002). BRs play critical roles in various aspects of plant growth and development, including germination, photomorphogenic responses, cell elongation, xylem differentiation, and male fertility (Bajguz, 2007; Kim et al., 2009). BAK1 was found to be identical to AtSERK3, and it is now known that other BAK1 homologs also belong to AtSERK family, including AtSERK2/BAK2, AtSERK4/BAK7, and AtSERK5/ BAK8 (Jeong et al., 2010). AtSERK1 and At- SERK2 have been pulled down in complexes with BRI1 in total protein fractions from Arabidopsis (Karlova et al., 2006), and AtSERK4 rescues bri1-5 and bri1-301 mutants (Albrecht et al., 2008), further indicating that these proteins are involved in BR signaling and are redundant with BAK1/AtSERK3.
SERK proteins in various species have been reported to play roles in plant innate immunity and senescence, too. BAK1 can form a complex with FLS2, an another LRR-RLK, initiating PAMP triggered immunity (PTI) with flagellin binding, and the bak1 mutant showed reduced responsiveness to flagellintriggered immunity (Chinchilla et al., 2007). BAK1-defecient plants exhibited broader necrotic region upon microbial infection, accompanied by increase of ROS production (Kemmerling et al., 2007). Interestingly, a different set of genes seem to be engaged in BAK1-mediated BR signaling and innate immunity, as genes induced by brassinolide (BL) do not overlap with the genes induced by infection (Kemmerling et al., 2007). Double mutant plants lacking BAK1 and BKK1/AtSERK4/BAK7 displayed a spontaneous cell death phenotype with constitutive defense gene expression (He et al., 2007), and transgenic plants expressing BAK7 RNA interference (RNAi) constructs showed early senescence with premature expression of senescence- associated genes (Jeong et al., 2010), suggesting additional function of SERK family proteins. Taken together, the pleiotropic functions of BAK1 open new possibilities for genetic engineering, because manipulation of a single gene such as BAK1 may result in the production of plants with both higher yield and improved disease resistance.
Although most of studies on BAK1 have been in Arabidopsis, studies of SERK proteins, including an ortholog of AtBAK1, are currently being extending to monocotyledonous plants. The first SERK gene in rice, OsSERK1, was screened from the rice cDNA library using a rice EST-clone probe derived from the amino acid sequence of DcSERK1 as a query (Ito et al., 2005), and it was also independently identified as a gene induced by the infection of Magnaporthe grisea (Hu et al., 2005). OsSERK2 was found in the data-base using OsSERK1 as a query (Ito et al., 2005). OsSERK1 shows more than 85% homology with OsBISERK1, benzothiadiazole (BTH)-induced SERK1 (Song et al., 2008). In addition to OsSERK1 and OsSERK2, OsSERK3 and OsSERK4 were also identified as homologs of BAK1/At- SERK3. The OsSERK1-4 genes all rescued bri1-5 mutant phenotypes, indicating that these proteins are functionally redundant with AtSERKs, including BAK1. Because the degree of phenotypic rescue of bri1-5 was highest with OsSERK1, OsSERK1 is believed to be the functional Arabidopsis BAK1/AtSERK3 ortholog in rice and has been renamed OsBAK1 (Li et al., 2009). However, phylogenetic analysis shows that OsSERK1/OsBAK1 is an ortholog of AtSERK1, not BAK1/AtSERK3. Moreover, recent sequence analysis has revealed nine more SERL (SERK-like) genes in rice (Singla et al., 2009). Therefore, it has been suggested that gene duplication occurred in genes encoding OsSERKs after the evolutionary divergence of monocots and dicots. As there are still many annotated partial cDNAs encoding only kinase domains that are very similar to the kinase domain of OsSERKs and OsSERLs in rice, it may be difficult to gain insight into the specific functions of each OsSERK gene.
In this study, our aim was to functionally characterize OsSERKs. Because we could not determine which gene is the true ortholog of AtBAK1 in rice, we generated transgenic rice transformed with OsBAK1RNAi construct to investigate the function of the OsSERK family, including OsBAK1. We found that most of the OsBAK1RNAi transgenic plants showed defective development of bulliform cells in the leaf epidermal layer and increased susceptibility to the rice blast-causing fungal pathogen Magnaporthe oryzae. These results indicate that a subset of OsSERK genes, including OsBAK1, play multiple roles in growth and development as well as in defense signaling in rice.
MATERIALS AND METHODS
Plant materials and growth conditions
Rice seeds (Oryza sativa, japonica cv. Nakdong) were used for transformation of all the genes in this study. Transgenic rice plants were grown in square containers with Murashige and Skoog (Duchefa) agar medium in growth chambers set with 16 h/8 h (L/D) photoperiod at 28℃/25℃ and 80% relative humidity with/without phosphinothricin selection. Then, 15-day-grown plants were transferred to large containers with rice paddy soil and grown in the same conditions or under sunlight.
Plasmid constructions and rice transformation
To generate the OsBAK1RNAi construct, a 250-bp genomic fragment that encodes the kinase domain of OsSERK1 was PCR-amplified using primer pairs of RBAK1CF/RBAK1BR and RBAK1KF/RBAK1XR to fit in the RNA interference cassette pHannibal. When designing the primers, we added NotI and XmaI to the RBAK1BR and RBAK1XR, respectively, to clone them into pSB505 binary vector later. The resulting plasmid, pHannibal-OsBAK1RNAi, was digested with NotI and XmaI and ligated into pSB505 binary vector to generate pSB505- OsBAK1RNAi. All fragments produced by PCR were confirmed by sequencing. Detailed primer sequences are listed in Supplementary Table 1.
Table 1.
Comparison of amino acids homologies among SERK and SERK-like proteins in rice and Arabidopsis
| OsSERK1 | OsSERK2 | OsSERK3 | OsSERK4 | OsSERL1 | OsSERL2 | OsSERL3 | OsSERL4 | OsSERL5 | OsSERL6 | OsSERL7 | OsSERL8 | OsSERL9 | OsBISERK1 | AtSERK1 | AtSERK2 | AtSERK3 | AtSERK4 | AtSERK5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OsSERK1 | 100 | 87 | 60 | 53 | 46 | 26 | 30 | 45 | 52 | 60 | 9 | 56 | 53 | 99 | 83 | 85 | 77 | 72 | 68 |
| OsSERK2 | 100 | 58 | 54 | 46 | 28 | 31 | 47 | 52 | 58 | 6 | 55 | 54 | 87 | 82 | 81 | 77 | 72 | 67 | |
| OsSERK3 | 100 | 42 | 41 | 25 | 29 | 38 | 44 | 100 | 5 | 43 | 42 | 60 | 57 | 59 | 56 | 55 | 52 | ||
| OsSERK4 | 100 | 56 | 23 | 27 | 42 | 48 | 42 | 11 | 87 | 99 | 53 | 53 | 52 | 49 | 48 | 47 | |||
| OsSERL1 | 100 | 25 | 25 | 40 | 44 | 41 | 9 | 57 | 56 | 46 | 46 | 45 | 45 | 43 | 43 | ||||
| OsSERL2 | 100 | 61 | 29 | 33 | 25 | 10 | 23 | 23 | 26 | 29 | 26 | 26 | 23 | 23 | |||||
| OsSERL3 | 100 | 34 | 42 | 29 | 7 | 22 | 27 | 30 | 31 | 30 | 30 | 30 | 26 | ||||||
| OsSERL4 | 100 | 54 | 38 | 10 | 45 | 42 | 45 | 45 | 44 | 44 | 42 | 42 | |||||||
| OsSERL5 | 100 | 44 | 7 | 50 | 48 | 52 | 52 | 51 | 49 | 48 | 48 | ||||||||
| OsSERL6 | 100 | 5 | 43 | 42 | 60 | 57 | 59 | 56 | 55 | 52 | |||||||||
| OsSERL7 | 100 | 8 | 11 | 9 | 6 | 9 | 8 | 9 | 2 | ||||||||||
| OsSERL8 | 100 | 86 | 55 | 55 | 55 | 52 | 51 | 49 | |||||||||||
| OsSERL9 | 100 | 53 | 52 | 52 | 49 | 48 | 47 | ||||||||||||
| OsBISERK1 | 100 | 83 | 84 | 77 | 71 | 67 | |||||||||||||
| AtSERK1 | 100 | 89 | 77 | 73 | 67 | ||||||||||||||
| AtSERK2 | 100 | 77 | 74 | 69 | |||||||||||||||
| AtSERK3 | 100 | 81 | 78 | ||||||||||||||||
| AtSERK4 | 100 | 85 | |||||||||||||||||
| AtSERK5 | 100 | ||||||||||||||||||
Full length of amino acids sequences of OsSERK1 (624 aa), OsSERK2 (628 aa), OsSERK3 (616 aa), OsSERK4 (607 aa), OsSERL1 (608 aa), OsSERL2 (644 aa), OsSERL3 (324 aa), OsSERL4 (620 aa), OsSERL5 (628 aa), OsSERL6 (616 aa), OsSERL7 (453 aa), OsSERL8 (543 aa), OsSERL9 (607 aa), OsBISERK1 (624 aa), AtSERK1 (625 aa), AtSERK2 (628 aa), AtSERK3 (615 aa), AtSERK4 (620 aa), and AtSERK5 (601 aa) were analyzed by pair-wise alignment using the CLUSTAL 2.1 Multiple Sequence Alignments. Amino acid sequences of OsSERK1-OsSERK4 were pulled out Li et al. (2009), and those of OsSERL1-OsSERL9 were from Single et al. (2009). All other amino acid sequences obtained from the data base NCBI.
All constructs including the pSB505 vector plasmid alone were introduced into wild-type rice (Oryza sativa, japonica cv. Nakdong) by Agrobacterium-mediated transformation as described by Hiei et al. (1994).
Expression by RT-PCR analysis
RNA was purified from the frozen tissues with extraction buffer (0.2 M Tris-Cl, pH 9.0, 0.4 M LiCl, 25 mM EDTA, 1% SDS) and aqua phenol (Q-biogene), and treated with RNase-free RQ1 DNase (Promega). First-strand cDNA synthesis was performed with SuperscriptIII-MMLV reverse transcriptase (Invitrogen) using oligo d(T15) as a primer. The same aliquot of first-strand cDNA was then used as a template for the second PCR step using Real Taq polymerase (RBC). The sequences of the primers and use of them are described in Supplementary Table 1.
Lamina joint assay
Imbibed seeds were sown on plates containing 1/2 MS media (100 mg/L myo-inositol, 125 mg/L cefotaxime, 0.6% agar, pH 5.6) and grown for 6 days described as above. Then, 1 μl of 70% ethanol containing 0, 1, 10, or 100 ng of BL was applied to the lamina joint of the second leaf. After incubation for 3 days, the bending angles between the leaf blade and leaf sheath were measured.
Treatment of rice blast fungus
Inoculation with Magnaporthe oryzae was performed as described (Park et al., 2009). The M. oryzae strain KJ201 was obtained from the Center for Fungal Genetic Resources. Conidia were harvested from fungal cultures grown on V8 agar medium (V8 80 ml/L, agar 15 g/L, pH 6.5) for 7 days under constant fluorescent light at 25℃. After counting the number of conidia in a small aliquot of conidial suspension using a hemocytometer, the concentration was adjusted to 4 × 104 spores/ml. The final conidial suspensions were sprayed onto the young leaves of transgenic plants grown for 7 weeks. Infected plants were incubated in a chamber with 100% humidity in the dark for 24 h and then, were placed in a growth chamber with normal conditions. Leaf samples were analyzed after four days.
Microscopic observation
A stereo microscope (Leica MZ 12.5) and Image Manager 5.0 software were used to observe the surfaces and internal structure of the leaves of transgenic plants.
RESULTS
Reduced expression of OsBAK1 by RNA interference caused inhibition of growth, abnormal development, and loss of BL sensitivity in rice
As there are no reports of natural mutants for OsBAK1, we searched for AtBAK1 orthologs among the rice genes that were reported to be as OsSERKs or OsSERLs in the public database using the amino acid sequence of AtBAK1 as a query. It was difficult to determine which gene was an exact AtBAK1 ortholog, because many similar genes were identified. OsSERKs and OsBISERK1 cluster together, closer to each other than to any other AtSERKs, based on amino acid sequence homology (Fig. 1A). However, OsSERK1, OsSERK2, and OsBISERK1 still showed the considerably high amino acid sequence homology to AtBAK1 (77%) and AtSERK4/AtBAK7 (71-72%). The amino acid sequence homologies between AtBAK1 and other OsSERKs or OsSERLs were in broad ranges (Fig. 1A and Table 1). Therefore, we used an RNA interference approach to knock down OsBAK1 and highly homologous genes simultaneously.
Fig. 1. Sequences and constructs for RNA interference for OsBAK1. (A) Phylogenetic analysis of SERK proteins in Arabidopsis and rice. Amino acid sequences encoded by AtSERK1 (At1g71830), AtSERK2 (At1g34210), AtBAK1/AtSERK3 (At4g33430), AtSERK4/ BAK7/BKK1 (At2g13790), AtSERK5 (At-2g13800) in Arabidopsis and OsSERK1 (Os08g0174700), OsBISERK1 (AY463361), OsSERK2 (Os04g0457800), OsSERK3 (Os- 06g0225300), OsSERK4 (Os02g- 0283800) in rice (Oryza sativa) were used for CLUSTAL 2.1 multiple sequence alignment. Phylogenetic trees were generated by PHYML and MEGA (Guindon and Gascuel, 2003). (B, C) The nucleotide (B) and amino acid (C) sequences of a partial kinase domain encoded by OsSERKs for RNA interference for OsBAK1. Alignments were drawn by the ClustalW program. (D) RT-PCR analysis showing reduced expression of OsBAK1 by OsBAK1RNAi in several independent transgenic lines. (E) RT-PCR analysis showing reduced expression of ORK1 in the OsBAK1RNAi#5 and OsBAK1RNAi#11 transgenic rice plants.
To clone the OsBAK1 RNAi construct, we first amplified a 250-bp fragment encoding part of the OsBAK1 kinase domain based on its ability to rescue bri1-5 (Li et al., 2009). The resulting 250-bp amplified region was identical to the corresponding region of OsSERK1 and OsSERK2 (Fig. 1B). Another three genes, OsBISERK1, Os08g0176200, and ORK1, showed 99%, 93%, and 93% identity in the corresponding regions, respectively (Fig. 1C). We inserted this fragment into the pSB505 binary vector with an inverted position to generate pSB505-OSBAK1RNAi and transformed into rice (Supplementary Fig. 1). We obtained 43 primary transformants in the T0 generation. Even in this T0 generation, most of the transgenic plants showed abnormal growth patterns, both in vegetative and reproductive growth. These disrupted growth patterns reappeared in the next generation (data not shown). We performed RTPCR analysis from the RNA of several independent T1 transgenic lines. While the expression level of OsBAK1 in OsBAK1RNAi #11 plants was similar to that of control plant containing empty vector plasmid, the OsBAK1 expressions level of OsBAK1RNAi #5, #7, and #14 plants were dramatically reduced (Fig. 1D). We selected two lines, one showing approximately normal OsBAK1 expression, #11, and another showing greatly reduced expression of OsBAK1, #5, for further analyses. In these two lines of transgenic plants, we tried to confirm the down-regulation of other homologous genes, OsBISERK1 and ORK1. However, it is very hard to design the primer set of OsBISERK1 due to high nucleotide sequence homology to OsSERK1. Only ORK1 was confirmed to be inhibited its expression in both transgenic rice plants (Fig. 1E).
Although there were some variations in growth, growth in most of the OsBAK1RNAi #11 plants were not much reduced in several criteria including total height and leaf elongation (Figs. 2A-2C). However, many OsBAK1RNAi #5 plants exhibited broad ranges of growth defects, severe to mild (Fig. 2A). Throughout the vegetative growth period, OsBAK1RNAi #5 transgenic plants displayed semi-dwarf stature, with fewer internodes than control plants containing the empty vector (Figs. 2A and 2B). Leaf elongation was clearly reduced in the OsBAK1RNAi #5 transgenic plants. However, leaf width of the OsBAK1RNAi #5 leaves showed no significant reduction compared with that of controls, but clearly wider than that of OsBAK1RNAi #11 plants (Fig. 2C). In addition, we observed several other unusual phenotypes in the OsBAK1RNAi #5 line, including corrugated leaves at the early stage of growth (Figs. 2D and 2L). Most of the other leaves of the OsBAK1RNAi #5 plants were dry and twisted (Figs. 2E and 2F), and the leaves were easily bent (Fig. 2H). We often observed that parts of the blades in the whole leaves were not fully opened and that side edges of leaf were confronted and fused together (Figs. 2G and 2I). We also observed abnormal plait-like lumps in the stem, which may indicate a failure in new leaf initiation (Figs. 2J and 2K). These abnormally looking phenotypes in leaves were not only limited in the OsBAK1RNAi #5 plants. Although the degree of them were weaker than in OsBAK1RNAi #5 plants, many of the leaves of the OsBAK1RNAi #11 plants also displayed dry and twisted as well as easily bent leaf shape (data not shown).
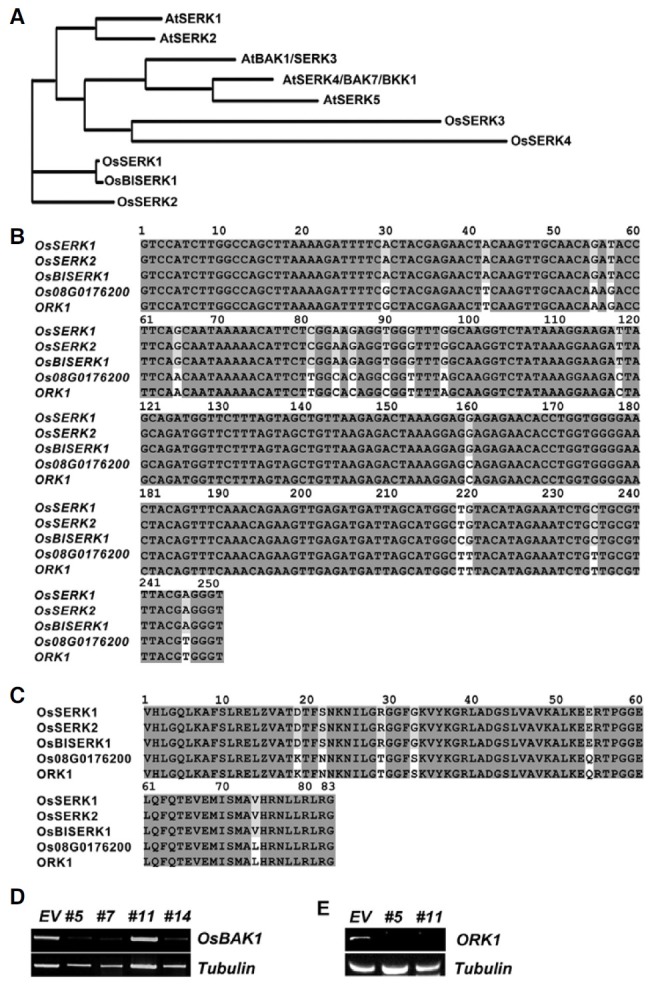
Fig. 2. Morphological analyses of transgenic plants expressing the OsBAK1RNAi construct. (A) Gross morphology of transgenic plants grown for 6 weeks in the greenhouse. EV empty vector containing transgenic plant, #11 OsBAK1RNAi #11 plant, and three representative OsBAK1RNAi #5 transgenic plants (#5-1, #5-9, and #5- 10) showing different degrees of growth defects. (B) Plant height measured at the 4th week and 6th week of growth. (C) The length and width of each leaf were measured in the transgenic plants grown for 4 weeks. Data presented in (B) and (C) are averages for four different plants in each specific line. In (B) and (C), statistical test was performed by ANOVA analysis and significance in differences was indicated by P value (*; P≤0.01, **; P≤ 0.05, and ***; P ≤0.005). (DL) Abnormal leaf features seen in the OSBAK1RNAi #5 plants. Corrugated leaves appeared at an early stage (D) and remained through later stages of development, as well as when grown in tissue culture media (L). Dried and twisted leaves (E, F), folded or fused leaf blades (G, I), bent leaves (H), and plait-like lumps in the nodes of stem (J, K) were also frequently observed.
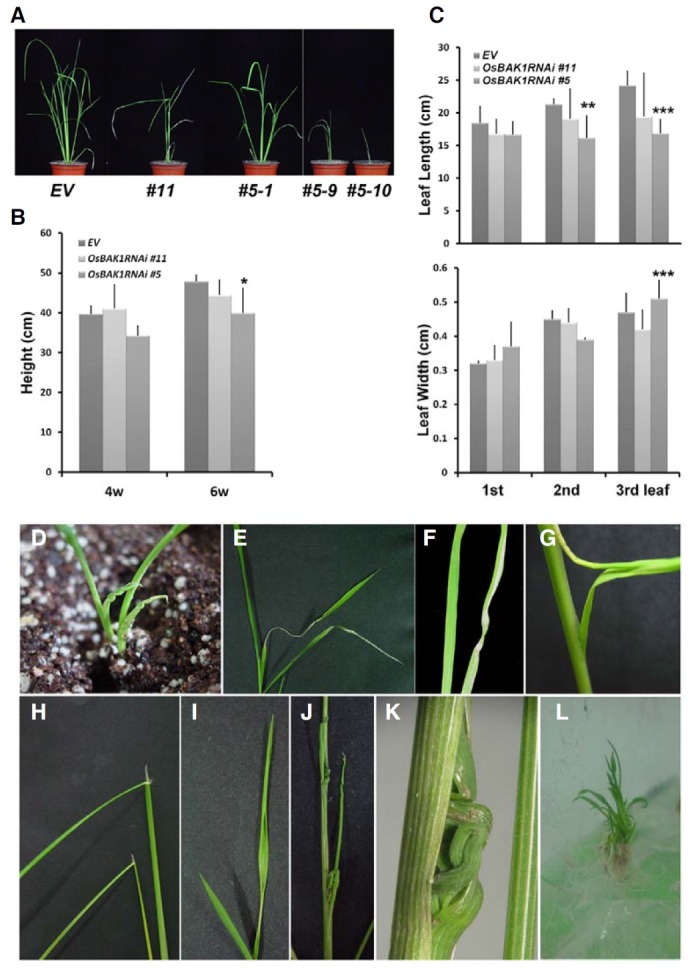
We next examined whether reduced OsBAK1 expression caused decreased sensitivity to BL in OsBAK1RNAi transgenic rice (Fig. 3). We observed that the OsBAK1RNAi #11 plants exhibited little bit less degrees of laminar inclination to the control plants with the increasing concentrations of BL. On the contrary, we detected little change in the angle even at high concentrations of BL in the OsBAK1RNAi #5 plants, indicating that these plants had dramatically reduced sensitivity to BL. Taken together, these results suggest that the reduced expression of OsBAK1 led to phenotypic alterations, including inappropriate leaf development, growth inhibition over the whole life time of rice plants, and BL insensitivity.
Fig. 3. BL responsiveness of OsBAK1RNAi transgenic plants. (A) Lamina joint assay. The 1 μl of BL containing indicated amount was applied to the lamina joint of the second leaf of each transgenic plant. Pictures were taken after three days. (B) Quantification of lamina joint assays in (A). The experiment was repeated four times.
Reduced OsBAK1 expression caused impairment in leaf development and increased susceptibility to biotic stress in rice
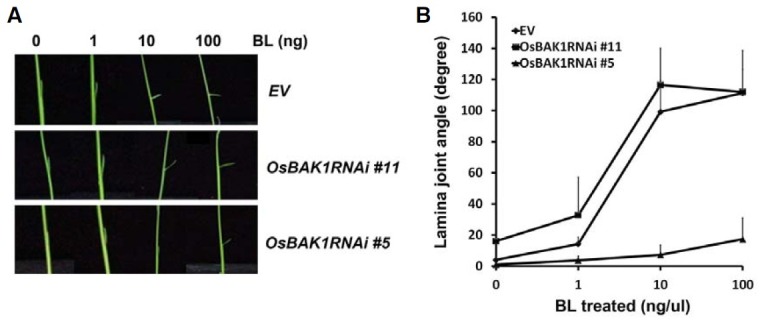
Abnormal leaf development and growth were distinct phenotypes in most of the transgenic plants expressing the OsBAK1RNAi construct, including the #5 lines. We next focused on characterizing the effect of OsBAK1 knockdown leaves. We first examined the surface of the OsBAK1RNAi #5 leaves using a stereomicroscope, and observed that the color of the leaves was pale green with dried, yellowish regions and necrotizing spots, which were not observed on the surface of wild-type rice leaves (Fig. 4A). We also examined the internal leaf structure by performing transverse sectioning of the leaves of wild-type and OsBAK1RNAi #5 plants. The leaves of the OsBAK1RNAi #5 were much thicker and contained more mesophyll cells which extended in the abaxial side. The aerenchyma in the leaves of OsBAK1RNAi transgenic plants was also larger than in wild-type plants. Moreover, whereas the flanking sides between the longitudinal veins were flat in wild-type leaf blades, the blades of the OsBAK1RNAi #5 leaves were not fully opened. This feature explained why we observed that parts of the blades in the OsBAK1RNAi #5 leaves were not fully opened (Figs. 2H and 2J). Interestingly, we noticed that there were no bulliform cells in the adaxial surface of the OsBAK1RNAi #5 leaves, unlike the wild-type leaves (Fig. 4B). Bulliform cells are larger than typical epidermal cells and are colorless, because they contain highly developed vacuoles and no chloroplasts (Becraft, 1999). The appropriate distribution of bulliform cells is known to be important for leaf curling in monocot plants (Becraft et al., 2002; Zhang et al., 2009).
Fig. 4. Defective leaf development of OsBAK1RNAi #5 plants. (A) Surface of leaves of an OsBAK1RNAi #5 plants (right) and control plant (left) grown for four weeks. Pictures in the lower panel show higher magnification of the boxed region. White bars indicate 300 μm. (B) Internal structures of leaves of an OsBAK1RNAi #5 plant (right) and a wild-type plant (left) grown for five weeks. Leaves containing veins were transversely sectioned by hand and observed under a compound microscope. Bulliform cells shown in wild-type plants are marked with arrow heads. Bar indicates 500 μm.
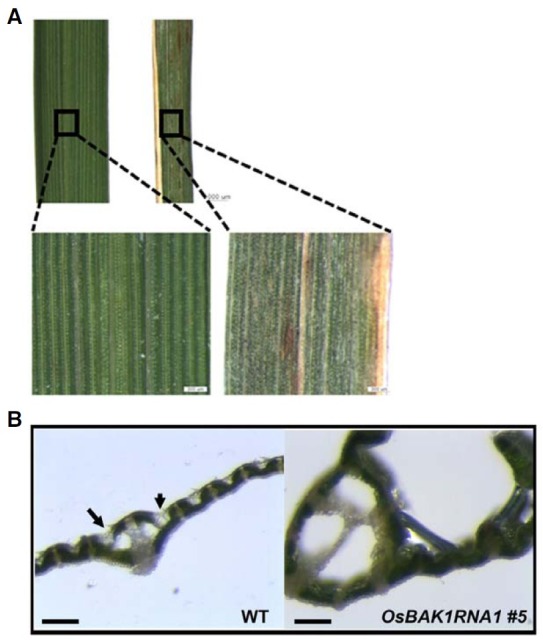
Furthermore, we observed that the leaves of OsBAK1RNAi #5 transgenic plants contained several necrotic spots, reminiscent of being infected, even under normal growth conditions (Fig. 5A). We cannot exclude the possibility that excessive dryness of the leaves, as shown in Fig. 4A, was the ultimate cause of this phenotype. However, it is also possible that OsBAK1RNAi #5 transgenic rice is less resistant to infections that would be easily overcome under normal conditions by plants. To test this we examined basal expression level of PR-1a, one of the marker genes of the defense for pathogenesis in rice as well as in Arabidopsis (Park et al., 2009). We found that the expression of PR-1a was greatly increased in the leaves of OsBAK1RNAi #5 plants, while it was similar in the leaves of OsBAK1RNAi #11 plants compared in wild type plants (Fig. 5B). To continuously know the defense capacity of the rice to disease- causing pathogen in vivo, we sprayed a conidial suspension of Magnaporthe oryzae, known as rice blast fungus onto OsBAK1RNAi #5 and control transgenic rice. At five days after infection, we observed that the rice blast disease had been spread more severely throughout the leaves and culms in OsBAK1RNAi #5 plants compared to the control plants, indicating that the reduced level of OsBAK1 also affects plant immunity (Fig. 5C). Taken together, these results suggested that reduced OsBAK1 expression caused increased susceptibility to biotic stress in rice as well as impairment in leaf development.
Fig. 5. Higher susceptibility to biotic stress of leaves of OsBAK1RNAi #5 plants. (A) The outer appearance of OsBAK1RNAi #5 line leaves (right) showing constitutive sickness compared with those of control plants (left) grown for two weeks. (B) RTPCR analysis showing increased expression of PR-1a in the OsBAK1RNAi transgenic lines. (C) Extent of OsBAK1RNAi #5 line (right) infection with Magnaporthe oryzae compared to wild-type plant (left) grown for 7 weeks. Enlarged leaf features of them are shown in the lower panel.

DISCUSSION
It seems that more OsSERKs and OsSERLs were generated by gene duplication after the evolutionary divergence of dicots and monocots. Although we cannot exclude the possibility that each gene has specific functions in the developmental process of rice, it is possible that each gene functions redundantly with other genes, making it difficult to investigate the specific roles of each gene by mutant analysis. The RNA interference approach to knock-down several related-genes together is often the most effective strategy in such studies (Blokland et al., 1994; Ecker and Davis, 1986). In this investigation, more than three OsBAK1-related genes, including OsSERK1, OsSERK2, and OsBISERK1, were targeted by our RNAi construct (Fig. 1). The resulting OsBAK1RNAi transgenic rice plants with reduced levels of OsBAK1 (Fig. 1D) exhibited intermediate growth retardation compared to strong BR-biosynthetic mutants, such as brd1-1 and brd1-2 (Hong et al., 2002). Similarly, BR-biosynthetic or BR-signaling mutants, such as d2, d11, brd2, and d61 display mild dwarfism in the vegetative stage but are still fertile in reproductive development (Jeon et al., 2010). These results indicate that a subset of OsSERK genes, including OsBAK1 is important for proper growth in rice mediated by BR signaling.
Another interesting feature of the OsBAK1RNAi transgenic plants is abnormal leaf development. Leaf morphology is one of the important parameters of rice architecture. In addition to growth inhibition, several abnormalities in leaf development (Figs. 2E-2L), was reported in the d1/d61 double mutant, in which rice Gα (RGA1) and OsBRI1 are defective, whereas this finding was not observed in the d61 single mutant (Oki et al., 2009). These data indicate that the reduced expression of OsBAK1 may modulate the expression of RGA1. Fused blades in several regions of a leaf, very dried and yellowish parts, and a constitutively wilted appearance, were often observed in the leaves of the OsBAK1RNAi transgenic plants. These defects could be partly attributable to the lack of bulliform cells as shown in Fig. 4B. Although there is some controversy about the exact function of bulliform cells in the grass family, their rapid expansion during a specific stage of leaf development has been proposed to be responsible for the unfolding of the leaf blade (Alvarez et al., 2008). The size and placement of bulliform cells are important factors that determine the direction of leaf curling. Usually, bulliform cells exist mainly in the adaxial surface of the leaves and are larger than neighboring epidermal cells. Increased numbers of bulliform cells in the abaxial side of sll1 (shallot-like1) mutant leaves and smaller adaxial bulliform cells in the leaves of nr7 (narrow leaf7) and nrl1-1 (narrow and rolled leaf 1) mutants have been shown to affect adaxial leaf curling (Fujino et al., 2008; Hu et al., 2010; Zhang et al., 2009). In comparison, overexpression of ACL1 (abaxially curled leaf 1) produced more and larger bulliform cells, leading to the abaxial leaf curling (Li et al., 2010). Therefore, it is possible that the fused and confronted leaf blades in the OsBAK1RNAi transgenic plants represent an extreme case, in which the lack of bulliform cells in the adxial side of the leaf, led to tight folding of leaf blade. In mature leaves, bulliform cells can act as motors adjacent to the mid-vein of leaf through changes in turgor. This mechanism provides plants with an advantage in resisting environmental stresses, especially drought and heat conditions. The constitutively wilted leaves of OsBAK1RNAi transgenic plants may be due to malfunction of bulliform cells at maturity.
In Arabidopsis, spontaneous cell death phenotypes were observed only in the double mutant plants lacking both BAK1 and BKK1/AtSERK4/BAK7 (He et al., 2007), or transgenic plants expressing BAK7 RNA interference (RNAi) constructs (Jeong et al., 2010). There are no reports about these kinds of phenotypes in single mutants of BAK1 or BKK1/AtSERK4/BAK7. In this sense, several necrotic spots in the leaves of OsBAK1RNAi #5 transgenic plants in normal condition (Fig. 5A) might be caused by the reduced expression of OsBAK1 and its homologs together through the RNA interference, resulting in higher level of basal of PR-1a expression (Fig. 5B). There are many reports to show induced expressions of pathogenesisrelated (PR) genes upon various stressful stimuli including infection of pathogens, wounding, and treatment of hormones, such as salicylic acid or jasmonate in rice as well as in Arabidopsis (Mitsuhara et al., 2008). However, no consistent correlation exists whether the higher expression of PR gene means the plants to have higher capacity to overcome these stresses or not. In our result, OsBAK1RNAi #5 transgenic rice plants, showing increased level of PR-1a, exhibited increased susceptibility to infection by M. oryzae KJ201 (Fig. 5C). The fungal pathogen M. oryzae uses a hemibiotrophic strategy for initial infection and proliferation and then switches to necrotrophy, resulting in necrotic lesions on the infected rice leaves (Park et al., 2009). Similar phenomena were reported in many cases. In Arabidopsis, although bak1 mutant showed increased expression of pathogen infection and senescence-related genes, such as PR1, PR2, PR5, Cf-like, and PTR3, it also showed enhanced susceptibility to necrotropic fungal infection (Kemmerling et al., 2007). It was also reported that lesion-like necrosis along the central leaf vein was developed in the transgenic rice overexpressing (At)NPR1 in which PR-1b transcript abundance was increased (Fitzgerald et al., 2008).
OsSERK1 has been independently isolated from a rice cDNA that was inducible by blast fungus infection. Expression of OsSERK1 is also activated by defense molecules, including salicylic acid, jasmonic acid, and abscisic acid (Hu et al., 2005). Another homolog, OsBISERK1, identified as a BTH-inducible gene is also induced by M. grisea (Song et al., 2008). Lettuce plants, in which the LsSERK was knocked down by posttranscriptional gene silencing, were also reported to have reduced somatic embryogenesis ability as well as increased susceptibility to Sclerotinia (Santos et al., 2009). These studies and our current findings raise the possibility of expanding the range of OsSERK gene function to include responses to biotic and abiotic stresses, as well as developmental processes, such as somatic regeneration and leaf cell specification.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, grant #: PJ008003), Rural Development Administration, Republic of Korea, and by the SRC Research Center for Women’s Diseases of Sookmyung Women’s University (2009) (grant # 3-0903-0022 to K.H.N.).
References
- 1.Albrecht C., Russinova E., Kemmerling B., Kwaaitaal M., de Vries S.C. Arabidopsis somatic embryogenesis receptor kinase proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. (2008);148:611–619. doi: 10.1104/pp.108.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez J.M., Rocha J.F., Machado S.R. Bulliform cells in Loudetiopsis chrysothrix (Nees) Conert and Tristachya leiostachya Nees (Poaceae): structure in relation to function. Braz. Arch. Biol. Technol. (2008);51:113–119. [Google Scholar]
- 3.Bajguz A. Metabolism of brassinosteroids in plants. Plant Physiol. Biochem. (2007);45:95–107. doi: 10.1016/j.plaphy.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Becraft P.W. Development of the leaf epidermis. Curr. Top. Dev. Biol. (1994);45:1–40. doi: 10.1016/s0070-2153(08)60313-0. [DOI] [PubMed] [Google Scholar]
- 5.Becraft P.W., Li K., Det N., Asuncion-Crabb Y. The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development. (2002);129:5217–5225. doi: 10.1242/dev.129.22.5217. [DOI] [PubMed] [Google Scholar]
- 6.Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., Jones J.D., Felix G., Boller T. A flagellininduced complex of the receptor FLS2 and BAK1 initiates plant defense. Nature. (2007);448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 7.Ecker J.R., Davis R.W. Inhibition of gene expression in plant cells by expression of antisense RNA. Proc. Natl. Acad. Sci. USA. (1986);83:5372–5376. doi: 10.1073/pnas.83.15.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald H.A., Chern M.S., Navarre R., Ronald P.C. Overexpression of (At)NPR1 in rice leads to a BTH-and environmental- induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. (2008);17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- 9.Fujino K., Matsuda Y., Ozawa K., Nishimura T., Koshiba T., Fraaije M., Sekiguchi H. NARROWLEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genomics. (2008);279:499–507. doi: 10.1007/s00438-008-0328-3. [DOI] [PubMed] [Google Scholar]
- 10.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. (2003);52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 11.He K., Gou X., Yuan T., Lin H., Asami T., Yoshida S., Russell S.D., Li J. BAK1 and BKK1 regulate brassinosteroid- dependent growth and brassinosteroid-independent celldeath pathways. Curr. Biol. (2007);17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 12.Hecht V., Veille-Calzada J.P., Hartog M.V., Schmidt E.D., Boutilier K., Grossniklaus U., de Vries S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. (2001);127:803–816. [PMC free article] [PubMed] [Google Scholar]
- 13.Hiei Y., Ohta S., Komari T., Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. (1994);6:70–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 14.Hong Z., Ueguchi-Tanaka M., Shimizu-Sato S., Inukai Y., Fujioka S., Shimada Y., Takatsuto S., Agetsuma M., Yoshida S., Watanabe Y., et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. (2002);32:495–508. doi: 10.1046/j.1365-313x.2002.01438.x. [DOI] [PubMed] [Google Scholar]
- 15.Hu H., Xiong L., Yang Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta. (2005);222:107–117. doi: 10.1007/s00425-005-1534-4. [DOI] [PubMed] [Google Scholar]
- 16.Hu J., Zhu L., Gao Z., Guo L., Fang Y., Zhang G., Dong G., Yan M., Liu J., Qian Q. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. (2010);73:283–292. doi: 10.1007/s11103-010-9614-7. [DOI] [PubMed] [Google Scholar]
- 17.Ito Y., Takaya K., Kurata N. Expression of SERK family receptor-like protein kinase genes in rice. Biochim. Biophys. Acta. (2005);1730:253–258. doi: 10.1016/j.bbaexp.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Jeon J.S., Jung K.H., Kim H.B., Suh J.P., Khush G.S. Genetic and molecular insight into the enhancement of rice yield potential. J. Plant Biol. (2011);54:1–9. [Google Scholar]
- 19.Jeong Y.J., Shang Y., Kim B.H., Kim S.Y., Song J.H., Lee J.S., Lee M.M., Li J., Nam K.H. BAK7 displays unequal genetic redundancy with BAK1 in brassinosteroid signaling and early senescence in Arabidopsis. Mol. Cell. (2010);29:259–266. doi: 10.1007/s10059-010-0024-0. [DOI] [PubMed] [Google Scholar]
- 20.Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. (2006);18:625–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemmerling B., Schwedt A., Rodriguez P., Mazzotta S., Frank M., Qamar S.A., Mengiste T., Betsuyaku S., Parker J.E., Mussig C., et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. (2007);17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 22.Kim T.W., Guan S., Sun Y., Deng Z., Tand W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. (2009);11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. (2002);110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 24.Li D., Wang L., Wang M., Xu Y., Liu Y., Xu Z., Li J., Chong K. Engineering of OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotech. J. (2009);7:791–806. doi: 10.1111/j.1467-7652.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- 25.Li L., Shi Z., Li L., Shen G., Wang X., An L., Zhang J. Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant. (2010);3:807–817. doi: 10.1093/mp/ssq022. [DOI] [PubMed] [Google Scholar]
- 26.Mitsuhara I., Iwai T., Seo S., Yanagawa Y., Kawahigasi H., Hirose S., Ohkawa Y., Ohashi Y. Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compound (121/180). Mol. Genet. Genomics. (2008);279:415–427. doi: 10.1007/s00438-008-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam K.H., Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. (2002);110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 28.Oki K., Inaba N., Kitagawa K., Fujioka S., Kitano H., Fujisawa Y., Kato H., Iwasaki Y. Function of the α-subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol. (2009);50:161–172. doi: 10.1093/pcp/pcn182. [DOI] [PubMed] [Google Scholar]
- 29.Park J.Y., Jin J., Lee Y.W., Kang S., Lee Y.H. Rice blast fungus (Magnaporthe oryzae) Infects Arabidopsis via a mechanism distinct from that required for the infection of rice. Plant Physiol. (2009);149:474–486. doi: 10.1104/pp.108.129536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos M.O., Romano E., Vieira L.S., Baldoni A.B., Aragao F.J.L. Suppression of SERK gene expression affects fungus tolerance and somatic embryogenesis in transgenic lettuce. Plant Biol. (2009);11:83–89. doi: 10.1111/j.1438-8677.2008.00103.x. [DOI] [PubMed] [Google Scholar]
- 31.Singla B., Khurana J., Khurana P. Structural characterization and expression analysis of the SERK/SERL gene family in rice (Oryza sativa). Int. J. Plant Genomics. (2009):539402. doi: 10.1155/2009/539402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song D., Li G., Song F., Zheng Z. Molecular characterization and expression analysis of OsBISERK1, a gene encoding a leucine-rich repeat receptor-like kinase, during disease resistance responses in rice. Mol. Biol. Rep. (2008);35:75–283. doi: 10.1007/s11033-007-9080-8. [DOI] [PubMed] [Google Scholar]
- 33.Toonen M., Schmidt E., Hendriks T., Verhoeven H., de Vries S.C. Identification of single embryo-forming cells in carrot suspension cultures. Acta Bot. Neerl. (1993);42:518–519. [Google Scholar]
- 34.Van Blokland R., Van der Geest N., Mol J.N.M., Kooter J.M. Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J. (1994);6:61–877. [Google Scholar]
- 35.Zhang G., Xu Q., Zhu X., Qian Q., Xue H. SHALLOT-LIKE1 is a KANADI transcriptional factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell. (2009);21:719–735. doi: 10.1105/tpc.108.061457. [DOI] [PMC free article] [PubMed] [Google Scholar]


