Abstract
Most Arabidopsis ecotypes display tolerance to the Tobacco ringspot virus (TRSV), but a subset of Arabidopsis ecotypes, including Estland (Est), develop lethal systemic necrosis (LSN), which differs from the localized hypersensitive responses (HRs) or systemic acquired resistance (SAR) characteristic of incompatible reactions. Neither viral replication nor the systemic movement of TRSV was restricted in tolerant or sensitive Arabidopsis ecotypes; therefore, the LSN phenotype shown in the sensitive ecotypes might not be due to viral accumulation. In the present study, we identified the Est TTR1 gene (tolerance to Tobacco ringspot virus 1) encoding a TIR-NBS-LRR protein that controls the ecotype-dependent tolerant/sensitive phenotypes by a map-based cloning method. The tolerant Col-0 ecotype Arabidopsis transformed with the sensitive Est TTR1 allele developed an LSN phenotype upon TRSV infection, suggesting that the Est TTR1 allele is dominant over the tolerant ttr1 allele of Col-0. Multiple sequence alignments of 10 tolerant ecotypes from those of eight sensitive ecotypes showed that 10 LRR amino acid polymorphisms were consistently distributed across the TTR1/ttr1 alleles. Site-directed mutagenesis of these amino acids in the LRR region revealed that two sites, L956S and K1124Q, completely abolished the LSN phenotype. VIGS study revealed that TTR1 is dependent on SGT1, rather than EDS1. The LSN phenotype by TTR1 was shown to be transferred to Nicotiana benthamiana, demonstrating functional conservation of TTR1 across plant families, which are involved in SGT-dependent defense responses, rather than EDS1-dependent signaling pathways.
Keywords: Arabidopsis, lethal systemic necrosis, TIR-NBS-LRR, tobacco ringspot virus, tolerance to tobacco ringspot virus 1
INTRODUCTION
Plants have evolved an array of defense mechanisms to counter challenges from a wide range of pathogens. As an initial defense, plants express proteins that recognize and induce nonspecific resistance responses to pathogen-associated molecular patterns (PAMP), similar to the innate immune response of animals (Jones and Dangl, 2006). Next, plants express resistance (R) genes that directly or indirectly recognize the products of pathogen-specific avirulence (Avr) genes to mediate gene-for-gene resistance (Flor, 1971). R gene-mediated pathogen recognition triggers downstream defense responses, which commonly include “rapid” apoptotic death of infected plant cells, manifesting as a hypersensitive response (HR) to minimize the spread of disease local (Hulbert et al., 2001), and ultimately leads to whole plant resistance response, systemic acquired resistance (SAR). SAR is associated with the induction of a pathogen-related (PR) genes and accumulation of endogenous salicylic acid (SA). Systemic necrosis, on the other hand, often found as sensitive responses in both plants and animal systems to pathogens, instead of HR. A phenomenon of lethal systemic necrosis (LSN) that R gene induces systemic cell deaths even with the induction of PR gene expression has not been reported yet.
Most characterized plant R genes show similar structures of nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains (Bent and Mackey, 2007; Jones and Dangl, 2006). The Arabidopsis thaliana genome contains approximately 149 NBS-LRR-encoding genes (Meyers et al., 2003). Some plant species with larger genomes (e.g., Medicago truncatula, Oryza sativa, and Populus trichocarpa) have more than 400 NBSLRR encoding sequences in their genomes (Kohler et al., 2008; Zhou et al., 2004). NBS-LRR type R genes can be grouped into two classes: the CC-NBS-LRR (CNL) type with a coiled-coil (CC) domain and the TIR-NBS-LRR (TNL) type with an orthologous N-terminal domain consisting of Drosophila Toll and mammalian Interleukin-1 receptor (TIR). Fifty-five “CNL” type R genes and 94 “TNL” type R genes in Arabidopsis have been reported (Hulbert et al., 2001), and each type of R protein interacts with different downstream signaling components (NDR1 and EDS1, respectively) to trigger defense responses (Aarts et al., 1998). NBS-LRR domains of RPS5 gene in Arabidopsis are thought to play a role of recognizing and interacting with AvrPphB, a bacterial protease (Ade et al., 2007). AVR-Pita protein binds specifically to the Leucine-rich domain (LRD) at the C-terminus of the Pi-ta protein, which approximately corresponds to the LRR domain of the RPM1 protein, both in the yeast two-hybrid system and in an in vitro binding assay (Jia et al., 2000). The full-length RRS1-R, a TIR-NBS-LRR protein conferring resistance to bacterial wilt, interacts with PopP2, a type III effector of R. solanacearum GMI1000, and NBS-LRR domain may intercepts the PopP2 effector to fulfill the function as a cytoplasmic guard protein (Deslandes et al., 2003). The tobacco N gene, well-known TIR-NBS-LRR class of R gene, confers resistance to TMV (Dinesh-Kumar et al., 1995). Some loss-of-function N alleles such as the TIR deletion and point mutations in the NBS region were found to induce systemic hypersensitive response (SHR), similar phenotype as LSN, by allowing TMV to move systemically within tobacco plants (Dinesh-Kumar et al., 2000). Recently, Stange et al. (2008) reported that five non-conservative substitutions in the LRR regions of Tobacco N gene and N homolog (NH) gene result in differential ligand recognition properties.
Although NBS-LRR proteins are mostly associated with disease resistance, they have also been linked to disease sensitivity in plants as well as hypersensitive human genetic disorders. In humans, a family of proteins containing nucleotide-binding oligomerization domains (NODs) is involved in disease responses and apoptosis (Inohara et al., 1999). Mutations in the NBS region of Nod2 induce the constitutive activation of NF-κB, which is responsible for Blau syndrome, an auto-inflammatory disease (Chamaillard et al., 2003). However, when Nod2 is over-stimulated, it is associated with the chronic intestinal inflammatory disorder Crohn’s disease (Wilmanski et al., 2008; Ye and Ting, 2008). These studies suggest that NBS-LRR proteins can initiate signaling cascades that result in disease, rather than resistance. The innate immune system is the most ancestral response against a variety of microbial challenges and acts as a basal defense system in both plants and animals (Bent and Mackey, 2007; Jones and Dangl, 2006; Medzhitov, 2007).
Tobacco ringspot virus (TRSV) is a member of the Nepovirus genus in the family Comoviridae, and it infects a wide range of dicotyledonous plants (Chu and Francki, 1979). Of 97 Arabidopsis ecotypes tested, most are tolerant and show no obvious disease symptoms to TRSV infection, but a subset of ecotypes are sensitive and develop lethal systemic necrosis (LSN), leading rapidly to death (Lee et al., 1996). A single gene, tolerance to tobacco ringspot virus 1 (TTR1), is thought to be responsible for both the common tolerant phenotype and the necrotic responses seen in the sensitive phenotype (Lee et al., 1996). In this study, we isolated TTR1, which encodes a “TNL”-type protein. Molecular mechanisms of R gene-mediated tolerance and/or sensitivity in the Arabidopsis-TRSV interaction were studied and we show evidence that TTR1-elicited responses are the result of impaired R gene-mediated disease responses.
MATERIALS AND METHODS
Plant materials and TRSV inoculations
Arabidopsis thaliana and Nicotiana benthamiana plants were grown in a growth chamber (23℃, 16 h light/8 h dark) for 3 weeks prior to TRSV inoculation. Disease phenotypes against TRSV were evaluated at 5-15 dpi (days post inoculation) and 5- 7 dpi, respectively. TRSV cultures were maintained and inoculations performed as previously described (Lee et al., 1996; Steere, 1956). The Arabidopsis ecotypes Aa-0, Bsch-0, Bur-0, Cal-0, Co-0, Col-0, Col-4, Cvi-0, Est, Ha-0, H55, Ka-0, Kn-0, Lc-0, Ms-0, Nd-0, Np-0, and Ws-0 lines were obtained from the Arabidopsis Biological Resource Center (ABRC).
Genetic analysis
Reciprocal crosses were made between Est and Col-0 Arabidopsis ecotypes. F1 and F2 progenies of each cross were examined for the disease phenotypes following TRSV inoculation. The segregation ratios of disease phenotypes from each cross were recorded and confirmed by PCR using SSLP markers, which were designed within the region flanking the TTR1 sequence based on information from the Arabidopsis Information Resource (TAIR).
PCR amplification and map-based cloning
Polymerase chain reaction (PCR) was performed using a PTC- 200 cycler (MJ Research, USA) with the following conditions: one cycle at 95℃ for 2 min, followed by 35 cycles at 94℃ for 30 s, 55℃ for 30 s, and 72℃ for 30 s, and finishing at 72℃ for 5 min. TTR1 was mapped between the nga129 and DFR markers on chromosome V, and co-segregated with the yUP5F5LE marker on the yUP5F5 YAC clone (Lee et al., 1996). Approximately 1,500 Col-0 × Est F2 plants were screened for recombinants between the X15S and SMS markers. Subsequently, the markers Y68C and Z67S between X15S and SMS were used to identify polymorphisms between Col-0 and Est. CAPS and SSLP markers used in this study are listed in the Supplementary Table 1. The genotypes of F2 individuals at the TTR1 locus were determined by TRSV inoculation.
Construction of fosmid and subclone libraries
Genomic DNA was isolated from the 3-week-old Est Arabidopsis ecotype as described (Sambrook et al., 2001) and partially digested with Sau3AI for size fractionation (Takara, Japan). End-repaired dephosphorylated 30-40-kb DNA fragments were ligated into the BamHI site of the fosmid vector pCC1FO (Epicentre Biotechnologies, USA). Packaging into phages and transfection into Escherichia coli EPI300-T1 were performed according to the manufacturer’s protocol. Fosmid library were screened and four fosmid clones (418-17, 942-19, 2411-19, 1506-2) were selected. The digested fragments of these fosmid clones were subcloned into a pPZP211 binary vector for plant transformation (Hajdukiewicz et al., 1994).
The nucleotide sequence of Arabidopsis thaliana ecotype Estland (Est) TTR1 has GenBank accession number FJ613384. The nucleotide sequence of various Arabidopsis ecotypes TTR1 partial mRNA described in this study has GenBank accession numbers FJ613385-FJ613402.
Agrobacterium-mediated plant transformation
Arabidopsis Col-0 ecotype plants were transformed with 27 genomic subclones of Est ecotypes using an Agrobacterium tumefaciens-mediated transformation method (Clough and Bent, 1998). N. benthamiana plants were also transformed with a clone (7-21) by A. tumefaciens-mediated transformation (Gallois and Marinho, 1995).
Northern blot analysis
Three-week-old Col-0 and Est Arabidopsis were inoculated with TRSV, and two leaves per ecotype were harvested from 0 to 7 dpi. Total RNA was extracted from these leaf tissues using TRI reagent (Molecular Research Center, Inc., USA), according to the manufacturer’s protocol. TRSV viral CP encoding DNA and PR1-encoding DNA were prepared by PCR and labeled with 32[P]-dCTP using the Multiprime DNA Labeling System (Amersham Pharmacia, USA). Northern blotting and hybridization were performed as described (Sambrook et al., 2001).
Sequence analyses
Amino acid sequences of the tolerant and sensitive Arabidopsis ecotypes were analyzed using the DNAMAN software (Lynnon Co., Canada). The LRR regions of the Arabidopsis ecotypes were PCR-amplified by 3′-RACE (Gibco-BRL, USA). The genespecific primers used to amplify the LRR regions of various ecotypes are shown in Supplementary Table 1.
Site-directed mutagenesis
Single amino acid substitutions in the LRR regions of TTR1 were generated using the QuickChange kit (Stratagene, USA) according to the manufacturer’s protocol. A SpeI/SalI fragment of about 2 kb of the LRR region of TTR1 was subcloned into a T vector (RBC Co., Taiwan) to create a master clone (~5 kb). PCR was performed to introduce a point mutation at each desired site in the LRR region using two synthetic oligonucleotide primers (Supplementary Table 1). Single amino acid substitutions of all 10 TTR1M clones were confirmed by direct DNA sequencing.
Virus-induced gene silencing (VIGS)
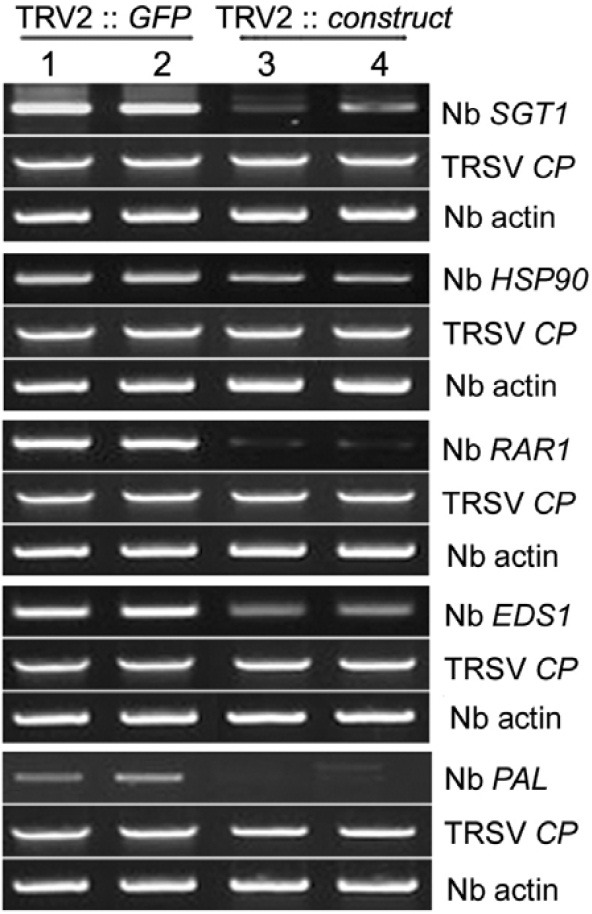
Three-week-old N. benthamiana plants expressing TTR1 were agroinfiltrated with TRV derivatives as previously described (Ratcliff et al., 2001). Agrobacterium with TRV2:pPZP211 vector, TRV2:PDS, and TRV2:GFP constructs were mixed with TRV at a 1:1 ratio and infiltrated individually on the cotyledons of transgenic N. benthamiana as experimental controls (Chung et al., 2004). NbSGT1, NbHSP90, NbRAR1, NbEDS1, and NbPAL were individually silenced by VIGS for 7 days and TRSV was challenged on the upper leaves of the plants with specific genes silenced for evaluation of the disease phenotypes at 5 dpi. Total RNA was isolated from the N. benthamiana leaf and each target mRNA expression level was determined by RT-PCR. The N. benthamiana actin mRNA was used as a positive control.
RESULTS
TRSV disease symptoms in Arabidopsis ecotypes
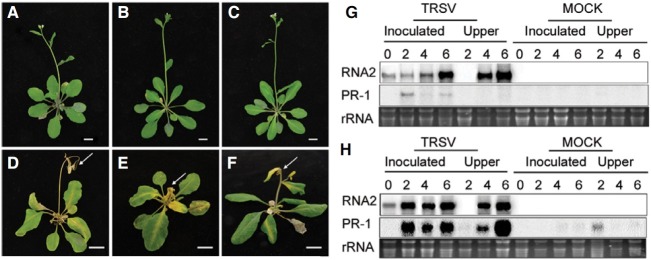
Three-week-old Arabidopsis ecotypes Aa-0, Bsch-0, Col-0, H55, Est, and Ka-0 were inoculated with TRSV. Aa-0, Col-0, and Ka- 0 exhibited tolerance toward TRSV infection displaying no obvious disease symptoms on the inoculated or systemic upper leaves (Figs. 1A-1C). However, ecotypes Bsch-0, Est, and H55 showed necrotic symptoms 4 dpi. Within 2 weeks of TRSV inoculation, shoot apical meristems and flower buds of these sensitive ecotypes completely withered, and growing tips of plants were characteristically hooked (Figs. 1D-1F), indicating that lethal systemic necrosis (LSN) occurred. The LSN response shown in ecotypes Bsch-0, Est, and H55, was different from the local lesions often associated with a localized HR in resistant plants, or from plants expressing systemic acquired resistance (SAR).
Fig. 1. Arabidopsis ecotype-specific tolerant/sensitive responses upon TRSV inoculations. The tolerant Col-0 (A), Aa-0 (B), Ka-0 (C), and sensitive Est (D), Bsch-0 (E), and H55 (F) exhibited distinct disease phenotypes. Lethal systemic necrosis (LSN) phenotype are shown in all inoculated leaves (arrows), systemic upper leaves (*), and shoot apical meristems of the sensitive ecotypes. Northern blot analysis reveals TRSV RNA2 and Arabidopsis PR1 gene expression after TRSV inoculation in tolerant Col-0 (G) and sensitive Est ecotypes (H). Scale bars = 1 cm.
Reciprocal crosses made between the tolerant ecotype Col-0 and the sensitive ecotype Est showed that all of the F1 progeny developed LSN following inoculation with TRSV. In F2 populations, tolerance and sensitivity to TRSV infection segregated in a 1:3 ratio (tolerant:sensitive; Supplementary Table 2). Thus we conclude that the allele for sensitivity to TRSV infection (TTR1) from Est was dominant over the allele for tolerance to TRSV infection (ttr1) from Col-0.
To determine whether the LSN responses of the sensitive Arabidopsis ecotypes were associated with the movement of TRSV, the movement of the virus was monitored over time in TRSV-inoculated Col-0 (Fig. 1G) and Est (Fig. 1H) plants by northern blot analysis using a probe corresponding to the region of RNA 2 (encoding the coat protein). In TRSV-inoculated plants, TRSV RNA was detected by 4 dpi in systemic upper leaves, and the viral RNA accumulation increased to comparable levels in inoculated and systemic upper leaves of both tolerant Col-0 and sensitive Est until 6 dpi, when the TRSVinfected Est plants became necrotic (Figs. 1G and 1H). The mRNA encoding pathogenesis-related protein 1 (PR-1) accumulated to much higher levels in the sensitive ecotype Est than in the tolerant ecotype Col-0, in both inoculated and systemic leaves (Figs. 1G and 1H). At 6 dpi, PR-1 expression was even greater in the systemic upper leaves than in inoculated leaves (Fig. 1H). Thus, the LSN phenotype was associated with the induction of plant defense pathways.
TTR1 encodes a TIR-NBS-LRR protein
A map-based cloning approach was used to identify the gene that confers sensitivity (LSN) to TRSV infection in Est ecotype Arabidopsis. TTR1 was previously mapped to chromosome V, between nga129 and DFR (Lee et al., 1996). For this genetic interval, cleaved amplified polymorphic sequence (CAPS) and simple sequence length polymorphism (SSLP) markers (Supplementary Table 1) were designed between X15S and SMS, and used to fine-map TTR1 in a population of 1,500 F2 plants from crosses between Col-0 and Est. From this population, 30 F2-derived lines were identified that contained recombinations between TTR1/SSLP and CAPS markers, and TTR1 was mapped between the markers Y68C and Z67S in the TAC clones (K23L20 and K21C13). A fosmid library of Est genomic DNA was constructed, and four fosmid clones (418-17, 942-19, 1506-2, and 2411-19) proximal to the TAC clones (K23L20 and K21C13) were selected by PCR using CAPS and SSLP markers (Supplementary Table 1). These fosmid clones were subcloned into the pPZP211 vector (Hajdukiewicz et al., 1994), and Col-0 plants transformed with one subclone, 7-21, responded to TRSV inoculation with the LSN phenotype, indicating that the clone contains the gene representing TTR1 (Fig. 2A). Sequence analysis of the 7-21 subclone showed high sequence identity to a complete open reading frame of Col-0, At5g44870 (1,170 amino acids), with five exons, four introns, and a truncated transposable element. The ORF of the 7-21 clone (TTR1) was re-transformed into Col-0, and LSN phenotype was reproduced upon TRSV infection (Fig. 2B), concluding that this ORF (Est TTR1) is responsible for LSN. Est TTR1 encodes a putative protein of 1,163 amino acids and resembles the “TNL”-type R genes (Fig. 3).
Fig. 2. Map-based cloning of TTR1 of Arabidopsis ecotype Est. (A) Schematic representations of the map-based cloning of TTR1. Contigs of three TAC clones and four fosmid clones were first identified and aligned near the DFR and nga129 markers. Subclones (n = 27) were generated from these fosmid clones, and each subclone was transformed into Col-0. Plants of each transgenic line were evaluated for the disease phenotype following TRSV inoculation. (B) Subclone 7-21, containing a single ORF of Est ecotype (TTR1) and a truncated transposable element, made the normally tolerant Col-0 sensitive to TRSV, whereas Col-0 transformed with the pPZP211 binary vector retained its tolerant phenotype. A single ORF TTR1 was reintroduced into the Col-0 background, and the LSN phenotype was confirmed in T1 plants.
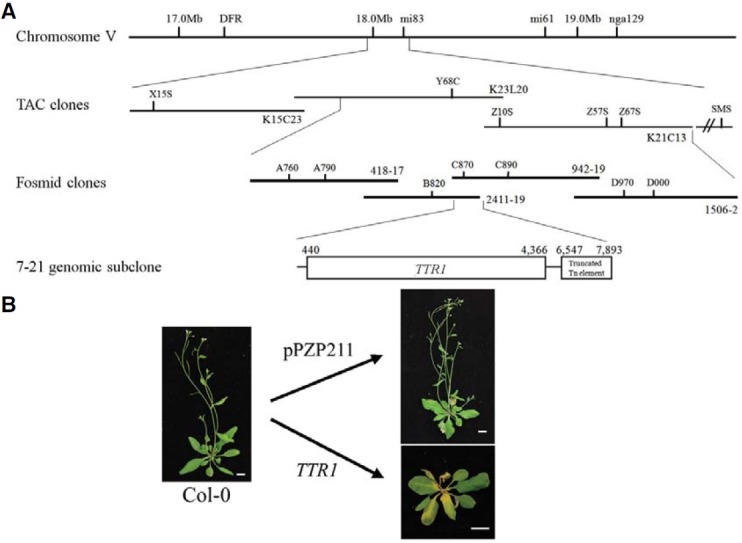
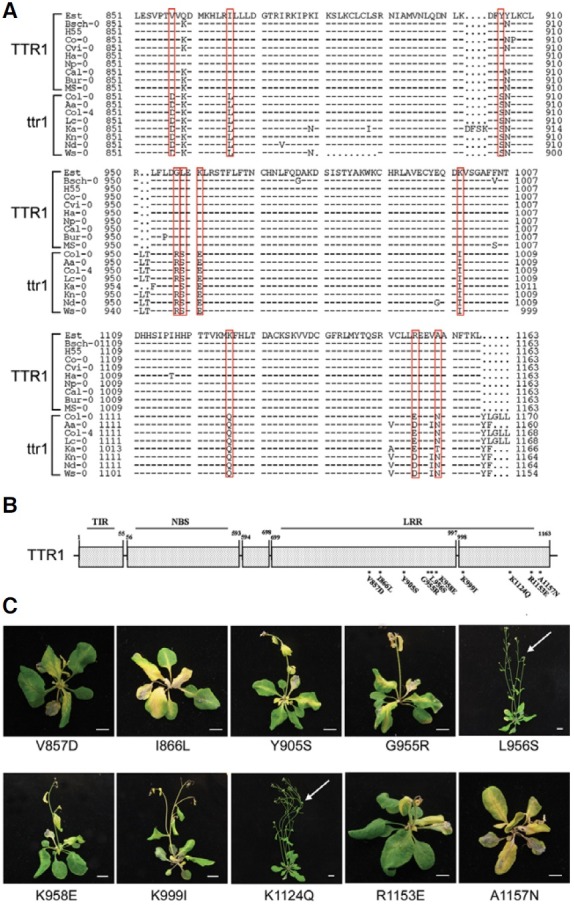
Fig. 3. Comparison of the TTR1 (Est) and ttr1 (Col-0) amino acid sequences. TTR1 encodes a “TNL”-type gene. The predicted sequences of TTR1 in Est and ttr1in Col-0 were aligned and compared using the DNAMAN software. The sequences of the TIRNBS regions in Est and Col-0 were identical, but 21 amino acid differences were observed in the LRR region. Identical amino acids are marked by dashes (-), and amino acid differences are indicated by asterisks (*).

Tolerance/sensitivity determined by the TTR1 LRR amino acid sequence
The predicted amino acid sequences of the TIR and NBS regions of the Est and Col-0 TTR1 proteins were identical, but the LRR regions differed at 21 positions (Fig. 3), indicating that the LRR region are responsible for the sensitivity/tolerance responses of each ecotype to TRSV.
We further evaluated 18 Arabidopsis ecotypes, 10 sensitive (Bsch-0, Bur-0, Cal-0, Co-0, Cvi-0, Est, H55, Ha-0, Ms-0, Np-0) and 8 tolerant (Aa-0, Col-0, Col-4, Ka-0, Kn-0, Lc-0, Nd-0, Ws- 0), for comparison of the predicted LRR regions. Partial amino acid sequences of these 18 ecotypes were amplified by 3′-rapid amplification of cDNA ends (3′-RACE), and direct DNA sequencing of the LRR regions (amino acid positions 851-1,163) revealed 10 amino acid changes that were conserved in all sensitive ecotypes and absent from all tolerant ecotypes (Fig. 4A). These changes are considered as potentially responsible for the LSN response to TRSV infection. A site-directed mutagenesis approach was taken to replace the unique 10 amino acid residues of sensitive TTR1 individually with those from tolerant ttr1 (Figs. 4A and 4B). One or more of each mutated TTR1M clone was introduced into tolerant Col-0 and the phenotype was evaluated after TRSV infection. As shown in Fig. 4C, Col-0 plants transformed with any one of the eight TTR1M clones (V857D, I866L, Y905S, G955R, K958E, K999I, R1153E, A1157N) showed the LSN phenotype upon TRSV inoculation, suggesting that these eight residues of ttr1 are not directly involved in LSN (Fig. 4C). However, Col-0 plants transformed with the L956S or K1124Q clones remained tolerant to TRSV (Fig. 4C), indicating that these two amino acid residues are essential for TTR1 function.
Fig. 4. The LRR region of TTR1/ttr1 determines ecotype-specific responses to TRSV. (A) Multiple sequence alignments of the LRR regions of TTR1/ttr1 loci from multiple Arabidopsis ecotypes, separating A. thaliana ecotypes into two groups. (B) Schematic representation of TTR1, a “TNL”-type gene with five exons, and point mutations used to determine the role of TTR1 in causing LSN. (C) Each mutated clone (TTR1M) was introduced into Col-0 by Agrobacterium-mediated transformation. Plants transformed with L956S and/or K1124Q showed tolerance to TRSV, indicating the loss of TTR1 function. Scale bars = 1 cm.
TTR1 is functional across plant families
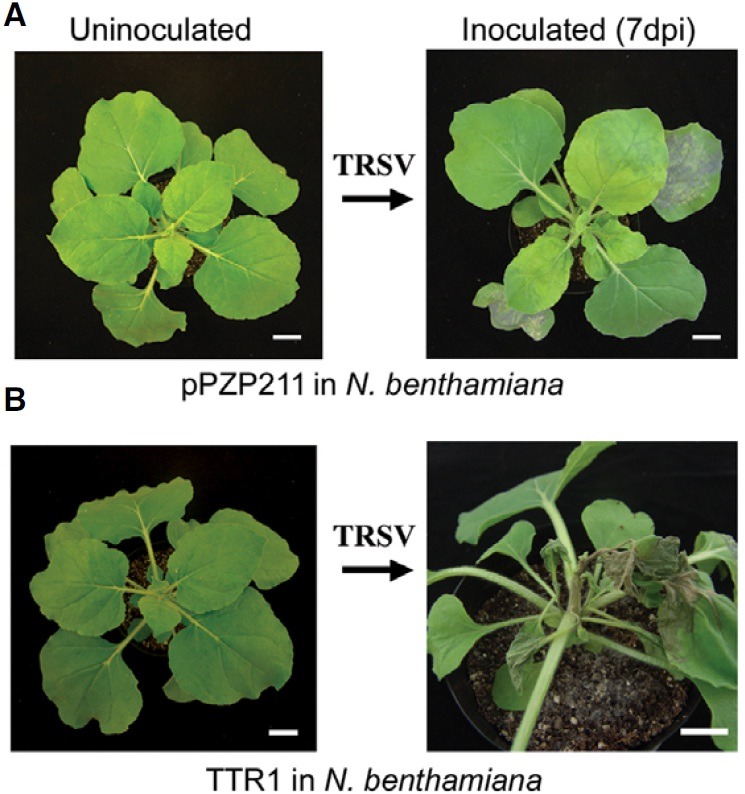
A. thaliana is in the family Brassicaceae, and we decided to investigate TTR1 function in N. benthamiana, which is in the family Solanaceae and is a model system for studying plantvirus interactions (Goodin et al., 2008). We found that wild-type N. benthamiana and N. benthamiana transformed with the pPZP211 binary vector were tolerant to TRSV (Fig. 5A), but N. benthamiana plants transformed with subclone 7-21 (Est TTR1) showed the LSN phenotype upon infection with TRSV (Fig. 5B), suggesting that TTR1 detrimentally interacted with downstream defense pathways in N. benthamiana, as seen in A. thaliana.
Fig. 5. Arabidopsis TTR1 causes LSN in N. benthamiana upon TRSV inoculation. (A) N. benthamiana transformed with the empty pPZP211 binary vector displays no obvious disease symptoms in response to TRSV. (B) N. benthamiana transformed with pPZP211 expressing Est TTR1 exhibited LSN (7 dpi), as seen in sensitive Arabidopsis ecotypes. Scale bars = 2 cm.
We further tested the roles of five genes (NbEDS1, NbSGT1, NbRAR1, NbHsp90, and NbPAL) in different defense-related signaling pathways for TTR1-mediated LSN via a Tobacco rattle virus (TRV) virus-induced silencing (VIGS) in N. benthamiana expressing Arabidopsis TTR1 with a pPZP211 vector and two foreign genes, phytoene desaturase (PDS) and green fluorescence protein (GFP) as VIGS control (Fig. 6). Plastid localized enzyme, PDS converts phytoene to colored ξ-carotene in carotenoid biosynthesis pathway (Wetzel and Rodermel, 1998). VIGS with pPZP211 vector (Fig. 6A) showed no phenotypical changes on growth of N. benthamiana, while silencing of PDS (Fig. 6B) exhibited whitening of leaves indicating successful VIGS of a target gene. Silencing of each defense related gene (NbSGT1, NbRAR1, NbEDS1, and NbPAL1) was also shown by significant reductions in the accumulation of the cognate mRNAs, implying successful silencing of the target (Fig. 7). Notably, no tolerant responses were observed in plants coinfected with TRSV and TRV2 containing GFP, NbEDS1, NbHsp90, NbPAL, or NbRAR1 (Figs. 6C, and 6E-6H). However, all plants co-inoculated with TRSV and TRV2 containing SGT1 were tolerant to TRSV infection (Fig. 6D), indicating that only SGT1 is involved in LSN. If this is true, the Arabidopsis TTR1 may operate to induce LSN via defense-related signaling pathways that exclusively involve SGT1, not Hsp90/RAR1. TTR1 mediated LSN is still remained to be elucidated, especially how to relay information leading to LSN with potential downstream components.
Fig. 6. The effects of various defense-related genes on the SA- and SGT/Hsp90/RAR1-mediated signaling pathways on the LSN phenotype in N. benthamiana. (A) pPZP211 vector control, (B) PDS, (C) GFP, (D) NbSGT1, (E) NbHSP90, (F) NbRAR1, (G) NbEDS1, and (H) NbPAL. Arrows indicate a withered shoot apical meristems, a sign of LSN by TTR1 in response to TRSV. Scale bars = 2 cm.

Fig. 7. Semiquantitative RT-PCR analyses to examine endogenous target gene expression levels. At 5 dpi, total RNA was purified from N. benthamiana leaf disks and used as a template for RT-PCR (25 cycles) to amplify N. benthamiana homologs of various plant defense-related genes to confirm silencing of target mRNAs. The N. benthamiana actin mRNA was used as a positive control.
DISCUSSION
To understand the molecular mechanisms underlying the ecotype- specific tolerance/sensitivity to TRSV, the TTR1 gene from the Arabidopsis ecotype Est, which induces LSN, was isolated by map-based cloning. We confirmed that the “TNL”-type Est TTR1 (a counterpart of Col-0 At5g44870, ttr1) indeed induces LSN by over-expression of the ORF of Est TTR1 with native promoter into the tolerant ecotype Col-0 in response to TRSV. Distinct amino acid changes were found between positions 823 and 1,170 of the LRR region of TTR1/ttr1, clearly dividing the Arabidopsis ecotypes into sensitive and tolerant subgroups (Fig. 2A). Amino acid sequence variations in the LRR regions of R proteins are believed to facilitate protein recognition of a variety of pathogen-derived molecules (Hulbert et al., 2001; Meyers et al., 2003).
In this study, we identified two amino acids in the LRR region that are responsible for sensitive ecotype-specific LSN responses to TRSV. Stange et al. (2008) reported that five nonconservative substitutions in the LRR regions of Tobacco N gene and NH gene (Gly81 → Arg80, Leu153 → Asp152, Arg198 → Glu197, Val99 → Glu198 and Ile221 → Lys220) result in differential ligand recognition properties. Comparative three dimensional models of N-LRR and NH-LRR proposed that the LRR region is important in pathogen recognition (Stange et al., 2008). Thus, we could postulate that two residues in the LRR region of TTR1, Leu956 and Lys1124, identified in this study as responsible amino acids for LSN phenotype of TTR1 might also be important for the proper LRR domain folding, leading to stable or transient recognition of TRSVderived molecules.
SAR is an inducible broad spectrum plant defense response that requires the accumulation of SA. Many “TNL”-type R proteins require EDS1, not NDR1, for defense response activities (Lee et al., 1996; Peart et al., 2002), leading to HR or SAR. As TTR1 function was shown to be transferred to another plant family, the Solanaceae (N. benthamiana), we utilized VIGS to further characterize the relationship between TTR1 and five known signaling components downstream. Interestingly, the VIGS results showed that TTR1 was not regulated by NbEDS1-dependent manner, but NbSGT1-dependent, suggesting that TTR1 might utilize unique regulatory systems to respond TRSV, as opposed to use the SA mediated signaling pathway by most known R genes. It has been reported that Hsp90, RAR1 and SGT1 play essential roles in development and disease resistance triggered by a number of R genes in plants (Hubert et al., 2003; Liu et al., 2004; Tai, 2008; Takahashi et al., 2003). But, in some cases, effector triggered immunity (ETI) and SAR requires only GmRAR1 and GmSGT1, but not GmHsp90, suggesting that RAR1- or SGT1 dependent signaling pathway does not always require Hsp90 in disease resistance in Soybean (Fu et al., 2009). This may be the case for TTR1 in Est ecotype Arabdiospsis, since we found that silencing of NbSGT1 abrogated LSN in TTR1 expressing N. benthamiana in response to TRSV infection.
Several lines of evidence have also demonstrated that innate immune responses are conserved in plants and animals (Burch-Smith et al., 2007). Many reports have shown that the Nod protein is a member of a family that includes the apoptosis regulator APAF1, a mammalian Nod-LRR (NLR) protein (Inohara et al., 1999). Two members of NLR family, Nod1 and Nod2, are reported to function as sensors to recognize distinct substructures of intracellular bacteria, activating diverse signal pathways (da Silva Correia et al., 2007). As the mutations of the LRR region of ttr1 induced LSN following TRSV infection, mutation of the LRR region of Nod2 is also believed to be responsible for Crohn’s disease, a chronic inflammatory disease (Chamaillard et al., 2003; Hugot et al., 2001). Nod1 and Nod2 interact with endogenous SGT1 and HSP90, which are components of the protein degradation and folding machinery, and participate in early resistance gene signaling pathways (Austin et al., 2002; Azevedo et al., 2002). Therefore, the LRR regions of both Nod2 and TTR1 may play similar roles with unique three dimensional structures recruiting unique regulatory systems to respond against pathogens. In light of mammalian Nod proteins, we propose that the Arabidopsis TTR1 supports the concept that innate immunity is conserved across different kingdoms. LSN phenotype observed in Est ecotype (sensitive to TRSV) should be differentiated from SAR of the Col-0 ecotype (tolerance to TRSV). Further investigation of the involvement of SGT1 in the unique TTR1-mediated disease response mechanism could provide more evidence to support conserved innate immunity across different kingdoms.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
This work was supported by the Crop Functional Genomics Center (grant number CG2132), the Korea Research Foundation Grant funded by the Korean Government (KRF-2008-532-F00001) and by a grant (Project No. 609002-5) from the Screening Center for Disease Resistant Vegetable Crops of TDPAF funded by Ministry for Food, Agriculture, Forestry and Fisheries of Korean government and 2011 Post Doctoral Course Program of National Academy of Agricultural Science, Rural Development Administration, Republic of Korea.
References
- 1.Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA. (1998);95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ade J., DeYoung B.J., Golstein C., Innes R.W. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA. (2007);104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin M.J., Muskett P., Kahn K., Feys B.J., Jones J.D., Parker J.E. Regulatory role of SGT1 in early R genemediated plant defenses. Science. (2002);295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- 4.Azevedo C., Sadanandom A., Kitagawa K., Freialdenhoven A., Shirasu K., Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. (2002);295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 5.Bent A.F., Mackey D. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. (2007);45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 6.Burch-Smith T.M., Schiff M., Caplan J.L., Tsao J., Czymmek K., Dinesh-Kumar S.P. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. (2007);5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamaillard M., Girardin S.E., Viala J., Philpott D.J. Nods, Nalps and Naip: intracellular regulators of bacterialinduced inflammation. Cell. Microbiol. (2003);5:581–592. doi: 10.1046/j.1462-5822.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- 8.Chu P.W., Francki R.I. The chemical subunit of tobacco ringspot virus coat protein. Virology. (1979);93:398–412. doi: 10.1016/0042-6822(79)90244-7. [DOI] [PubMed] [Google Scholar]
- 9.Chung E., Seong E., Kim Y.C., Chung E.J., Oh S.K., Lee S., Park J.M., Joung Y.H., Choi D. A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol. Cells. (2004);17:377–380. [PubMed] [Google Scholar]
- 10.da Silva Correia J., Miranda Y., Leonard N., Ulevitch R. SGT1 is essential for Nod1 activation. Proc. Natl. Acad. Sci. USA. (2007);104:6764–6769. doi: 10.1073/pnas.0610926104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deslandes L., Olivier J., Peeters N., Feng D.X., Khounlotham M., Boucher C., Somssich I., Genin S., Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA. (2003);100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinesh-Kumar S.P., Whitham S., Choi D., Hehl R., Corr C., Baker B. Transposon tagging of tobacco mosaic virus resistance gene N: its possible role in the TMV-N-mediated signal transduction pathway. Proc. Natl. Acad. Sci. USA. (1995);92:4175–4180. doi: 10.1073/pnas.92.10.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinesh-Kumar S.P., Tham W.H., Baker B.J. Structure- function analysis of the tobacco mosaic virus resistance gene N. Proc. Natl. Acad. Sci. USA. (2000);97:14789–14794. doi: 10.1073/pnas.97.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flor H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. (1971);9:275–296. [Google Scholar]
- 15.Fu D.Q., Ghabrial S., Kachroo A. GmRAR1 and GmSGT1 are required for basal, R gene-mediated and systemic acquired resistance in soybean. Mol. Plant Microbe Interact. (2009);22:86–95. doi: 10.1094/MPMI-22-1-0086. [DOI] [PubMed] [Google Scholar]
- 16.Gallois P., Marinho P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Methods Mol. Biol. (1995);49:39–48. doi: 10.1385/0-89603-321-X:39. [DOI] [PubMed] [Google Scholar]
- 17.Goodin M.M., Zaitlin D., Naidu R.A., Lommel S.A. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. (2008);21:1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 18.Hajdukiewicz P., Svab Z., Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. (1994);25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 19.Hubert D.A., Tornero P., Belkhadir Y., Krishna P., Takahashi A., Shirasu K., Dangl J.L. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. (2003);22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cezard J.P., Belaiche J., Almer S., Tysk C., O’Morain C.A., Gassull M., et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. (2001);411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 21.Hulbert S.H., Webb C.A., Smith S.M., Sun Q. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. (2001);39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 22.Inohara N., Koseki T., del Peso L., Hu Y., Yee C., Chen S., Carrio R., Merino J., Liu D., Ni J., et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. (1999);274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 23.Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. (2000);19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones J.D., Dangl J.L. The plant immune system. Nature. (2006);444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.M., Hartman G.L., Domier L.L., Bent A.F. Identification and map location of TTR1, a single locus in Arabidopsis thaliana that confers tolerance to tobacco ringspot nepovirus. Mol. Plant Microbe Interact. (1996);9:729–735. doi: 10.1094/mpmi-9-0729. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y., Burch-Smith T., Schiff M., Feng S., Dinesh-Kumar S.P. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. (2004);279:2101–2108. doi: 10.1074/jbc.M310029200. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. (2007);449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 28.Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. (2003);15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peart J.R., Cook G., Feys B.J., Parker J.E., Baulcombe D.C. An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. (2002);29:569–579. doi: 10.1046/j.1365-313x.2002.029005569.x. [DOI] [PubMed] [Google Scholar]
- 30.Ratcliff F., Martin-Hernandez A.M., Baulcombe D.C. Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. (2001);25:237–245. doi: 10.1046/j.0960-7412.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 3rd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor: (2001). [Google Scholar]
- 32.Stange C., Matus J.T., Dominguez C., Perez-Acle T., Arce-Johnson P. The N-homologue LRR domain adopts a folding which explains the TMV-Cg-induced HR-like response in sensitive tobacco plants. J. Mol. Graph. Model. (2008);26:850–860. doi: 10.1016/j.jmgm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Tai Y.S. Interactome of signaling networks in wheat: the protein-protein interaction between TaRAR1 and TaSGT1. Mol. Biol. Rep. (2008);35:337–343. doi: 10.1007/s11033-007-9091-5. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi A., Casais C., Ichimura K., Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA. (2003);100:11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wetzel C.M., Rodermel S.R. Regulation of phytoene desaturase expression is independent of leaf pigment content in Arabidopsis thaliana. Plant Mol. Biol. (1998);37:1045–1053. doi: 10.1023/a:1006021522259. [DOI] [PubMed] [Google Scholar]


