Abstract
IQ motif-containing GTPase-activating protein 1 (IQGAP1), which is a well-known calmodulin (CaM) binding protein, is involved in a wide range of cellular processes including cell proliferation, tumorigenesis, adhesion, and migration. Interaction of IQGAP1 with CaM is important for its cellular functions. Although each IQ domain of IQGAP1 for CaM binding has been characterized in a Ca2+-dependent or -independent manner, it was not clear which IQ motifs are physiologically relevant for CaM binding in the cells. In this study, we performed immunoprecipitation using 3xFLAGhCaM in mammalian cell lines to characterize the domains of IQGAP1 that are key for CaM binding under physiological conditions. Interestingly, using this method, we identified two novel domains, IQ(2.7-3) and IQ(3.5-4.4), within IQGAP1 that were involved in Ca2+-independent or -dependent CaM binding, respectively. Mutant analysis clearly showed that the hydrophobic regions within IQ(2.7-3) were mainly involved in apoCaM binding, while the basic amino acids and hydrophobic region of IQ(3.5-4.4) were required for Ca2+/CaM binding. Finally, we showed that IQ(2.7-3) was the main apoCaM binding domain and both IQ(2.7-3) and IQ(3.5-4.4) were required for Ca2+/CaM binding within IQ(1- 2-3-4). Thus, we identified and characterized novel direct CaM binding motifs essential for IQGAP1. This finding indicates that IQGAP1 plays a dynamic role via direct interactions with CaM in a Ca2+-dependent or -independent manner.
Keywords: calcium, calmodulin, in vitro, IQ motif, IQGAP1
INTRODUCTION
IQ motif-containing GTPase-activating protein 1 (IQGAP1), which is a scaffolding protein, has multi-functional roles, including cell proliferation, tumorigenesis, adhesion, and migration (Noritake et al., 2005; White et al., 2009). These physiological roles are mainly mediated by dynamic association and dissociation of IQGAP1 with numerous binding proteins, such as calmodulin (CaM), Cdc42, Rap1, ERK2, E-cadherin, and CLIP- 170 (Briggs and Sacks, 2003). For instance, overexpression of IQGAP1 can alter the subcellular distribution of Cdc42 from the cytoplasm to plasma membrane via direct binding of GTPbound Cdc42, consequently leading to induction of microspikes and filopodia (Swart-Mataraza et al., 2002). IQGAP1 also regulates cellular signaling including MEK-ERK signaling via direct binding to MEKs and ERK2 (Roy et al., 2004; 2005).
The function of IQGAP1 can be regulated by CaM, a key player in Ca2+ signaling. The binding of CaM to IQGAP1 can induce conformational changes of IQGAP1, leading to regulation of IQGAP1-protein interactions or alternation of IQGAP1’s subcellular localization (Briggs et al., 2002; Ho et al., 1999; Li et al., 1999; Mateer et al., 2002). For example, Ca2+ increased the binding between CaM and IQGAP1 with a concomitant attenuation in the binding between IQGAP1 and GTP-bound Cdc42, which eliminated the effects of IQGAP1 on activated Cdc42 (Ho et al., 1999).
CaM binding of IQGAP1 is mainly mediated by four IQ motifs, which are known collectively as the Ca2+-independent CaM binding domain (Bahler and Rhoads, 2002; Rhoads and Friedberg, 1997). Usually, an IQ motif comprises 20-25 amino acids, with generalized consensus sequences ([I,L,V]QXXXRXXXX[R,K]). For example, the IQ motif of neuromodulin and neurogranin can bind to CaM in a Ca2+-independent manner (Alexander et al., 1987; Baudier et al., 1991). However, the IQ motif also can associate with CaM in a Ca2+-dependent manner. For example, calcium vector protein target and NinaC myosin can associate with CaM in a Ca2+-dependent manner (Petrova et al., 1996; Porter et al., 1995). In addition, many IQ containing proteins have multiple repeated IQ motifs including IQGAP1. Repeated IQ motifs play roles in anchoring CaM within the cells, because the nonconventional myosin family usually contains repeated IQ motifs (Bahler and Rhoads, 2002). For example, human myosin IIa or myosin V has five repeated or 2-6 repeated IQ motifs, respectively. A scaffolding protein, IQGAP1, also has four IQ motifs. Interestingly, length of the linker in repeated IQ motifs is similar between each IQ motif, suggesting the possibility that the linkers might have other functions than just connecting the different IQ motifs.
It has been shown that all four IQ motifs of IQGAP1 can bind to Ca2+/CaM, while the third and fourth IQ motifs of IQGAP1 can bind to apoCaM using a CaM 4B pull-down assay (Li and Sacks, 2003). Although the CaM 4B pull-down assay is normally used for CaM binding, this approach cannot be used to reliably discriminate physiologically relevant CaM binding proteins. For example, when proteomics was combined with CaM 4B pull-down assay, several unrelated proteins including ribosomal proteins were identified in HeLa cells (Jang et al., 2007).
In this study, a new CaM binding strategy that overcomes the limitations of the CaM 4B pull-down assay was used to characterize the essential domain of IQGAP1 for CaM binding under physiological conditions. Using 3xFLAG-hCaM immunoprecipition (IP) combined with 3xFLAG peptide elution, we found novel CaM binding domains within the IQ motif of IQGAP1, IQ(2.7-3) and IQ(3.5-4.4), which were shown to be Ca2+- independent or -dependent CaM binding motifs, respectively. We further showed that the hydrophobic residues within IQ(2.7- 3) were critical for apoCaM binding, and both basic amino acids and a hydrophobic residue within IQ(3.5-4.4) were critical for Ca2+/CaM binding. Finally, mutant analysis clearly showed that IQ(2.7-3) was required for apoCaM binding and both IQ(2.7-3) and IQ(3.5-4.4) were required for Ca2+/CaM binding. Thus, our study implies that IQGAP1 plays a dynamic role by interacting with CaM in a Ca2+-dependent or -independent manner.
MATERIALS AND METHODS
DNA construction
To clone human CaM (hCaM) and human IQGAP1 (hIQGAP1), we performed polymerase chain reaction (PCR) using the HeLa cDNA library. The following primers were used: hCaM, hCaM-D3-S/hCaM-XbaI-A; hIQGAP1, hIQGAP1-S/hIQGAP1-A (Table 1). The PCR products of hCaM or hIQGAP1 were subcloned into the p3xFLAG-CMV-7.1 vector (Sigma) using HindIII/XbaI restriction sites for hCaM or the pEGFP-C3 vector by using HindIII/BamHI for hIQGAP1, respectively.
Table 1.
List of oligonucleotides and their sequences
| Name | Sequence (5′ → 3′) |
|---|---|
| hCaM-D3-S | CGCCCAAGCTTATGGCTGACCAGGCTGACT |
| hCaM-BamHI-A | CGTCTAGATCACTTTGCAGTCATCATC |
| hCaM-XhoI-A | TGCCTCGAGTCACTTTGCAG TCATCAT |
| hCaM-XbaI-A | GCAGATCTTCACTTTGCAG TCATCAT |
| 3xFLAG-NdeI-S | TACATATGGACTACAAAGACCATGACGGT |
| hIQGAP1-S | AATGAAGGCCTGATCACCA |
| hIQGAP1-A | GTCACTTTGGTCCAGCAGGT |
| IQ1-D3-S | CGCCCAAGCTTGGCCTGATCACC AGGCTG |
| IQ2-D3-S | CGCCCAAGCTTCAAATCCCTGCCATCACC |
| IQ3-D3-S | GCAAGCTTGATGAAGTTGTAAAGATT |
| IQ4-D3-S | CGAAGCTTCGGGACCATATAAATGAC |
| IQ2.7-D3-S | CGCCCAAGCTTGCATATCAAGATCGGTTA |
| IQ3.5-D3-S | CGCCCAAGCTTCGAAAGCGCTATCGAGAT |
| IQ3.5(AAA)-D3-S | CGCCCAAGCTTGCTGCTGCTTATCGAGATCGC CTGCAG |
| IQ2-BamHI-A | CGCGGATTCCTTCATCTTTGTGGGAGCG |
| IQ3-BamHI-A | CGCGGATCCTCAGTCATTTATATGGTCCC |
| IQ4-BamHI-A | CGCGGATCCTCAGTCATTTATATGGTCCCG |
| IQ4.4-BamHI-A | CGCGGATCCTCATGCCCGAATAAAAGCCTG |
| IQ3(Δ10)-BamHI-A | CGCGGATCCTCAGCGATCTCGATAGCGCTT |
| IQ2-BamHI-S | CGCGGATCCCAAATCCCTGCCATCACC |
| IQ2.7-BamHI-S | CGCGGATCCGCATATCAAGATCGGTTA |
| IQ3.5-BamHI-S | CGCGGATCCCGAAAGCGCTATCGAGAT |
| IQ3-XhoI-A | CGACCGCTCGAGTCAGTCATTTATATGGTCCC |
| IQ4.4-XhoI-A | CGACCGCTCGAGTCATGCCCGAATAAAAGCCTG |
| IQ3(D1/2)-S | AAGATTCAGTCCCTGGATAGGGATCACCAAGCT CGA |
| IQ3(D1/2)-A | TCGAGCTTGGTGATCCCTATCCAGGGACTGAAT CTT |
| IQ2(D3)-S | TTAGCTTACGATCGCTCCCAC |
| IQ2(D3)-A | GTGGGAGCGATCGTAAGCTAA |
| IQ4(D4)-S | GACATTATCAAAGACCAGGCTTTTATT |
| IQ4(D4)-A | AATAAAAGCCTGGTCTTTGATAATGTC |
The serial constructs of IQGAP1 for EGFP-fusion constructs were obtained by performing PCR using the following specific primer set: IQ2, IQ2-D3-S/IQ2-BamHI-A; IQ3, IQ3-D3-S/IQ3- BamHI-A; IQ4, IQ4-D3-S/IQ4-BamHI-A; IQ(1-2), IQ1-D3-S/IQ2- BamHI-A; IQ(2-3), IQ2-D3-S/IQ3-BamHI-A; IQ(3-4), IQ3-D3- S/IQ4-BamHI-A; IQ(1-2-3-4), IQ1-D3-S/IQ4-BamHI-A; IQ(2.7-3), IQ2.7-D3-S/IQ3-BamHI-A; IQ(3.5-4.4), IQ3.5-D3-S/IQ4.4- BamHI-A; IQ(2.7-3(Δ10)), IQ2.7-D3-S/IQ3(Δ10)-BamHI-A; IQ (3.5-4.4, AAA), IQ3.5-AAA-S/IQ4.4-BamHI-A (Table 1). The PCR products were separately subcloned into HindIII–BamHIdigested pEGFP-C3 to create pEGFP-C3-IQs serial constructs. The mutant fragments of IQ(2.7-3, D1/2), IQ(2.7-3, D3), IQ(3.5- 4.4, D4), IQ(1-2-3-4, D1/2), IQ(1-2-3-4, D4), and IQGAP1 (1-2-3-4, D1/2/4) were generated by recombinant PCR using specific sense and antisense primers. These were a mutant containing D1/2, IQ3(D1/2)-S/IQ3(D1/2)-A; a mutant containing D3, IQ2(D3)- S/IQ2(D3)-A; a mutant containing D4, IQ4(D4)-S/IQ4(D4)-A. PCR products were separately subcloned into HindIII-BamHI-digested pEGFP-C3 to create pEGFP-C3-IQs serial mutants.
The serial constructs of IQGAP1 for the GST-fusion constructs were obtained by performing PCR using the following specific primer set: IQ(2.7-3), IQ2.7-BamHI-S/IQ3-XhoI-A; IQ (3.5-4.4), IQ3.5-BamHI-S/IQ4.4-XhoI-A; IQ(2-3), IQ2-BamHIS/ IQ3-XhoI-A (Table 1). The PCR products were separately subcloned into BamHI/XhoI-digested pGEX 4T-1 to create pGEX4T-1-IQs serial mutants.
The 3xFLAG-hCaM construct for E. coli expression was obtained by performing PCR using the following specific primer set: 3xFLAG-NdeI-S/hCaM-BamHI-A (Table 1). The PCR products were separately subcloned into NdeI–BamHI-digested pET21a to create the pET21a-3xFLAG-hCaM construct.
FLAG IP and Western blotting
For FLAG IP, transfected cells were lysed with a buffer containing 1% Triton X-100, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and protease inhibitor cocktails (Roche). The lysates were centrifuged at 15,000 rpm for 15 min at 4℃ in a microcentrifuge. The protein concentration of the supernatants was quantified using the BCA assay (Thermo Scientific). Resulting cell lysates were equally separated and added to 2 mM CaCl2 and 5 mM EGTA. The cell lysate was incubated with 10 μl of mouse anti- FLAG M2-agarose beads (Sigma) at 4℃ for 2 h. Beads were washed three times with the lysis buffer. Beads were incubated with 100 μM 3xFLAG peptides (Sigma) containing lysis buffer for an additional 30 min at 4℃. Finally, the supernatant for 3xFLAG-hCaM IP was mixed with 5X SDS sample buffer and analyzed by Western blotting.
For Western blotting, proteins separated on 10% gels were transferred to a 0.45 μm nitrocellulose membrane. The membrane was blocked with 5% non-fat milk and incubated with the primary antibody in TBST buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.01% Tween 20) containing 5% non-fat milk. The bound primary antibodies were detected using a goat anti-mouse or a goat anti-rabbit IgG-horseradish peroxidase conju gate and an ECL detection system (Millipore). The intensity of the protein bands was quantified using the ImageJ program (National Institutes of Health).
The rabbit polyclonal anti-IQGAP1 antibody was purchased from Santa Cruz Biotechnologies. The rabbit polyclonal anti- CaM antibody was obtained from Chemicon. The rabbit polyclonal anti-GFP antibody was purchased from Invitrogen. The mouse anti-FLAG M2 antibody was obtained from Sigma.
CaM binding using CaM 4B or 3xFLAG-hCaM in vitro
For in vitro 3xFLAG-hCaM binding, 3xFLAG-hCaM proteins were purified from E. coli using M2-agarose. GST-IQs fusion proteins were purified from E. coli using a general purification method (GE Healthcare). Each purified GST-IQs protein was incubated at 4℃ for 1 h with 30 μl CaM 4B beads or 3xFLAGhCaM charged M2-agarose in a binding solution containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% BSA, 1% Triton-X 100, protease inhibitor cocktails (Roche) in the presence of 2 mM CaCl2 or 5 mM EGTA. Beads were then washed three to four times with binding buffer. In the case of 3xFLAG-hCaM IP, beads were incubated with 100 μM 3xFLAG peptides (Sigma) containing lysis buffer for an additional 30 min at 4℃. Finally, proteins captured on the beads were eluted with 1X SDS sample buffer, resolved by SDS-PAGE, and subjected to Coomassie blue staining. Each experiment was typically repeated at least three times to obtain significant results.
Immunocytochemistry
The cultures were fixed with 4% paraformaldehyde in 1X PBS. After blocking nonspecific binding by preincubatiing cells with 3% BSA in 1X PBS, the cells were incubated with the monoclonal mouse anti-FLAG antibody (1:250, Sigma) and the secondary cyanine 5 (Cy5)-conjugated anti-mouse antibody (1:1000, Amersham Biosciences, USA) to detect 3xFLAG-hCaM. For IQGAP1, the cells were incubated with the polyclonal rabbit anti-IQGAP1 antibody (1:250, Santa Cruz) and the secondary cyanine 3 (Cy3)-conjugated anti-rabbit antibody (1:1000, Amersham Biosciences, USA). We obtained and analyzed fluorescence images using a confocal laser-scanning microscope (Radiance 2000; Zeiss, Germany) and NIH ImageJ software (National Institutes of Health, USA), respectively.
RESULTS
Generation of 3xFLAG-hCaM
To overcome the limitation of the CaM 4B pull-down assay, we applied IP using a FLAG antibody (FLAG IP) for CaM binding, since this approach is useful for identifying physiologically relevant binding proteins within the cells. Previous work has shown that an N-terminal fused HA epitope does not interfere with CaM binding (Szymanska et al., 1997). Therefore, we fused a 3xFLAG epitope to the N-terminus of hCaM, and generated 3xFLAG-hCaM for FLAG IP.
To mimic endogenous CaM binding, we expressed 3xFLAG-hCaM, which was similar to endogenous CaM, in HEK293T cells, as shown in Fig. 1A. We then examined the cellular localization of 3xFLAG-hCaM in HeLa cells. As shown in Fig. 1B, 3xFLAG-hCaM was co-localized with endogenous IQGAP1 under physiological conditions in HeLa cells. This result is consistent with previous reports, which showed that IQGAP1 can act as a reservoir for CaM within cells in resting states (Ho et al., 1999; Joyal et al., 1997). These results suggested that ectopically expressed 3xFLAG-hCaM could associate with endogenous IQGAP1 in HeLa cells in resting states.
Fig. 1. Expression of 3xFLAG-hCaM and its binding to IQ(1-2-3-4) motif of IQGAP1. (A) Immunoblotting of endogenous CaM or overexpressed 3x FLAG-hCaM in HEK293T cells. (B) Colocalization of 3XFLAG-hCaM with IQGAP1 in HeLa cells. Immunocytochemistry analysis was performed to detect endogenous IQGAP1 and exogenous 3xFLAG-hCaM 24 h after transfection. Scale bar, 20 μm. (C) CaM binding of overexpressed IQ(1- 2-3-4) motif of IQGAP1 fused to EGFP in HEK293T cells using 3xFLAGhCaM IP or CaM 4B pull-down. EGFPIQ( 1-2-3-4) was equally bound to Ca2+/ CaM and apoCaM in both 3xFLAGhCaM IP and CaM 4B pull-down. Bound proteins were visualized by immunoblotting using an anti-GFP antibody. As a loading control, the amount of 3xFLAG-hCaM was detected using an anti-FLAG antibody.
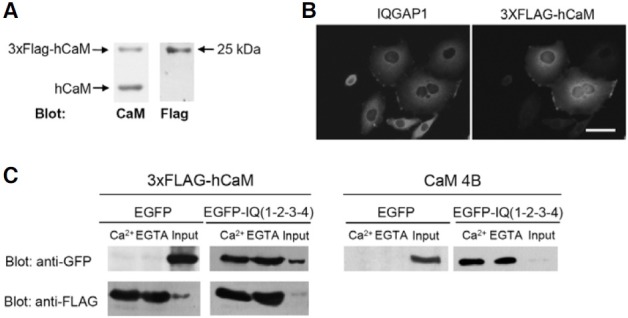
We then examined the interaction between 3xFLAG-hCaM and IQGAP1. It has been shown using the CaM 4B pull-down assay that CaM binding of IQGAP1 is primarily mediated by four IQ motifs. Therefore, we cloned the IQ motifs of human IQGAP1 from HeLa cDNA, and generated EGFP-IQ(1-2-3-4). After overexpressing 3xFLAG-hCaM and EGFP-IQ(1-2-3-4) in HEK293T cells, we performed 3xFLAG-hCaM IP. To exclude non-specific binding, 3xFLAG-hCaM binding complexes were eluted using 3xFLAG-peptides as described in the materials and methods. As a result, EGFP-IQ(1-2-3-4) was successfully and equally immunoprecipitated in the presence or absence of Ca2+ (Fig. 1C). As a control, EGFP alone could not bind to 3xFLAG-hCaM (Fig. 1C). Similar results were observed in the CaM 4B pull-down assay (Fig. 1C). Overall, these results suggested that 3xFLAG-hCaM could bind to IQ(1-2-3-4) in a Ca2+- independent manner as was observed in the CaM 4B pull-down assay. Overall, these results indicate that 3xFLAG-hCaM retains its ability as a CaM, and 3xFLAG-hCaM IP can be useful for investigating CaM binding in the cells.
CaM binding of each IQ motif of IQGAP1
Previously, it was shown that all IQ motifs of IQGAP1 could bind to Ca2+/CaM, while only the third and fourth IQ motifs of IQGAP1 could bind to apoCaM in the CaM 4B pull-down assay (Li and Sacks, 2003). However, it was not clear which IQ motifs were physiologically relevant for CaM binding in the cells. Therefore, we decided to examine the CaM binding domain of each IQ motif using 3xFLAG-hCaM IP in HEK293T cells.
EGFP-IQ(1-2), EGFP-IQ(2-3), and EGFP-IQ(3-4) were generated by PCR using specific primers (Table 1) and CaM binding was examined by 3xFLAG-hCaM IP in HEK293T cells. In these experiments, EGFP-IQ(2-3) could bind to 3xFLAG-hCaM in a Ca2+-independent manner, but the binding to 3xFLAGhCaM was greatly decreased in the presence of Ca2+ (Fig. 2). In the case of EGFP-IQ(3-4), it could bind to 3xFLAG-hCaM in a Ca2+-dependent manner (Fig. 2). In contrast, EGFP-IQ(1-2) could not bind to 3xFLAG-hCaM (Fig. 2). Taken together, these results suggest that IQ2, IQ3 and IQ4 but not IQ1 might be involved in CaM binding.
Fig. 2. Binding of IQ motifs of IQGAP1 to 3xFLAG-hCaM in HEK 293T cells. CaM binding of overexpressed IQ motifs of IQGAP1 fused to EGFP in HEK293T cells using 3xFLAG-hCaM IP. EGFP-IQ(2-3) showed obvious apoCaM binding. EGFP-IQ(3-4) showed significant Ca2+/CaM binding in 3xFLAG-hCaM IP. However, EGFP-IQ(1-2), EGFP-IQ2, EGFP-IQ3 and EGFP-IQ4 displayed no CaM binding in 3xFLAG-hCaM IP. Bound proteins were visualized by immunoblotting using an anti-GFP antibody (upper) or using an anti-FLAG antibody (lower).
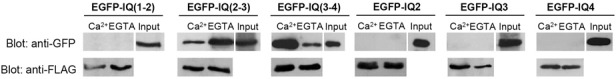
To further examine the CaM binding of IQ2, IQ3 or IQ4 motif individually, CaM binding of EGFP-IQ2, -IQ3 or -IQ4 was evaluated using 3xFLAG-hCaM IP. However, unexpectedly, IQ2, IQ3 or IQ4 itself failed to bind to 3xFLAG-hCaM (Fig. 2). These results suggested that IQ3 and IQ4 of IQGAP1 were not functional motifs for CaM binding. Instead, considering the CaM binding of IQ(2-3) and IQ(3-4), other CaM binding domains might be present within IQ(2-3) or IQ(3-4), respectively.
Novel Ca2+-independent CaM binding domains within IQ motifs of IQGAP1
To test this hypothesis, we examined CaM binding motifs within IQ(2-3) of IQGAP1. Interestingly, EGFP-IQ(2.7-3) including Cterminal IQ2, full length of IQ3 and a linker, showed Ca2+- independent CaM binding similar to EGFP-IQ(2-3) (Fig. 3). To further narrow down the binding motif, we deleted 10 amino acids from the C-terminus of IQ(2.7-3), and generated EGFP-IQ( 2.7-3(Δ10)). However, EGFP-IQ(2.7-3(Δ10)) did not bind to CaM in 3xFLAG-hCaM IP, indicating that the 10 amino acids were necessary for binding (Fig. 3).
Fig. 3. Elucidation of apoCaM binding domain within IQ motifs of IQGAP1 in HEK293T cells. (A) A schematic representation of the IQ motif of IQGAP1. Alignment of amino acid sequences of each constructs. The arrow indicates a point mutation to Asp (D). (B) CaM binding of overexpressed IQ motifs of IQGAP1 using 3xFLAG-hCaM IP in HEK293T cells. EGFPIQ( 2.7-3) showed significant apoCaM binding in 3xFLAG-hCaM IP. On the other hand, EGFP-IQ[2.7-3(Δ10)], IQ(2.7-3, D1/2), and IQ(2.7-3, D3) did not show any CaM binding in 3xFLAG-hCaM IP. Bound proteins were visualized by immunoblotting using an anti-GFP antibody. Each experiment was repeated three times.
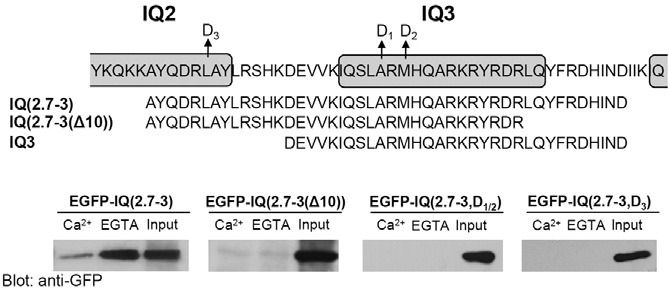
Next, to further characterize the CaM binding sites within this motif, we generated a point mutation construct in which two hydrophobic residues within the IQ3 motif were mutated from ARM to DRD. Previously, it was shown that mutations in selected hydrophobic residues in all IQ motifs of IQGAP1 completely abolished CaM binding (Li and Sacks, 2003). In this study, we mutated two hydrophobic residues from ARM to DRD within IQ3 motif, and generated EGFP-IQ(2.7-3, D1/2) (Fig. 3). Intriguingly, these mutations abolished CaM binding of EGFPIQ( 2.7-3) in 3xFLAG-hCaM IP (Fig. 3). This result suggests that these hydrophobic sites are critical for Ca2+-independent CaM binding.
We then determined whether a partially overlapped IQ2 motif was also involved in CaM binding in IQ(2.7-3). To address this question, we mutated RLA to RDA in the hydrophobic residues within the IQ2 motif overlapped in IQ(2.7-3). This mutant also abolished CaM binding of EGFP-IQ(2.7-3) in 3xFLAG-hCaM IP (Fig 3). This result indicates that this hydrophobic site within IQ2 motif is also essential for CaM binding of IQ(2.7-3).
Novel Ca2+/CaM binding domains within IQ motifs of IQGAP1
We then examined the Ca2+-dependent binding motif within IQ(3-4) of IQGAP1. We analyzed the amino acid sequence of the linker between IQ3 and IQ4, which may be involved in Ca2+/CaM binding. Interestingly, amino acid sequences including the linker between IQ3 and IQ4 contained the 1-8-14 CaM binding motif, which is known as a Ca2+/CaM binding motif [(FILVW)xxxxxx(FAILVW)xxxxx(FILVW)] (Bahler and Rhoads, 2002) (Fig. 4). In addition, basic amino acids were also shown to be important for CaM binding. For example, the Ca2+/CaM binding domain of smooth muscle myosin light chain kinase has basic amino acids in front of the 1-8-14 motif (Blumenthal et al., 1985). Interestingly, IQ3 in IQGAP1 contained a basic rich sequence (RKRYR) within the IQ3 motif in front of the 1-8-14 motif.
Fig. 4. Elucidation of a novel Ca2+/CaM binding domain within IQ motifs of IQGAP1 in HEK293T cells. (A) A schematic representation of the IQ motif of IQGAP1. Alignment of amino acid sequences of each constructs. The arrow indicates a point mutation to Asp (D) or Ala (A). (B) CaM binding of overexpressed IQ motifs of IQGAP1 using 3xFLAG-hCaM IP. EGFPIQ( 3.5-4.4) showed Ca2+-dependent CaM binding. On the other hand, EGFP-IQ(3.5-4.4, D4), and IQ(3.5-4.4, AAA) did not show any CaM binding in 3xFLAGhCaM IP. Bound proteins were visualized by immunoblotting using an anti-GFP antibody. Each experiment was repeated three times.
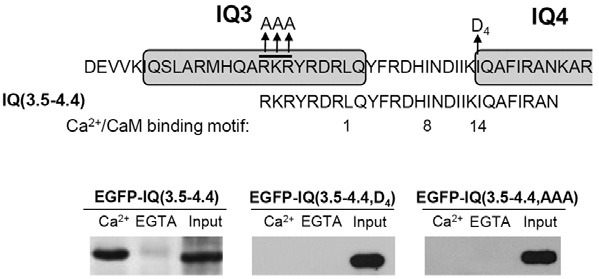
To examine whether the basic rich sequence (RKRYR) within the IQ3 motif and 1-8-14 motif are required for binding, we generated EGFP-IQ(3.5-4.4) containing both 1-8-14 CaM binding motif and basic rich sequence (Fig. 4). Interestingly, as shown in Fig. 4, 3xFLAG-hCaM could bind to IQ(3.5-4.4) in a Ca2+- dependent manner similar to IQ(3-4).
Next, to examine whether the hydrophobic residues in IQ4 was involved in CaM binding, we mutated the hydrophobic residues from IQ to DQ within IQ(3.5-4.4), and generated EGFP-IQ(3.5-4.4, D4). This mutant abolished Ca2+/CaM binding of EGFP-IQ(3.5-4.4) in 3xFLAG-hCaM IP (Fig. 4). To examine the role of the basic rich sequences, we also mutated RKR to AAA in IQ(3.5-4.4). This mutant also abolished Ca2+/CaM binding of EGFP-IQ(3.5-4.4) in 3xFLAG-hCaM IP (Fig. 4). Overall, these results suggest that the hydrophobic site and basic amino acids play important roles in Ca2+/CaM binding of IQ(3.5-4.4).
CaM binding of novel CaM binding motifs of IQGAP1 in vitro
Although we showed the binding of novel CaM binding motifs in HEK293T cells, these results could be due to indirect association of other CaM binding proteins within the cells. Therefore, we determined whether novel CaM binding motifs can directly bind to CaM in vitro. For these experiments, we purified IQ(2.7- 3) or IQ(3.5-4.4) fused to GST in E. coli and performed Fig. CaM binding experiments using CaM 4B in vitro. As shown in Fig. 5, purified IQ(2.7-3) and IQ(3.5-4.4) had two bands. Considering the length of IQ(2.7-3) or IQ(3.5-4.4) compared to that of GST, the upper bands might correspond to the full-length protein and the lower band might be a degradation product. As shown in Fig. 5A, we found that GST-IQ(2.7-3) or IQ(3.5-4.4) could bind to CaM 4B in a Ca2+-independent or -dependent manner, respectively (Fig. 5). As a control, GST alone did not bind to CaM 4B. Overall, our results suggest that IQ(2.7-3) or IQ(3.5-4.4) could directly bind to CaM in a Ca2+-independent or -dependent manner in vitro.
Fig. 5. CaM binding of IQ(2.7-3) and IQ(3.5-4.4) in vitro. (A) CaM binding of purified GST-IQ(2.7-3), GST-IQ (3.5-4.4), or GST-IQ(2-3) motif of IQGAP1 from E. coli using CaM 4B (A). GSTIQ( 2.7-3) and GST-IQ(2-3) displayed CaM binding to CaM 4B in a Ca2+- independent manner. On the other hand, GST-IQ(3.5-4.4) displayed Ca2+- dependent CaM binding. GST, which does not bind CaM, was used as a control. (B) CaM binding of purified GST-IQ(2-3) using purified 3xFLAGhCaM. GST-IQ(2-3) displayed Ca2+-independent CaM binding with 3xFLAG-hCaM. The amount of 3xFLAG-hCaM, which was used as a loading control, was also shown. GST, which does not bind CaM, was used as a control. Bound protein levels were visualized using coomassie blue staining. Each experiment was typically repeated at least three times.
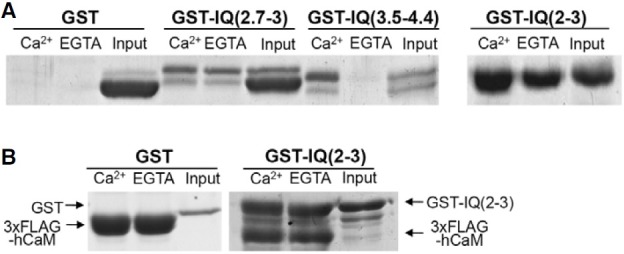
The CaM in vitro binding pattern of IQ(2.7-3) was different from that in 3xFLAG-hCaM IP in HEK293T cells (Figs. 3 and 5). IQ(2.7-3) equally bound to Ca2+/CaM and apoCaM in vitro, while IQ(2.7-3) bound to apoCaM more than to Ca2+/CaM in 3xFLAG-hCaM IP in HEK293T cells. To further confirm the CaM binding of IQ(2.7-3), we examined the CaM binding of IQ(2-3), which preferred apoCaM binding in 3xFLAG-hCaM IP in HEK293T cells. As was observed for IQ(2.7-3), IQ(2-3) also showed equal binding to apoCaM and Ca2+/CaM using CaM 4B in vitro (Fig. 5).
A plausible explanation is that 3xFLAG-hCaM has a different binding property compared to CaM 4B due to the N-terminal fusion of the 3xFLAG into CaM. To test this, we examined the CaM binding of IQ(2-3) in vitro using purified 3xFLAG-hCaM. We purified 3xFLAG-hCaM in E. coli, and measured binding between GST-IQ(2-3) and 3xFLAG-hCaM. However, GST-IQ( 2-3) could also equally bind to 3xFLAG-hCaM in the presence or absence of Ca2+ (Fig. 5B). As a control, GST alone did not bind to 3xFLAG-hCaM. Thus, this data indicates that IQ(2.7-3) and IQ(2-3) equally bound to proCaM and Ca2+/CaM in vitro using 3xFLAG-hCaM.
Disruption of CaM binding by serial mutations within IQ(1-2-3-4)
Finally, to clearly demonstrate CaM binding to the novel CaM binding domains within the IQ motifs of IQGAP1, we generated several point mutants within IQ(1-2-3-4), and examined CaM binding using 3xFLAG-hCaM IP.
First, we examined apoCaM binding of IQ(1-2-3-4). As described above, IQ(2-7-3) is a unique apoCaM binding motif within IQ(1-2-3-4). Therefore, we generated IQ(1-2-3-4, D1/2), and examined CaM binding. Similar to the IQ(2.7-3, D1/2), IQ(1- 2-3-4, D1/2) mutant completely abolished apoCaM binding in 3xFLAG-hCaM IP, but Ca2+/CaM binding remained intact (Fig. 6). This result strongly suggests that the IQ(2.7-3) motif is a unique apoCaM binding site within IQ(1-2-3-4).
Fig. 6. Effect of selective mutations in IQ(1-2-3-4) of IQGAP1 on CaM binding in HEK293T cells. CaM binding of selective mutants of EGFP-IQ(1-2-3-4) using 3xFLAGhCaM IP in HEK293T cells. EGFP-IQ(1-2- 3-4, D1/2) showed impaired apoCaM binding, but intact Ca2+/CaM binding. EGFP-IQ(1-2-3-4, D4) showed partial reduced Ca2+/CaM binding. On the other hand, EGFP-IQ(1-2-3-4, D1/2/4) showed the complete impaired CaM binding. Bound proteins were visualized by immunoblotting using an anti-GFP antibody. Each experiment was typically repeated at least three times.
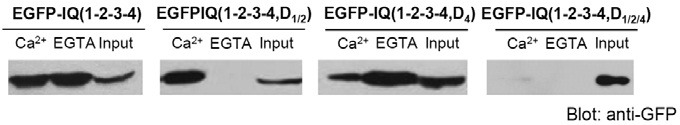
Second, we examined Ca2+/CaM binding of IQ(1-2-3-4). As shown above, IQ(2.7-3) and IQ(3.5-4.4) were involved in Ca2+/ CaM binding. Therefore, we generated IQ(1-2-3-4, D4) and IQ(1-2-3-4, D1/2/4). Our western blot data showed that IQ(1-2-3- 4, D4) partially attenuated Ca2+/CaM binding and intact apoCaM binding was retained (Fig. 6). Meanwhile, IQ(1-2-3-4, D1/2/4) completely abolished CaM binding of IQGAP1(Fig. 6). Taken together, these results indicate that IQ(3.5-4.4) and IQ(2.7-3) are the major Ca2+/CaM binding domains within the IQ motif of IQGAP1.
DISCUSSION
In this study, we discovered novel CaM binding domains within the IQ motifs of IQGAP1 including IQ(2.7-3) and IQ(3.5-4.4), which are believed in more physiologically relevant CaM binding motifs than each IQ motif itself of IQGAP1. We further characterized their binding properties and the specific residues required for CaM binding within the motifs. This study will provide a better understanding of IQGAP1’s binding properties with CaM.
3xFLAG-hCaM IP as a novel CaM binding strategy
In this study, we used 3xFLAG-hCaM IP to identify CaM binding proteins in the cells. This methods was shown to be superior to the CaM 4B pull-down assay in regards to CaM binding specificity. First, we expressed 3xFLAG-hCaM in cells (Fig. 2A). This strategy allowed the expressed 3xFLAG-hCaM to compete with endogenous CaM for association with endogenous CaM binding proteins in the cells. Second, 3xFLAG-peptide elution was used to further exclude non-specific binding in 3xFLAGhCaM IP, which was not available in the CaM 4B pull-down assay. This approach allowed us to assess physiologically relevant CaM binding of 3xFLAG-hCaM in the cells. Using this strategy, we could identify novel and physiologically relevant CaM binding domains within the IQ motif of IQGAP1. Although the length of the 3xFLAG peptide is short, fusion of 3xFLAG to the N-terminus of CaM might distort the binding capacity of Ca2+/CaM or apoCaM. However, we were able to exclude this possibility because various constructs including IQ(1-2-3-4), IQ(2-3), IQ(2.7-3) and IQ(3.5-4.4) showed similar binding properties with CaM 4B. Thus, our approach using 3xFLAG-hCaM IP combined with 3xFLAG elution will be useful for examining CaM binding of various CaM binding proteins in vivo.
Novel apoCaM binding sites within IQ motifs of IQGAP1
Previously, it was shown that four IQ motifs of IQGAP1 were the major CaM binding domains, and the calponin homologue domain had weak CaM binding (Ho et al., 1999; Li and Sacks, 2003). However, we discovered novel CaM binding sites within the IQ motifs of IQGAP1, IQ(2.7-3) and IQ(3.5-4.4).
IQ(2.7-3) showed significant apoCaM binding in vitro and in HEK293T cells. Within the IQ motifs of IQGAP1, this domain is a unique and significant apoCaM binding site, because IQ(1-2- 3-4, D1/2) selectively abolished apoCaM in 3xFLAG-hCaM IP (Fig. 6). Previously, it was shown that IQ3 and IQ4 were involved in apoCaM binding within IQGAP1, since mutations of the Arg residues in IQ3 and IQ4 domains impaired apoCaM binding in the CaM 4B pull-down assay (Li and Sacks, 2003). However, we showed that point mutations of only two hydrophobic residues within IQ3 motif completely impaired apoCaM binding within IQ(1-2-3-4) of IQGAP1 in 3xFLAG-hCaM IP (Fig. 6). Considering that Arg residues of IQ3 belong to a part of IQ(2.7-3), apoCaM binding of IQ3 in a previous report is believed to be mediated by IQ(2.7-3), which is consistent with our result. On the other hand, we found that GST-IQ4 could not bind to CaM 4B in vitro (data not shown). Therefore, the binding of IQ4 to CaM 4B in the previous report might be mediated by an indirect association with other CaM 4B binding proteins in the COS cells; however, this hypothesis will need to be verified by further studies. Thus, our results implied that IQ(2.7-3) might be primarily involved in apoCaM binding in cells.
IQ(2.7-3) or IQ(2-3) could equally associate with CaM 4B or 3xFLAG-hCaM in vitro (Fig. 5). However, using 3xFLAG-hCaM IP in HEK293T cells, IQ(2-3) and IQ(2.7-3) could bind to apo-CaM more than Ca2+/CaM (Figs. 2 and 3). There are several possible reasons why 3xFLAG-hCaM IP binds less to Ca2+/ CaM in HEK293T cells, but equally in vitro. IQ(2-3) seems to have equal apoCaM and Ca2+/CaM binding capacity, because IQ(2-3) could equally and strongly bind to Ca2+/CaM and apo- CaM using CaM 4B or 3xFLAG-hCaM (Fig. 5). Within the cells, there are lots of Ca2+/CaM binding proteins with high affinities for Ca2+/CaM (Jang et al., 2007; Koo et al., 2009; Ruegg et al., 1989). For example, the peptide derived from MLCK binds to Ca2+/CaM with a Kd of 1 nM (Ruegg et al., 1989). Using mRNA display or proteomics combined with CaM pull-down, numerous Ca2+/CaM binding proteins were identified (Jang et al., 2007; Shen et al., 2005). Therefore, a plausible explanation is that although we could not exclude the effects of different tag such as GST and EGFP on CaM binding, the different binding patterns may have been primarily mediated by the different conditions during the binding experiments. In vitro, IQ(2.7-3) competes only with BSA for the binding to CaM 4B or 3xFLAGhCaM, leading to equal association with apoCaM or Ca2+/CaM. On the other hand, in 3xFLAG-hCaM IP, there are lots of Ca2+/CaM binding proteins in HEK293T cells. Several Ca2+/ CaM binding proteins will compete with IQ(2.7-3) or IQ(2-3) to recruit limited 3xFLAG-hCaM in HEK293T cells, resulting in less binding to Ca2+/CaM than apoCaM. Thus, 3xFLAG-hCaM IP will be useful for identifying binding preferences to physiologically relevant apoCaM or Ca2+/CaM.
Novel Ca2+/CaM binding sites within IQ motifs of IQGAP1
We discovered physiologically relevant novel Ca2+/CaM binding sites within the IQ motif of IQGAP1 including IQ(3.5-4.4). The 1-8-14 CaM binding motif [(FILVW)xxxxxx(FAILVW) xxxxx(FILVW)] is known as a Ca2+/CaM binding motif (Bahler and Rhoads, 2002). For example, smooth muscle MLCK, caldesmon, and titin belong to this family. Interestingly, the amino acid sequence of IQ(3.5-4.4) (RKRYRDRL(1)QYFRDHI(8)NDIIKI(14)QAFIRAN) contains this Ca2+/CaM binding motif. Consistent with this, a mutation of IQ to DQ within IQ4 impaired Ca2+/CaM binding of IQ(3.5-4.4). Interestingly, in some proteins, basic amino acids were located in front of the 1-8-14 motif in IQ(3.5-4.4). Usually basic amino acids within a CaM binding motif play a key role in binding. For example, MLCK, which is Ca2+-dependent CaM binding protein, has the basic 1-8-14 CaM binding motif (KRRW(1)KKNFIAV(8)SAANRF(14)KKISSSGAL) with a Kd of 1 nM (Blumenthal et al., 1985). CaMK IV also has the basic 1-8- 14 CaM binding motif (RRKL(1)KAAVKAA(8)VKAVVA(14)SSRLGSA). Although not exactly similar to MLCK or CaMK IV, IQ(3.5-4.4) has three basic aa (RKRYR) located in front of the 1-8-14 motif. This domain is involved in CaM binding, because replacement of RKR to AAA impaired CaM binding (Fig. 4).
In a previous report, only mutations of hydrophobic residues in all IQ motifs of IQGAP1 were required to impair Ca2+/CaM binding in the CaM 4B pull-down assay (Li and Sacks, 2003), suggesting that all IQ motifs were involved in Ca2+/CaM binding. In contrast, our results showed that both IQ(2.7-3) and IQ(3.5- 4.4) were involved in Ca2+/CaM binding, because IQ(1-2-3-4, D1/2/4) completely abolished Ca2+/CaM binding in 3xFLAGhCaM IP (Fig. 4). It might be possible that all IQ motifs are associated with Ca2+/CaM directly or through other binding proteins in the COS cells in the CaM 4B pull-down assay. However, our results showed that under physiological conditions, IQ(2.7- 3) and IQ(3.5-4.4) might be primarily involved in Ca2+/CaM binding in cells.
Overall, in this study, we discovered novel CaM binding motifs in the IQ motifs of IQGAP1. Our approach using 3xFLAGhCaM IP will be useful to examine CaM binding of various CaM binding proteins in vivo including IQGAP2 and IQGAP3. It will be interesting to examine the physiological roles of these novel CaM binding motifs in IQGAP1. Our study will provides a better understanding of the roles of Ca2+ in the regulation of IQGAP1 functions via CaM binding under physiologically relevant conditions.
Acknowledgments
J-A Lee was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010- 0010824), and by a grant of the Korean Health Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A100488).
References
- 1.Alexander K.A., Cimler B.M., Meier K.E., Storm D.R. Regulation of calmodulin binding to P-57. A neurospecific calmodulin binding protein. J. Biol. Chem. (1987);262:6108–6113. [PubMed] [Google Scholar]
- 2.Bahler M., Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. (2002);513:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 3.Baudier J., Deloulme J.C., Van Dorsselaer A., Black D., Matthes H.W. Purification and characterization of a brain-specific protein kinase C substrate, neurogranin (p17). Identification of a consensus amino acid sequence between neurogranin and neuromodulin (GAP43). that corresponds to the protein kinase C phosphorylation site and the calmodulin-binding domain. J. Biol. Chem. (1991);266:229–237. [PubMed] [Google Scholar]
- 4.Blumenthal D.K., Takio K., Edelman A.M., Charbonneau H., Titani K., Walsh K.A., Krebs E.G. Identification of the calmodulin-binding domain of skeletal muscle myosin light chain kinase. Proc. Natl. Acad. Sci. USA. (1985);82:3187–3191. doi: 10.1073/pnas.82.10.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs M.W., Sacks D.B. IQGAP1 as signal integrator: Ca2+ , calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. (2003);542:7–11. doi: 10.1016/s0014-5793(03)00333-8. [DOI] [PubMed] [Google Scholar]
- 6.Briggs M.W., Li Z., Sacks D.B. IQGAP1-mediated stimulation of transcriptional co-activation by beta-catenin is modulated by calmodulin. J. Biol. Chem. (2002);277:7453–7465. doi: 10.1074/jbc.M104315200. [DOI] [PubMed] [Google Scholar]
- 7.Ho Y.D., Joyal J.L., Li Z., Sacks D.B. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J. Biol. Chem. (1999);274:464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 8.Jang D.J., Guo M., Wang D. Proteomic and biochemical studies of calcium- and phosphorylation-dependent calmodulin complexes in Mammalian cells. J. Proteome Res. (2007);6:3718–3728. doi: 10.1021/pr0703268. [DOI] [PubMed] [Google Scholar]
- 9.Joyal J.L., Annan R.S., Ho Y.D., Huddleston M.E., Carr S.A., Hart M.J., Sacks D.B. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J. Biol. Chem. (1997);272:15419–15425. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- 10.Koo S.C., Choi M.S., Chun H.J., Shin D.B., Park S.B., Kim Y.H., Park H.M., Seo H.S., Song J.T., Kang K.Y., et al. The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Mol. Cells. (2009);27:563–570. doi: 10.1007/s10059-009-0081-4. [DOI] [PubMed] [Google Scholar]
- 11.Li Z., Sacks D.B. Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J. Biol. Chem. (2003);278:4347–4352. doi: 10.1074/jbc.M208579200. [DOI] [PubMed] [Google Scholar]
- 12.Li Z., Kim S.H., Higgins J.M., Brenner M.B., Sacks D.B. IQGAP1 and calmodulin modulate E-cadherin function. J. Biol. Chem. (1999);274:37885–37892. doi: 10.1074/jbc.274.53.37885. [DOI] [PubMed] [Google Scholar]
- 13.Mateer S.C., McDaniel A.E., Nicolas V., Habermacher G.M., Lin M.J., Cromer D.A., King M.E., Bloom G.S. The mechanism for regulation of the F-actin binding activity of IQGAP1 by calcium/calmodulin. J. Biol. Chem. (2002);277:12324–12333. doi: 10.1074/jbc.M109535200. [DOI] [PubMed] [Google Scholar]
- 14.Mizumoto T., Hashimoto Y., Hirokawa M., Ohkuma N., Iizuka H., Ohkawara A. Calmodulin activities are significantly increased in both uninvolved and involved epidermis in psoriasis. J. Invest. Dermatol. (1985);85:450–452. doi: 10.1111/1523-1747.ep12277177. [DOI] [PubMed] [Google Scholar]
- 15.Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. IQGAP1: a key regulator of adhesion and migration. J. Cell Sci. (2005);118:2085–2092. doi: 10.1242/jcs.02379. [DOI] [PubMed] [Google Scholar]
- 16.Petrova T.V., Takagi T., Cox J.A. Phosphorylation of the IQ domain regulates the interaction between Ca2+ -vector protein and its target in Amphioxus. J. Biol. Chem. (1996);271:26646–26652. doi: 10.1074/jbc.271.43.26646. [DOI] [PubMed] [Google Scholar]
- 17.Porter J.A., Minke B., Montell C. Calmodulin binding to Drosophila NinaC required for termination of phototransduction. EMBO J. (1995);14:4450–4459. doi: 10.1002/j.1460-2075.1995.tb00124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhoads A.R., Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. (1997);11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 19.Roy M., Li Z., Sacks D.B. IQGAP1 binds ERK2 and modulates its activity. J. Biol. Chem. (2004);279:17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- 20.Roy M., Li Z., Sacks D.B. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol. Cell. Biol. (2005);25:7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruegg J.C., Zeugner C., Strauss J.D., Paul R.J., Kemp B., Chem M., Li A.Y., Hartshorne D.J. A calmodulin-binding peptide relaxes skinned muscle from guinea-pig taenia coli. Pflugers Arch. (1989);414:282–285. doi: 10.1007/BF00584627. [DOI] [PubMed] [Google Scholar]
- 22.Shen X., Valencia C.A., Szostak J.W., Dong B., Liu R. Scanning the human proteome for calmodulin-binding proteins. Proc. Natl. Acad. Sci. USA. (2005);102:5969–5974. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swart-Mataraza J.M., Li Z., Sacks D.B. IQGAP1 is a component of Cdc42 signaling to the cytoskeleton. J. Biol. Chem. (2002);277:24753–24763. doi: 10.1074/jbc.M111165200. [DOI] [PubMed] [Google Scholar]
- 24.Szymanska G., O’Connor M.B., O’Connor C.M. Construction of an epitope-tagged calmodulin useful for the analysis of calmodulin-binding proteins: addition of a hemagglutinin epitope does not affect calmodulin-dependent activation of smooth muscle myosin light chain kinase. Anal. Biochem. (1997);252:96–105. doi: 10.1006/abio.1997.2319. [DOI] [PubMed] [Google Scholar]
- 25.White C.D., Brown M.D., Sacks D.B. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. (2009);583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


