Abstract
The entrance of influenza virus into host cells is facilitated by the attachment of the globular region of viral hemagglutinin to the sialic acid receptors on host cell surfaces. In this study, we have cloned the cDNA fragment encoding the entire globular region (residues 101-257) of hemagglutinin of the H9N2 type avian influenza virus (A/ck/Korea/ ms96/96). The protein segment (denoted as the H9 peptide), which was expressed and purified in E. coli, was used for the immunization of BALB/c mice to obtain the anti-H9 antiserum. To identify specific DNA aptamers with high affinity to H9 peptide, we conducted the SELEX method; 19 aptamers were newly isolated. A random mixture of these aptamers showed an increased level of binding affinity to the H9 peptide. The sequence alignment analysis of these aptamers revealed that 6 aptamers have highly conserved consensus sequences. Among these, aptamer C7 showed the highest similarity to the consensus sequences. Therefore, based on the C7 aptamer, we synthesized a new modified aptamer designated as C7-35M. This new aptamer showed strong binding capability to the viral particles. Furthermore, it could prevent MDCK cells from viral infection by strong binding to the viral particles. These results suggest that our aptamers can recognize the hemagglutinin protein of avian influenza virus and inhibit the binding of the virus to target receptors required for the penetration of host cells.
Keywords: anti-viral effect, DNA aptamer, H9N2 avian influenza virus, hemagglutinin
INTRODUCTION
Avian influenza, also known as bird flu, is an acute viral disease that severely infects a wide range of animal species, including humans. The natural reservoirs of avian influenza A viruses (AIVs) are wild birds, such as ducks, geese and shorebirds, and the viruses have rapidly spread to become one of the most prevalent diseases in domestic poultry worldwide (Choi et al., 2008; Thomas and Noppenberger, 2007). In addition, the AIVs have caused serious economic burdens in the poultry industry and constituted an important threat to human health (Du et al., 2008).
In general, the host immune system fights against the infection of AIVs. One of the induced immune responses is the production of neutralizing antibodies against the virus, and the consequent antibodies provide protection against further viral challenges and benefit the recovery process (Ada and Jones, 1986; Virelizier, 1975). The most significant protein that elicits effective antibodies against influenza virus is hemagglutinin (HA). HA plays a key role in the infection process; it mediates the attachment of the virus to host cells by binding to the sialic acid–containing oligosaccharide receptors on the surface of target cells. The subsequent viral entry and membrane fusion are also mediated through HA (Skehel and Wiley, 2000; Wilson and Cox, 1990).
HA exists as trimeric spikes on the viral membrane, and each monomer contains the globular region with a sialic acid receptor-binding pocket that is surrounded by variable antibody-binding sites. HA-specific antibodies can neutralize the infectivity of the virus and thus prevent the infection of antigenically related influenza viruses. Therefore, anti-HA antibodies are responsible for the inhibition of early phase viral infection and the elicitation of protective capacity (Gao et al., 1989). However, the antigenic variation of HA, referred to as antigenic shift and drift, interferes with effective immune responses and makes it difficult to develop anti-influenza agents by using this protein as a target in spite of its potential capability (Wilson and Cox, 1990). The receptor-binding pocket is the only highly conserved region maintained during the antigenic evolution of HA (Wilson et al., 1981), and blocking this region would prevent viral infection (Skehel and Wiley, 2000; Weis et al., 1988). However, this site is not likely exposed to the immune system because of the conformational restriction of the trimeric form of HA in native conditions (Jeon and Arnon, 2002). The domain corresponding to the 91-261 residues of HA protein (HA91-261) in the H3N2 subtype of the human influenza virus encompasses the entire globular region, including the receptor-binding pocket, and is responsible for the attachment of the virus to the host cell receptor (Jeon et al., 2004; Wilson et al., 1981). This domain is common in all H3 type strains, and other types of human influenza HA proteins (H1 and H2) might also contain this globular region as their mechanisms of host infection are the same. The mice immunized with this peptide were partially protected against the challenge infection with influenza virus (Jeon and Arnon, 2002).
Aptamers are artificial nucleic acid ligands that bind to their targets with high affinity and have the capacity to inhibit proteinprotein interactions with a potency similar to that observed in antibodies (Shamah et al., 2008). Aptamers are isolated from highly diverse (1015-1018) starting libraries of synthetic oligonucleotides by an in vitro selection process termed SELEX (systematic evolution of ligands by exponential enrichment) (Tuerk and Gold, 1990). SELEX involves the repetitive reduction of the library complexity via rounds of selective binding to the target molecule and re-amplification (Klug and Famulok, 1994). Aptamers with high specificity and affinity for their cognate proteins can be used as a functional mimicry of the original ligand protein to natural receptors. Moreover, they can replace specific antibodies due to their relative advantages over the use of anti-bodies. Currently, aptamers serve as useful tools to help analyze the mechanisms of viral replication and develop potential applications as diagnostic biosensors and antiviral agents (James, 2007).
Previously, we isolated a novel DNA aptamer, A22, for its capacity to bind to the HA91-261 peptide of the H3-type influenza virus (Jeon et al., 2004). We have demonstrated that this aptamer is capable of blocking the binding of the virus to host cells and consequently preventing viral infection. The protective effect of A22 is compatible with infection by other types of human influenza viral strains; however, the efficacy was lower than in the case of the H3-type influenza strain. Recently, subtype- specific RNA aptamers against human influenza virus have been reported (Gopinath et al., 2006a). These aptamers can distinguish closely related H3N2 strains and could be used in the genotyping of viruses. Currently, these applications have been extended to AIVs and influenza B viruses (Cheng et al., 2008; Gopinath et al., 2006b), and we previously attempted to isolate DNA aptamers that interact with the AIV H9 peptide (Park et al., 2008).
In this study, we have isolated new aptamers that bind directly to the globular region of the H9-type HA of AIV. We demonstrate here that these aptamers and the modified apta-mer are capable of preventing the viral infection of host cells.
MATERIALS AND METHODS
Virus, cells and mice
The avian influenza strain A/ck/korea/ms96/96 (H9N2) was used for the cloning of the HA gene and the infection of host cells. The virus was purified from allantoic fluid by a sucrose gradient, and used as an antigen in the aptamer-binding assay. Madin-Darby Canine Kidney (MDCK) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal calf serum (Wellgene, Korea) and used to measure the infectivity of the virus. The BALB/c mice (10-12 weeks old) were obtained from the Experimental Animal Center of Hallym University.
Cloning and preparation of H9 peptide
Total RNA from AIV was isolated and used for RT-PCR using the primers corresponding to residues 101 to 257 (designated as H9) encoding the globular region of the HA protein; 5′- CCGGATCCTCAGCTGTCAATGGGAT-3′ and 5′-GGAAGCTT GGGGCAATTAGATTCC-3′, respectively. The resultant PCR product was cloned into the pQE30 plasmid (Qiagen) for the expression of its gene product in E. coli. The overexpressed peptide, obtained by isopropyl-β-D-thiogalactopyranoside (IPTG) induction for 4 h, was purified on a Ni-NTA column (Qiagen) by the binding of the N-terminal histidine tag.
Immunization of mice with H9 peptide and serum preparation
To raise antibodies against the H9 peptide, each mouse was immunized in the route of footpads with 50 μg/μl of peptide in complete Freund’s adjuvant. The mice were boosted twice, at 2-week intervals, using the same amount of antigen in incomplete Freund’s adjuvant. The mice were sacrificed 1 week after the last immunization and the sera were collected. An immunoblot analysis and ELISA assay were conducted to determine the antigenicity of the anti-H9 antiserum.
Immunoblot analysis
Total cell lysates or the recombinant H9 peptide was separated by 15% SDS-PAGE, and was then electrotransferred and immobilized onto nitrocellulose membrane (Millipore). The membrane was blocked with 5% non-fat milk in TBS containing 0.1% Tween-20 (TBS-T) at 25℃ for 2 h, and incubated with mouse anti-H9 peptide antisera or chicken anti-virus antibody at 4℃ for 15 h. Biotin-conjugated C7-35M oligomer was also used for the primary binding of the blot. The membrane was rinsed ten times with TBS-T and blocked with 5% non-fat milk in TBS-T at 25℃ for 2 h. After washing, the membrane was further incubated with peroxidase-labeled rabbit anti-mouse IgG or goat anti-chicken IgY antibodies. The peroxidase-labeled streptavidin was used for the detection of biotinylated oligomers. The bound antibodies or streptavidin conjugates were detected by the addition of Immobilon™ Western Chemiluminescent HRP substrate (Millipore).
Enzyme-linked immunosorbent assay (ELISA)
ELISA microtiter plates (Nunc) were coated with H9 peptide (1 μg/100 μl per well) in PBS and incubated at 4℃ for 15 h. The plates were washed four times with PBS containing 0.1% (v/v) Tween-20 (PBS-T) and then blocked with 200 ml of blocking solution (1% BSA in PBS) at 25℃ for 2 h. After washing with PBS-T, serial dilutions of the chicken anti-virus or mouse anti- H9 antibodies were added and incubated for 2 h. After washing to remove the unbound antibodies, 50 μl of peroxidase-labeled goat anti-virus IgY or anti-mouse IgG-conjugated secondary antibodies were added. To examine the binding of the aptamers to the H9 peptide, 200 pmole of expectant biotinylated aptamers were mixed in 200 μl of selection buffer followed by denaturation at 80℃ for 10 min and then cooling on ice for 10 min. After the addition of 50 μl of serial-diluted aptamers to each well, the plates were incubated at 37℃ for 2 h. After washing four times, the peroxidase-conjugated streptavidin was added. Color development was performed by the addition of 50 μl of freshly prepared substrate solution (ABTS, Roche) for 10 min at 25℃. After stopping the reaction with 50 μl of 2 M H2SO4, the plates were read with a multichannel ELISA reader (Thermo Electron Corporation Multiskan Ascent) at 405 nm.
SELEX
The preparation and selection of aptamers were conducted using the method developed by Jeon et al. (2004), with modification. Briefly, the aptamer library containing a central randomized sequence of 28 nucleotides flanked by common primers, 5′-ATTAACCCTCACTAAAGGGA-(N)28-TATGGTCGAATAAGTTAACG-3′ was synthesized (Operon). The ssDNA aptamer library was denatured at 80℃ for 10 min and then cooled on ice for 10 min. For the selection process, 30 nmoles of this product was pre-treated with 25 μl Ni-NTA Superflow (Qiagen) in 500 μl of selection buffer (50 mM Tris-HCl; pH 7.4, 5 mM KCl, 100 mM NaCl, 1 mM MgCl2, 100 μg tRNA, 0.2% BSA) at 4℃ for 30 min. After the removal of non-specifically bound aptamers by centrifugation, the supernatant was mixed with 200 μg of the recombinant His-tagged H9 peptide in 500 μl of selection buffer at 37℃ for 30 min. The bound aptamer-H9 peptide complexes were purified by adding 25 μl of Ni-NTA Superflow. After washing three times with 1 ml of selection buffer, the complexes were resuspended in 50 μl of 10 mM Tris buffer and amplified by PCR using the previously mentioned 5′ and 3′ primers: 5′-ATTAACCCTCACTAAAGGGA-3′ and 5′-CGTTAA CTTATTCGACCATA-3′, respectively, under the condition of 100 pmol/primer in a final volume of 50 μl at 46℃ for 30 cycles. After repeating the SELEX procedure in quadruplicate, the amplified nucleotides were cloned into the pGEM-T Easy vector (Promega).
Reverse screening of aptamer
Biotinylated ssDNA of each plasmid from the individual clones was synthesized using the biotinylated 5′ primer, which has the same sequence as the 5′ primer in the aptamer. The solid phase ELISA plate was prepared by coating with 100 μl/well of 100 μg/ml streptavidin diluted in 0.1 M NaHCO3 and incubation overnight at 37℃. After washing four times with PBS, the wells were blocked with 200 μl of PBS containing 1% BSA at 25℃ for 2 h followed by washing three times with PBS-T. After washing, 100 μl of 2.5 pmol/100 μl of each biotinylated ssDNA was added to the wells and incubated at 37℃ for 2 h followed by washing four times with PBS-T. Subsequently, 100 μl/well of 10 HAU/well virus or 2 μg/well of histidine-labeled H9 peptide was added and incubated at 37℃ for 2 h. After washing the wells four times with PBS-T, anti-His antibodies or anti-virus sera were added to the corresponding wells. Standard ELISA procedures were performed to complete the reverse screening assay.
Viability of MDCK cell (MTT assay)
MDCK cells were plated onto a 96-well plate (0.5 × 104/well) 1 day before the experiments. The medium was aspirated, and the cells were infected with 10 μl of avian influenza virus at a MOI of 0.2, in the presence or absence of aptamers (100 to 1000 pmole) and then incubated in culture medium (DMEM supplemented with 2% fetal bovine serum) at 37℃ for 24 h. The viability assay was performed by adding 10 μl of MTT solution (Takara) to the cell culture and incubating at 37℃ for 2 h. The optical density was determined using an ELISA plate reader at 450 nm.
Statistical analysis
The p values were calculated using Student’s t-test, and p < 0.05 was considered statistically significant according to the Statistical Package for the Social Sciences (SPSS) version 10.0 software program.
RESULTS
Cloning of receptor binding pocket of type 9 HA protein
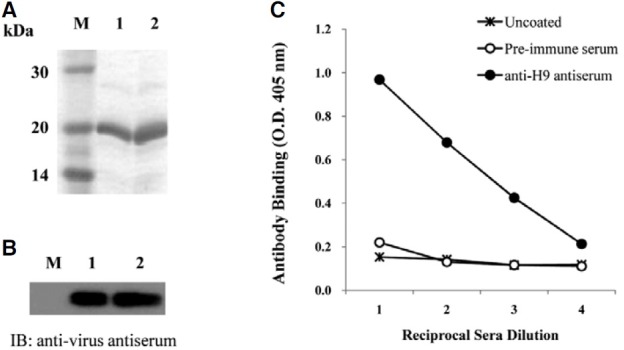
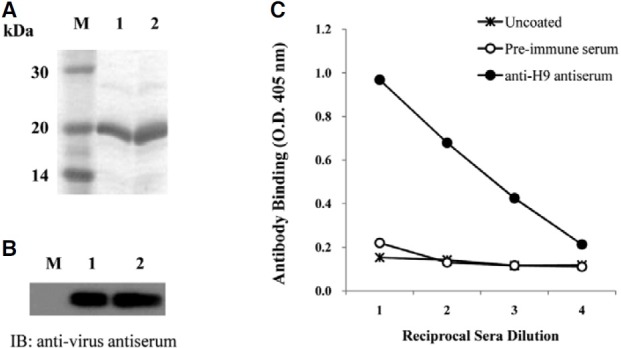
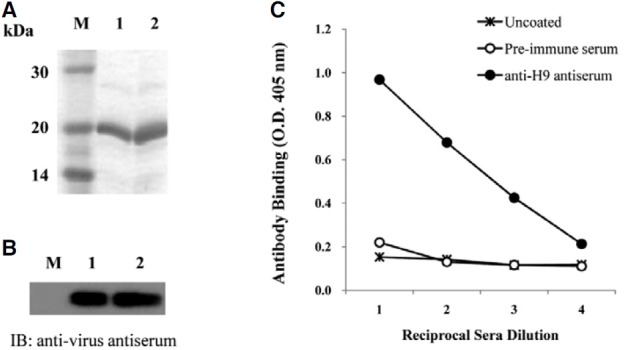
The membrane glycoprotein, HA, expressed on AIV, attaches to the sialic acid receptors on the surface of host cells and facilitates the entry of the virus into host cells. The globular region of the type 9 HA (H9), containing residues 101-257 of AIV, was amplified by RT-PCR. The amplified product was cloned into an expression vector, and confirmed by DNA sequencing analysis (Fig. 1). The IPTG-induced overexpressed H9 peptide was purified by its C-terminal His-tag and confirmed by SDS-PAGE (Fig. 2A). The antigenicity of the H9 peptide to the chicken anti-virus antibody was confirmed by immunoblot analysis (Fig. 2B). The purified H9 peptide was subsequently used to immunize mice for the production of anti- H9 antiserum. As shown in Fig. 2C, the anti-H9 antiserum prepared from the immunized mice showed an increased binding affinity to the H9 peptide as compared with the pre-immune serum as a control.
Fig. 1. The sequence of the H9 peptide, globular region (residue 101-257) of HA. Total RNA isolated from A/ck/Korea/ms96/ 96 avian influenza virus was used for cloning the H9 peptide using RT-PCR.

Fig. 2. Purification of H9 peptide and examination of its immunogenicity. (A) The H9 peptide was expressed by IPTG induction in E. coli and then analyzed by 15% SDS-PAGE. Lane M, molecular weight markers; lanes 1 and 2, 10 and 20 μg of purified protein, respectively. (B) The expression of the H9 peptide was confirmed using chicken anti-viral antibody. Total lysates prepared from the cells after IPTG induction (lane 1) and purified H9 peptides (lane 2) were analyzed by immunoblotting using chicken anti-virus antiserum. (C) Antiserum was prepared from the mice immunized with the H9 peptide and used for the ELISA assay to test the binding to the H9 peptide coated on the microtiter plate.
Selection of the aptamers having specific affinity to the H9 peptide
A nucleotide library was obtained from a pool of 1017 single-stranded DNA (ssDNA) molecules containing a random segment of 28 nucleotides flanked by 5′ and 3′ common primers (shown in “Materials and Methods”) as conserved linkers to amplify the selection process. This random library was screened with H9 peptide by a process termed SELEX, as described in “Materials and Methods”.
The DNA library was mixed with the H9 peptide, and the DNA-peptide complexes were purified on a Ni-NTA column. The aptamers were screened from 1015 highly diverse starting species. The aptamers that bound to the H9 peptide were purified and amplified by PCR. After four rounds of amplification, an ELISA assay was performed to further screen the ssDNA aptamers with high affinity to the H9 peptide. Previously, we isolated 10 aptamers (group A) using this same strategy (Park et al., 2008); however, we did not show significant protective activities against viral infection compared with the anti-virus antibody (data not shown). In this report, a total of 19 aptamers were selected by reverse screening from two independent experiments (groups B and C). The selected aptamers were cloned into the plasmid vector, and their sequences were analyzed by DNA sequencing (Table 1).
Table 1.
Nucleotide sequences of the aptamers screened according to their affinity to the H9 peptide. The specific nucleotide sequences of each aptamer were shown.
| Clone | Specific sequence (N28) | Clone | Specific sequence (N28) |
|---|---|---|---|
| B1 | TCGAGACTTGCTAGTGTCGAATTGGCG | C7 | GGTAGTTATAGTATATGGAAGGGGGTGT |
| B2 | GAACGTCGGTGAGCGGGTTGTGTGCCG | C12 | TTGGGTGGTGGTGGTAGGCAGCACTTGG |
| B3 | GGTGGGTCAGACGGAGTGTGGAGGTGC | C13 | GTGGGGGCGTCCAGTCGCACGGGATAGG |
| B5 | GGGCTGCCAGTGCTTGGATTGGCGTCGT | C14 | CGCGGGCAAAAGGGACGGAGAGCATCGA |
| B6 | CGCCTAGGGATACTGGTTACGGGCCGTA | C15 | TAGCAAGATCTGCAGGGGGACTCGTGTA |
| B7 | TGGACGGGCAGGAGTGTTTTGTGTGTCG | C16 | GGAGAGGGGACGCGGCTAACTACTAGGA |
| B8 | ATCCATCCAGGAGTTGCGGGCAGTGGCG | C17 | GAGGCGGGAGGGTAGTAGGGACTGAAGT |
| B9 | TTGCAGACTGATTCTGTGGTGTGACCGT | C18 | TGGGCGCGATAATCAGTGCCATGATGCA |
| C1 | GGGGGGGCGGGGAGGCCTACGCCTGCGC | C19 | ATGTGCCGCTAGATCAACCAGGGGTAGA |
| C5 | CGGTTTGGCGCTCTGGTATTCTTCGGTA | ||
Among the 3 groups, the aptamers of Group C persistently showed a high affinity for the H9 peptide; the relative binding affinity of the individual aptamers was about 0.5 to 0.82-fold as compared with that of the mouse anti-H9 antibody. While the mixture of randomly chosen five aptamers from Group C did not show a significant increase in binding affinity to the H9 peptide with high deviations for each experiment, the mixture of more than 5 components (6 to 8) of aptamers showed increasing levels of binding affinity to the H9 peptide as compared with the mouse anti-H9 antibody, respectively (Fig. 3). The highest binding affinity was achieved by the mixing 8 components (Cmix8 in Fig. 3B).
Fig. 3. Analysis of the binding affinity of selected DNA aptamer mixtures against H9 peptide or viral particles. (A) The relative binding affinity of aptamer mixtures compared to the H9 peptide. The binding reactions were performed with individual aptamers or mixtures of aptamers from group C. Cmix5 indicates the mixture of 5 aptamers containing C7, C14, C17, C18 and CX (CX is one of C5, C12, C13, C15, or C16 aptamers). Cmix6 is the mixture of Cmix5-15, which contains C15 for CX and the C19 aptamer. Cmix8 is the mixture of Cmix6, C5 and C12. The highest binding affinity was achieved using Cmix8. The non-specific 5′-PCR primer for SELEX was used as a control. (B) An ELISA assay was used to measure the relative binding affinity of the Cmix8 or the mouse anti- H9 peptide antibody as described in “Materials and Methods”. Cmix8 showed a higher binding affinity to the virus than the antibody.
Consensus sequence of aptamers to H9 peptide
The sequences of the random aptamers from each group with demonstrated binding affinity to the H9 peptide were analyzed, and a consensus sequence was identified among them (Fig. 4A). The sequences of the C7 aptamer showed the highest similarity to the consensus sequence. A 35-mer nucleotide, designated as C7-35M, was newly synthesized based on the sequences of the C7 aptamer and 6 other aptamers. From its sequence, the putative secondary structure of C7-35M could be predicted using the DNAdraw software program (mfold ver. 3.2) (Zuker, 2003) (Fig. 4B). The binding affinity of C7-35M against the H9 peptide was about half the affinity of mouse anti-H9 antibodies (Fig. 4C), which is consistent with ELISA results in Fig. 3. And, the C7-35M showed similar biding affinity to those of individual aptamers. Next, we investigated whether this aptamer could bind to the H9N2 type AIV. The biotinylated aptamer was added to the microtiter plate previously coated with the virus, and an ELISA assay was conducted. Interestingly, the C7-35M showed much more significant binding affinity to the H9 type AIV compared to the mouse antibody against H9 peptide (Fig. 4D).
Fig. 4. Analysis of consensus nucleotide sequences of specific aptamers that bind to the HA9 peptide. (A) The common consensus sequence was obtained by the arrangement of specific sequences of 6 aptamers from groups B and C. The bold letters are used to indicate the consensus sequences that are conserved in more than five aptamers, and the dashed letters indicate the sequences conserved in more than three aptamers. N represents the non-consensus sequence. (B) The predicted stem-and-loop structure of C7-35M aptamer containing the consensus sequences and a part of the 3′ primer region. The nucleotides in the circles indicate the conserved sequences represented as bold letters in (A). (C) The analysis of relative binding affinity to the H9 peptide. The equal amount of cell lysates and H9 peptide was separated in duplicate, and then hybridized with mouse anti-H9 peptide antibody or C7-35M oligomer for each duplicate. The relative density was electronically determined by ImageJ software from NIH Image. (D) The relative binding affinities of C7-35M, chicken anti-virus and mouse anti-H9 peptide antibody to the viral particles.

Inhibition of viral infectivity by aptamer
The anti-viral activities of the C7-35M aptamer against the H9N2 type AIV were measured by MTT assay in MDCK cells. The efficacy was determined by comparing the cell survival rates in aptamer-treated or untreated conditions after viral infection. We found that the exposure of the cells to the H9N2 influenza virus led to an almost total cell death. Egg supernatant was used as a control, which showed a 3-fold higher survival rate as compared with the virus treatment (Fig. 5). The cell viability was increased by the addition of C7-35M in a dose-dependent manner, indicating the capacity of C7-35M to prevent the viral infectivity of the cells. The treatment of the aptamer at 100 pmole could inhibit the viral infection by 13%, while the treatment of the aptamer at 1000 pmole could inhibit the viral infection by 55%. These results suggest that C7-35M can prevent and suppress viral infection.
Fig. 5. Anti-viral effects of C7-35M. The C7-35M treatment showed protective effects on the viability of MDCK cells infected with H9N2 influenza virus. After 24 h of incubation with virus, the aptamer was added at various concentrations (ranging 100-1,000 pmole). The cell viability was measured by MTT assay as described in Materials and Methods. The relative viability of the cells [(O.D. value of aptamer - virus)/(O.D. value of Egg - virus)] in different concentrations of aptamers is indicated.
DISCUSSION
AIV is classified into subtypes according to the combination of two types of surface glycoproteins that include 16 hemagglutinin (HA) and 9 neuraminidase (NA) molecules (Fouchier et al., 2005). Among them, 3 novel AIV strains, A/H5N1, A/H7N7 and A/H9N2, have emerged in the human population (Lipatov et al., 2004). Although there has been no evidence of person-toperson transmission of the AIVs, their significant genetic variability and the characteristics of the segmented genome provide the risk that they can undergo natural genetic reassortment and acquire the ability to spread from person-to-person (Du et al., 2008).
Global outbreaks of highly pathogenic avian influenza (HPAI) in domestic poultry have been reported since the last decade and are still increasing (He et al., 2007; Wong and Yuen, 2006). Meanwhile, low pathogenic avian influenza (LPAI) strains, such as H9N2, have become endemic and continue to spread over Asia, Middle Eastern countries and South Africa (Sorrell et al., 2007). The H9N2 subtype of AIV has widely circulated through the world since the first detection in turkeys in the United States in 1966 (Homme and Easterday, 1970); it has occasionally been transmitted to members of the mammalian species, including humans (Lin et al., 2000; Lu et al., 2001; Peiris et al., 1999). It was reported that an increasing number of contemporary H9N2 viruses have been modified to obtain the binding capacity for the receptor of human epithelial cells (Matrosovich et al., 2001; Wan and Perez, 2007). Therefore, although HPAI H5N1 has been the major health threat to humans since 2003, this event demonstrates that the H9N2 virus can also be a potential threat, leading to pandemics by improving their transmissibility for human infection (Sorrell et al., 2009; Wu et al., 2008). The use of antiviral drugs has been recognized as the primary public health strategy for mitigating the severity of a new influenza strain. However, the success of this strategy requires the prompt onset of therapy within 48 h of the appearance of clinical symptoms (Alexander et al., 2008). The development of an effective vaccine depends on the understanding of protective immune responses against natural infection, particularly those directed against specific viral constituents.
Neuraminidase (NA)-specific antibodies do not prevent influenza virus infection, but rather prevent the release of viruses from the infected cells. The antibodies to the matrix M2 protein, which is conserved within the A-type influenza viruses, are cross-protective against the viral infection of different subtypes, but the level of protection is low. The antibodies to the conserved nucleoprotein (NP) and matrix M1 protein can be induced, but they fail to contribute to protection (Cinatl et al., 2007). However, the anti-HA antibodies are the most significant in their protective capacity. HA is a major surface glycoprotein of the influenza virus and the principal target of the protective immune response.
In this report, we have isolated ssDNA aptamers that have significant binding capacities for the evolutionary conserved region of HA molecule. We have focused on the receptor-binding pocket that is not easily exposed to the outside, and obtained this region by using E. coli expression system. It is not verified whether the property of this partial peptide lacking N-glycosylation could be enhanced in eukaryotic expression system, though this modification might be important for the binding affinity of native whole protein to the host cell (Roth et al., 2010). This peptide was used for the initial screening of many possible candidates, though we could not catch sugar-binding aptamers. We suggest that there are several binding sites on H9 peptide that are recognized by these aptamers because the binding affinity was increased when each aptamer was applied as a mixture (Fig. 3), while the individual aptamer had a relatively low binding affinity as compared with the antibody obtained from a mouse immunized with the same peptide (Fig. 2). The features of multiple recognitions by the aptamer mixture might be similar to that of polyclonal antibodies generated from the immunization of animals. We have also confirmed the binding of selected aptamers to the viral particles. In these cases, several aptamers showed higher binding affinity to the virus rather than to the peptide or vice versa, which suggests the possible involvement of glycosylation in the recognition and binding.
We have identified a conserved sequence, present in several aptamers, that was obtained from two independent screening processes by SELEX. In addition to the random 28-mer region, this consensus sequence extends to the 3′ linker region, which parties potentially a part of the stem and loop secondary structure (Fig. 4). Interestingly, the 35-oligonucleotide C7 aptamer containing the consensus sequence (C7-35M) showed a much higher binding affinity for the H9 peptide than the original C7 aptamer. This increased binding affinity could result from the differences in their structures under natural conditions. The binding affinity of both aptamers to the H9 peptide was lower than that of antibody obtained from the mice immunized with the H9 peptide (Figs. 3A and 4C). However, the aptamer C7- 35M showed a significant binding affinity for the viral particles with native conformation of HA molecules, while the mouse antibody rarely recognized the virus (Fig. 4). This result can be explained by the fact that the mouse antibody was generated against the denatured form of H9 peptide. Indeed, we have attempted to investigate the qualitative binding affinity of our aptamers to the H9 peptide. However, unfortunately, we could see little binding activity of C7-35M to H9 peptide via surface plasmon resonance analysis as in ELISA (data not shown). We are planning to determine the binding activity of C7-35M to the virus particles themselves.
In summary, our aptamer, C7-35M, showed higher binding affinity to the virus particle regardless of its affinity to the H9 peptide originally used for the initial screening. In addition, perhaps more importantly, the C7-35M treatment significantly reduced the cell death invoked by viral infection (Fig. 5). These results demonstrate that the C7-35M aptamer might be a candidate for the development of anti-AVI drugs for poultry or the therapeutic use for possible human disease.
Acknowledgments
This research was supported by the Technology Development Program for Agriculture and Forestry, Ministry for Agriculture, Forestry and Fisheries, Republic of Korea (106126-3-HD110).
References
- 1.Ada G.L., Jones P.D. The immune response to influenza infection. Curr. Top Microbiol. Immunol. (1986);128:1–54. doi: 10.1007/978-3-642-71272-2_1. [DOI] [PubMed] [Google Scholar]
- 2.Alexander M.E., Moghadas S.M., Rost G., Wu J. A delay differential model for pandemic influenza with antiviral treatment. Bull. Math. Biol. (2008);70:382–397. doi: 10.1007/s11538-007-9257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng C., Dong J., Yao L., Chen A., Jia R., Huan L., Guo J., Shu Y., Zhang Z. Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX. Biochem. Biophys. Res. Commun. (2008);366:670–674. doi: 10.1016/j.bbrc.2007.11.183. [DOI] [PubMed] [Google Scholar]
- 4.Choi J.G., Lee Y.J., Kim Y.J., Lee E.K., Jeong O.M., Sung H.W., Kim J.H., Kwon J.H. An inactivated vaccine to control the current H9N2 low pathogenic avian influenza in Korean. J. Vet. Sci. (2008);9:67–74. doi: 10.4142/jvs.2008.9.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cinatl J., Jr., Michaelis M., Doerr H.W. The threat of avian influenza A (H5N1). Part IV: Development of vaccines. Med. Microbiol. Immunol. (2007);196:213–225. doi: 10.1007/s00430-007-0052-3. [DOI] [PubMed] [Google Scholar]
- 6.Du N., Li W., Li Y., Liu S., Sui Y., Qu Z., Wang Y., Du Y., Xu B. Generation and evaluation of the trivalent inactivated reassortant vaccine using human, avian, and swine influenza A viruses. Vaccine. (2008);26:2912–2918. doi: 10.1016/j.vaccine.2008.03.048. [DOI] [PubMed] [Google Scholar]
- 7.Fouchier R.A., Munster V., Wallensten A., Bestebroer T.M., Herfst S., Smith D., Rimmelzwaan G.F., Olsen B., Osterhaus A.D. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. (2005);79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X.M., Liew F.Y., Tite J.P. Identification and characterization of T helper epitopes in the nucleoprotein of influenza A virus. J. Immunol. (1989);143:3007–3014. [PubMed] [Google Scholar]
- 9.Gopinath S.C., Misono T.S., Kawasaki K., Mizuno T., Imai M., Odagiri T., Kumar P.K. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. (2006a);87:479–487. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]
- 10.Gopinath S.C., Sakamaki Y., Kawasaki K., Kumar P.K. An efficient RNA aptamer against human influenza B virus hemagglutinin. J. Biochem. (2006b);139:837–846. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 11.He Q., Velumani S., Du Q., Lim C.W., Ng F.K., Donis R., Kwang J. Detection of H5 avian influenza viruses by antigen-capture enzyme-linked immunosorbent assay using H5- specific monoclonal antibody. Clin. Vaccine Immunol. (2007);14:617–623. doi: 10.1128/CVI.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homme P.J., Easterday B.C. Avian influenza virus infections. I. Characteristics of influenza A-turkey-Wisconsin- 1966 virus. Avian Dis. (1970);14:66–74. [PubMed] [Google Scholar]
- 13.James W. Aptamers in the virologists’ toolkit. J. Gen. Virol. (2007);88:351–364. doi: 10.1099/vir.0.82442-0. [DOI] [PubMed] [Google Scholar]
- 14.Jeon S.H., Arnon R. Immunization with influenza virus hemagglutinin globular region containing the receptor-binding pocket. Viral. Immunol. (2002);15:165–176. doi: 10.1089/088282402317340314. [DOI] [PubMed] [Google Scholar]
- 15.Jeon S.H., Kayhan B., Ben-Yedidia T., Arnon R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. (2004);279:48410–48419. doi: 10.1074/jbc.M409059200. [DOI] [PubMed] [Google Scholar]
- 16.Klug S.J., Famulok M. All you wanted to know about SELEX. Mol. Biol. Rep. (1994);20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y.P., Shaw M., Gregory V., Cameron K., Lim W., Klimov A., Subbarao K., Guan Y., Krauss S., Shortridge K., et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA. (2000);97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipatov A.S., Govorkova E.A., Webby R.J., Ozaki H., Peiris M., Guan Y., Poon L., Webster R.G. Influenza: emergence and control. J. Virol. (2004);78:8951–8959. doi: 10.1128/JVI.78.17.8951-8959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X., Renshaw M., Tumpey T.M., Kelly G.D., Hu-Primmer J., Katz J.M. Immunity to influenza A H9N2 viruses induced by infection and vaccination. J. Virol. (2001);75:4896–4901. doi: 10.1128/JVI.75.10.4896-4901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matrosovich M.N., Krauss S., Webster R.G. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. (2001);281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 21.Park M., Jeon S.H., Mo I.P., Choi S.Y. Interaction of DNA Aptamers with Avian Influenza Virus H9 peptide. J. Biomed. Res. (2008);9:10. [Google Scholar]
- 22.Peiris M., Yuen K.Y., Leung C.W., Chan K.H., Ip P.L., Lai R.W., Orr W.K., Shortridge K.F. Human infection with influenza H9N2. Lancet. (1999);354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 23.Roth J., Zuber C., Park S., Jang I., Lee Y., Kysela K.G., Le Fourn V., Santimaria R., Guhl B., Cho J.W. Protein N-glycosylation, protein folding, and protein quality control. Mol. Cells. (2010);30:497–506. doi: 10.1007/s10059-010-0159-z. [DOI] [PubMed] [Google Scholar]
- 24.Shamah S.M., Healy J.M., Cload S.T. Complex target SELEX. Acc. Chem. Res. (2008);41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 25.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. (2000);69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 26.Sorrell E.M., Ramirez-Nieto G.C., Gomez-Osorio I.G., Perez D.R. Genesis of pandemic influenza. Cytogenet. Genome Res. (2007);117:394–402. doi: 10.1159/000103203. [DOI] [PubMed] [Google Scholar]
- 27.Sorrell E.M., Wan H., Araya Y., Song H., Perez D.R. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc. Natl. Acad. Sci. USA. (2009);106:7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas J.K., Noppenberger J. Avian influenza: a review. Am. J. Health Syst. Pharm. (2007);64:149–165. doi: 10.2146/ajhp060181. [DOI] [PubMed] [Google Scholar]
- 29.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. (1990);249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 30.Virelizier J.L. Host defenses against influenza virus: the role of anti-hemagglutinin antibody. J. Immunol. (1975);115:434–439. [PubMed] [Google Scholar]
- 31.Wan H., Perez D.R. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J. Virol. (2007);81:5181–5191. doi: 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weis W., Brown J.H., Cusack S., Paulson J.C., Skehel J.J., Wiley D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. (1988);333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 33.Wilson I.A., Cox N.J. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. (1990);8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 34.Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. (1981);289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 35.Wong S.S., Yuen K.Y. Avian influenza virus infections in humans. Chest. (2006);129:156–168. doi: 10.1378/chest.129.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu R., Sui Z.W., Zhang H.B., Chen Q.J., Liang W.W., Yang K.L., Xiong Z.L., Liu Z.W., Chen Z., Xu D.P. Characterization of a pathogenic H9N2 influenza A virus isolated from central China in 2007. Arch. Virol. (2008);153:1549–1555. doi: 10.1007/s00705-008-0139-1. [DOI] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. (2003);31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


