Abstract
Hydrogen peroxide (H2O2) functions as a ubiquitous intracellular messenger besides as an oxidative stress molecule. This dual role is based on the distinct cellular responses against different concentrations of H2O2. Previously, we demonstrated that both low (> 1 mM) and high (4–10 mM) doses of exogenous H2O2 induce filamentous growth with distinct cell morphology and growth rate in Candida albicans, suggesting the different transcription response. In this study, we revealed that the sub-toxic and toxic levels of H2O2 indeed induced pseudohyphae, but not true hyphae. Supporting this, several hyphae-specific genes that are expressed in true hyphae induced by serum were not detected in either sub-toxic or toxic H2O2 condition. A DNA microarray analysis was conducted to reveal the transcription profiles in cells treated with sub-toxic and toxic conditions of H2O2. Under the sub-toxic condition, a small number of genes involved in cell proliferation and metabolism were up-regulated, whereas a large number of genes were up-regulated in the toxic condition where the genes required for growth and proliferation were selectively restricted. For pseudohyphal induction by sub-toxic H2O2, Cek1 MAPK activating the transcription factor Cph1 was shown to be important. The absence of expression of several hyphae-specific genes known to be downstream targets of Cph1-signaling pathway for true hyphae formation suggests that the Cek1-mediated signaling pathway is not solely responsible for pseudohyphal formation by subtoxic H2O2 and, but instead, complex networking pathway may exists by the activation of different regulators.
Keywords: Candida albicans, H2O2, intracellular messenger, signaling pathway, transcription profiling
INTRODUCTION
Increasing evidence supports an alternative and beneficial function of reactive oxygen species (ROS) as an important regulator of nitric oxide- or calcium-mediated signal transduction (Thannickal et al., 2000). Phagocytic immune cells activate NADPH oxidase complexes to generate oxygen radical (O2−·), which is subsequently converted to hydrogen peroxide (H2O2), as a cytotoxic agent during the engulfment of microbes (Lorenz et al., 2004). In addition, studies conducted in non-immune cells have implicated the Nox family of NADPH oxidases with the generation of ROS in response to various extra-cellular stimuli including cytokines that regulate various cellular functions including immunity, cell proliferation, cell differentiation, signal transduction, and ion transport (Foreman et al., 2003; Reth, 2002; Rhee et al., 2000; 2005). Even though the chemical nature of ROS generated in response to the activation of various receptors has not been well-characterized, H2O2 represents a major component of ROS in cells activated by cytokine or growth factors (Ohba et al., 1994; Sundaresan et al., 1995). Thus, H2O2 is considered to function as a ubiquitous intracellular messenger whose localization, expression, and activity are tightly regulated. Moreover, different levels of H2O2 can induce distinct responses within a cell. For example, distinct transcriptional responses are induced in response to low (sub-toxic) and high (toxic) levels of H2O2 in mammalian cells, Saccharomyces cerevisiae, or Schizosaccharomyces pombe (Quinn et al., 2002; Vivancos et al., 2006). At low levels of H2O2, cells become mitogenic and proliferative, while pro-oxidants involved in apoptosis are expressed in cells exposed to high levels of H2O2 (Sablina et al., 2005), indicative of concentration-specific responses to H2O2. These contradictory roles of H2O2 have spurred cellular evolution of several powerful enzymatic systems to prevent excessive accumulation of H2O2, thereby maintaining homeostasis at the cellular regulatory level (Stone et al., 2006; Veal et al., 2007).
Candida albicans is a major human pathogen that causes diseases ranging from thrush and vaginal yeast infections in normal individuals to life-threatening systemic infections in immunocompromised individuals (Calderone et al., 2001). An important feature of C. albicans that is relevant to pathogenesis is its ability to switch from a budding yeast form to a filamentous form that includes both pseudohyphae and true hyphae (Lo et al., 1997; Sudbery et al., 2004). The morphological transition is triggered by various nutritional and environmental factors such as specific carbohydrates or amino acids, serum, high temperature, neutral pH, N-acetyl-glucosamine, high carbon dioxide, and starvation (Biswas et al., 2007). These various hyphal inducers trigger a wide range of signal transduction pathways involved in morphogenesis (Cottier et al., 2009; Dhillon et al., 2003). The well-characterized signaling pathways implicated in the morphological transition are the cyclic AMP (cAMP) pathway and the mitogen-activated protein kinase (MAP kinase) pathway (Roman et al., 2007). The cAMP pathway plays a major role in hyphal development and pathogenesis in C. albicans (Harcus et al., 2004; Rocha et al., 2001). Efg1, a basic helix-loop-helix protein, is the major transcription factor in the cAMP pathway (Stoldt et al., 1997) and plays a critical role in hyphal morphogenesis (Harcus et al., 2004). An efg1 mutant strain is defective in hyphal development under most hyphal-inducing conditions, including serum, and exhibits reduced virulence (Leng et al., 2001).
MAPK pathways drive a variety of mechanisms in eukaryotic cells to couple environmental responses to transcriptional regulation. In C. albicans, there are three different MAPK pathway routes involving Hog1, Cek1, and Mkc1. The Hog1 MAPK pathway is involved in at least three separate processes: response/adaptation to stress, morphogenesis, and cell wall formation. The Cek1 MAPK pathway of C. albicans includes Cst20 MAPK kinase kinase (MAPKKK), Hst7 MAPK kinase (MAPKK), Cek1 MAPK, and the transcription factor Cph1 (Leberer et al., 1996). Null mutants for any of these genes are defective in hyphal development on solid medium in response to inducers such as synthetic low ammonium; however, hyphae develop normally in response to serum (Csank et al., 1998; Leberer et al., 1996; Liu et al., 1994). Mkc1, the homologue of the S. cerevisiae Mpk1 MAPK, plays a role in maintaining cellular integrity and cell wall formation as deduced from the osmotically-remediable sensitivity of mutant cells to certain cell-wall-interfering compounds (Navarro-Garcia et al., 1995).
We previously demonstrated that exogenous H2O2 induces filamentous growth in C. albicans (Nasution et al., 2008). However, sub-toxic (1 mM) and toxic (10 mM) concentrations of H2O2 produced distinct cell morphologies and growth rate effects. Cells grown at the sub-toxic level of H2O2 exhibited a mixture of normal yeast and pseudohyphae forms, and a growth rate similar to untreated control cells. In contrast, most cells grown at the toxic concentration of H2O2 exhibited swollen pseudohyphae and severely impaired growth. The present study aimed to understand the mechanism that underlies these cellular and physiological differences. In response to various environmental stimuli, unicellular organisms like C. albicans undergo a substantial modulation of the gene expression pattern. DNA microarray-based transcriptome analyses of C. albicans cells treated with sub-toxic and toxic H2O2 were performed to investigate the difference in gene expression.
MATERIALS AND METHODS
Gene names
All C. albicans genes and proteins are hereafter named without prefix, while those of other organisms are prefixed (e.g., S. cerevisiae CPH1 and its protein product are designated ScCPH1 and ScPrx1p, respectively).
Strains and cell culture
The C. albicans strains are listed in Table 1. Unless mentioned otherwise, cells were cultured at 30°C in YPD medium (1% yeast extract, 2% peptone, and 2% glucose).
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SC5314 | Wild-type | W. Fonzi |
| JKC19 | Δura3::imm434/Δura3::imm434, Δcph1::hisG/Δcph1::hisG | S. Kang |
| HLC52 | Δura3::imm434/Δura3::imm434, Δefg1::hisG/Δefg1::hisG | S. Kang |
| HLC54 |
Δura3::imm434/Δura3::imm434, Δcph1::hisG/Δcph1::hisG Δefg1: hisG/Δefg1::hisG-URA3-hisG |
S. Kang |
| CK43B-16 | ura3/ura3 cek1Δ::hisG-URA3-hisG/cek1Δ::hisG | M. Whiteway |
| CK43B-RI | ura3/ura3 cek1Δ::hisG/cek1Δ::hisG::CEK1-URA3 | M. Whiteway |
Hyphal induction
Yeast cells were transferred to YPD supplemented with 10% fetal bovine serum (FBS) (Welgene) and further grown at 37°C for either 2 h for germ tube or 6 h for true hyphae. To assess the effect of H2O2, overnight cultures were diluted with YPD medium supplemented with various concentrations of H2O2 and further incubated at 30°C.
Apoptosis assay
Cellular integrity and externalization of phosphatidylserine (PS) were assessed using an apoptosis detection kit (Koma Biotech). Appropriately treated cells were washed twice in phosphate buffered saline (PBS), resuspended in 1 ml KS buffer (1 M sorbitol, 0.1 M potassium phosphate, pH 7.0) and digested for 30 min at 33°C with 10 U zymolyase (Zymo Research) in 10 mM 2-mercaptoethanol. The resulting protoplasts were washed with and resuspended in 0.2 ml annexin binding buffer (Zymo Research). Then, 3 μl of annexin V-fluorescein isothiocyanate (FITC) (200 μg ml−1) and 10 μl of propidium iodide (PI; 30 μg ml−1) were added and incubated for 30 min in the dark. Excitation and emission wavelengths were 488 and 518 nm, respectively, for FITC and 540 and 620 nm, respectively, for PI. Plain and fluorescent images were immediately captured with a fluorescence microscope (Carl Zeiss).
RNA preparation
Total RNA was prepared using an RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions.
Microarray and data analysis
Whole genome microarray slides for C. albicans were purchased from NRC Biotechnology Research Institute (Montreal). Labeled dCTP was prepared by conjugating Cy3 or Cy5 (Genechem) with dCTP. Cy3- or Cy5-labeled cDNAs were prepared by reverse transcription of total RNA from untreated control and H2O2-treated cells with Cy3- and Cy5-labled dCTP, respectively. Labeled cDNAs were ethanol-precipitated and resuspended in 30 μl of hybridization solution [1× SSC, 0.25 M Na2HPO4, 2× Denhardt, 1 mM EDTA, 4.5% sodium dodecyl sulfate (SDS)]. After the control Cy3-labeled cDNA was mixed with each of Cy5-labeled cDNAs, the mixture was placed on the C. albicans 14K chips in the MAUI AO chamber (BioMicro Systems). The slides were hybridized for 12 h at 62°C and then washed at room temperature with 2× SSC and 0.1% SDS for 2 min, 1× SSC for 3 min, and 0.2× SSC for 2 min. The slides were centrifuged at 3000 rpm for 20 s to dry. The hybridized slides were scanned using a GenePix 4000B scanner (Molecular Devices) and the images were analyzed using GenePix Pro 5.1 software (Molecular Devices) and GeneSpring GX 7.3.1 software (Agilent Technologies). Spots that were judged as substandard by visual examination of each slide and those that had dust artifacts or spatial defects were flagged and excluded for further analysis. To filter out unreliable data, spots with signal-to-noise (signal background standard deviation - background standard deviation) below 10 were not included in the data. Data were normalized by Global, LOWESS, print-tip, and scaled normalization for data reliability. Fold-change filters included the requirement that the genes be present in at least 200% of controls for up-regulated genes and lower than 50% of controls for down-regulated genes. Data were clustered in groups of genes that behaved similarly in a time course experiment using GeneSpring GX 7.3.1 software. An algorithm based on the Pearson correlation was utilized to separate genes of similar patterns. Data from four independent biological replicates were used for each analysis. To determine the degree of induction of gene expression, spot intensities were normalized against ACT1. The adjusted values were used to determine differential gene expression (Cy3/Cy5) for each spot. The values for each experiment corresponding to particular genes were averaged. The reproducibility of the microarray analysis was assessed by correlation of datasets. Functional classification of genes was carried out based on gene ontology (GO) terms addressed in the Candida Genome Database (www.candidagenome.org). The whole microarray data generated by this study has been submitted to ArrayExpress (http://www.ebi.ac.uk/microarry-as/ae/) under accession number E-MEXP-2680.
Polymerase chain reaction (PCR)
The utilized oligonucleotide PCR primers are listed in Supplementary Table 4. The amplification conditions were 95°C for 1 min, 55°C for 1 min, and 72°C for an appropriate period of time depending on the length of DNA to be amplified. If sequencing is necessary, PCR products were gel-purified and cloned into the pGEM-T easy vector (Promega).
Northern blot analysis
Fifteen micrograms of total RNA was used. DNA fragments used for probe were prepared by PCR from SC5314 genomic DNA with the appropriate gene-specific primers. Purified PCR fragments were radioactively labeled with 32P-dCTP (GE Healthcare) with random primers. The rest procedures were carried out as previously described (Sambrook et al., 2001). Actin was used as an internal control.
Western blot analysis
Total extracts were routinely prepared from mid-logarithmic growth phase cells as described previously (Shin et al., 2005). Proteins in 30 μg of total extract were resolved by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. The primary antibody was an anti-phospho-p44/p42 MAPK (Erk1/2) (Thr202/Tyr204) rabbit antibody (Cell Signaling Technology) and the secondary antibody was horseradish peroxidase (HRP)-conjugated goat antibody against rabbit IgG (Santa Cruz Biotechnology). Protein bands were visualized by using an enhanced chemiluminescence system (GE Healthcare).
Calcofluor staining
Cells were washed twice and resuspended in water and incubated for 30 min with 20 μl of calcofluor (1 mg ml−1 in water). Then, cells were washed twice with PBS. The fluorescence was observed at 365 nm with a LSM 510 Meta confocal laser scanning microscope (Carl Zeiss).
RESULTS
Toxicity of exogenous H2O2
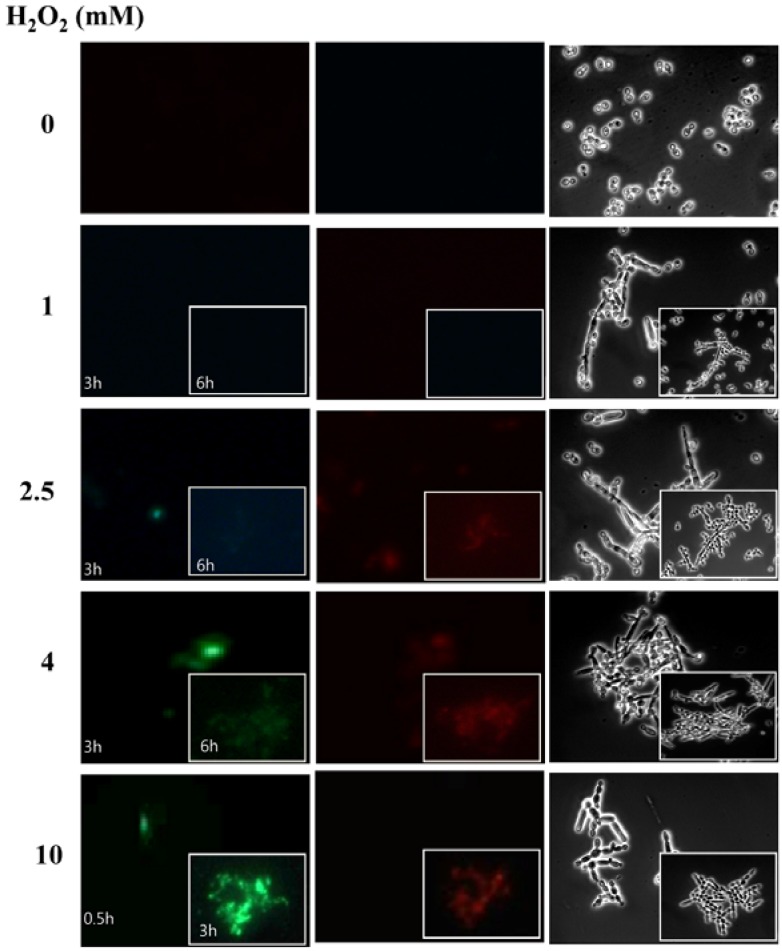
In response to low and high doses of H2O2, cells exhibit distinct biological responses (Martin et al., 2002). However, depending on the cell types examined, there is considerable variation in the concentration of exogenous H2O2 required to initiate a particular biological response (Buggisch et al., 2007; Li et al., 2006; Mesquita et al., 2009). We previously observed that C. albicans treated with a low dose (1 mM) of H2O2 for 3 h undergoes normal filamentation, whereas cells treated with a high dose (10 mM) of H2O2 for 30 min exhibit swollen filamentous growth (Nasution et al., 2008). Another study reported that C. albicans treated with 10 mM H2O2 for 200 min is arrested at G2/M and undergoes apoptosis (Phillips et al., 2003). Thus, 10 mM H2O2 for 30 min or longer is considered to be toxic to C. albicans. Apoptosis is also observed in a small portion (approximately 10%) of cells treated with as low as 2.5 mM H2O2 for 200 min (Phillips et al., 2003). Accordingly, the toxicity of exogenous of H2O2 needs to be clearly defined in C. albicans. In this study, the toxicity of various concentrations of exogenous H2O2 was determined at two time points on the basis of cellular integrity and externalization of PS. Compared to untreated control cells, no sign of apoptotic or necrosis was detectable at 1 mM H2O2 (Fig. 1). However, a larger portion of cells treated with ≥ 2.5 mM for 3 and 6 h underwent apoptosis indicated by annexin (+) PI (−), and necrosis annexin (+) PI (+) (Fig. 1). Thus the toxicity of H2O2 seems to be function of concentration and exposure time. So, we defined the sub-toxic condition as 1 mM for 3 h and the toxic condition as 10 mM for 30 min for pseudohyphal induction by H2O2.
Fig. 1.
H2O2 toxicity. Cells were treated with various concentrations of H2O2 for the indicated periods of time. Fluorescence microscopy examination revealed healthy cells with annexin V (−) PI (−), apoptotic cells with annexin V (+) PI (−), and necrotic cells with annexin V (+) PI (+). Annexin is indicated in green and PI in red.
Molecular and morphological characterization of H2O2-induced pseudohyphae
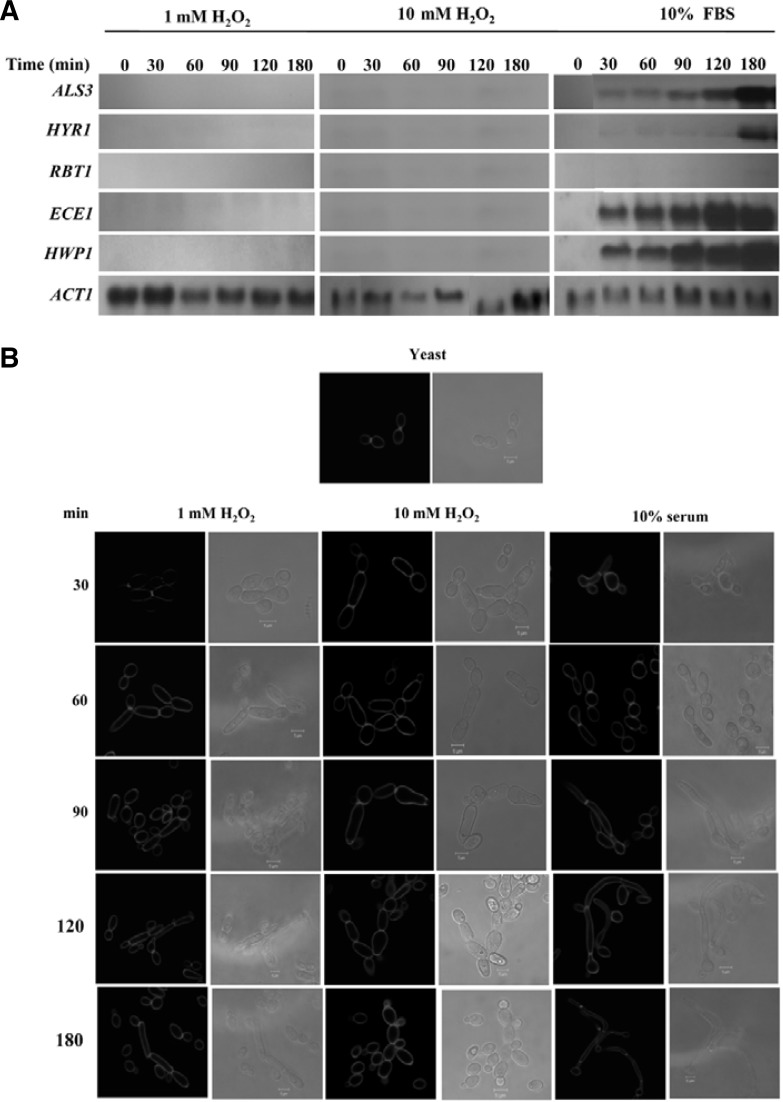
A set of genes are specifically expressed during hyphal differentiation in C. albicans, collectively called hyphae-specific genes (HSGs) including ALS3, ALS8, ECE1, HWP1, HYR1, IDH1, and HGC1 (Biswas et al., 2007; Nantel et al., 2002). Appropriately, the expression of ASL3, HYR1, RBT1, ECE1, and HWP1 was examined for up to 180 min in cells treated with 1 and 10 mM H2O2, and 10% FBS. As shown in Fig. 2A, none of the genes examined displayed activity in cells treated with 1 and 10 mM H2O2, while four genes except RBT1 were expressed in a time-dependent manner in cells treated with 10% FBS. These data suggest that the mechanism by which subtoxic H2O2 induces peusdohyphal differentiation differs at the molecular level from that induced by FBS. To assess this speculation, the filamentous growth in the presence of 1 and 10 mM H2O2, and 10% FBS were characterized by calcofluor staining. As shown in Fig. 2B, nearly all filamentous forms induced by both H2O2 displayed constrictions at the septa between individual cellular compartments, a typical characteristic of pseudohyphae, whereas filamentous growth induced by 10% FBS showed no such constrictions but possessed a distinct germ tube, which is a typical characteristic of true hyphae. Cells exposed to 10 mM H2O2 for 30 min became larger in width than cells exposed to 1 mM H2O2 for the same time. Exposure to the high dose for 180 min was associated with a loss of cell integrity, presumably due to the toxicity of H2O2. Thus, filamentous forms induced by H2O2 were distinct from those induced by 10% FBS in terms of HSG expression and morphology.
Fig. 2.
Characterization of pseudohyphae induced by H2O2. (A) Expression of hyphae-specific genes. Total RNAs were prepared from cells cultured in YPD containing 1 mM H2O2 for 180 min, 10 mM H2O2 for 30 min, and 10% serum for 180 min at 37°C. Fifteen micrograms of RNA were loaded per lane for Northern analysis. Transcripts for hyphae-specific genes were detected using probes amplified with gene-specific primers. Lanes: 1, 0 min; 2, 30 min; 3, 60 min; 4, 90 min; 5, 120 min; 6, 180 min. (B) Results of calcofluor staining. Above cells cultured for 30, 60, 90, 120, and 180 min were washed twice and resuspended in water, and incubated for 30 min with 20 μl of calcofluor (1 mg ml−1 in water). The fluorescence was observed at 365 nm with a confocal laser scanning microscope.
Transcription profiling of H2O2-induced pseudohyphae
Although distinct from FBS-induced true hyphae, no difference in HSGs expression and morphology was evident between 1 and 10 mM H2O2-induced pseudohyphae, except for cell disintegration in the presence of 10 mM H2O2 (Figs. 1 and 2B). Appropriately, the transcription profiles were compared by DNA microarrays between cells treated with a sub-toxic condition of H2O2 (1 mM) for 3 h and a toxic condition of H2O2 (10 mM) for 30 min. The DNA chips contained 6,320 C. albicans genes, and were considered to represent almost the complete transcriptome of C. albicans. Average values from four independent array results for each condition were used to analyze statistically significant changes in expression using the Significance Analysis of Microarrays software (http://www-stat.stanford.edu/~tibs/SAM/). The entire microarray data has been registered at ArrayExpress (www.ebi.ac.uk/arrayexpress) under the accession number E-MEXP-2680. The cutoff value for differentially regulated genes was a 2.0-fold change with a P-value ≤ 0.05. Based on these criteria, 10 and 270 genes were found to be up-regulated by the sub-toxic and toxic doses of H2O2, respectively (Table 2 and Supplementary Table 2, respectively). Since the number of genes up-regulated by the sub-toxic condition of H2O2 was too small to functionally compare with those up-regulated by the toxic condition of H2O2, an alternative approach was used, where 31 genes with P-value ≤ 0.1 for sub-toxic H2O2-regulated genes were selected (Supplementary Table 1). Seven genes were commonly up-regulated in both conditions (Supplementary Fig. 1A). Meanwhile, only three functionally unknown genes (FUR4, orf19.2034, and orf19.7151) were down-regulated by the sub-toxic condition of H2O2, whereas 151 genes were down-regulated by the toxic H2O2 condition (Supplementary Table 3). FUR4, orf19.2034, and orf19.7151 were among the toxic down-regulated genes.
Table 2.
Functional categories of up-regulated genes in the sub-toxic condition
| Category | Gene | ORF ID | Fold change
|
Function of gene product | |
|---|---|---|---|---|---|
| Sub-toxic | Toxic | ||||
| Metabolism | NA | orf19.2788 | 2.7 | 1.6 | Riboflavin biosynthetic process |
| Cell wall proteins | PSA2 | orf19.4943 | 3.5 | 1.5 | Cell wall mannoprotein biosynthetic process |
| ALS2 | orf19.2121 | 2.5 | 1.3 | ALS family protein; role in adhesion | |
| Response to oxidative | SOD3 | orf19.7111.1 | 3.6 | 1.9 | Cytosolic manganese-containing superoxide dismutase |
| Stress | PRX1 | orf19.5180 | 2.2 | 1.3 | Putative cysteine peroxidase |
| DNA replication | orf19.5614 | 2.0 | 1.7 | DNA replication | |
| Stress response | MRF1 | orf19.1149 | 2.7 | 51.0 | Mitochondrial respiratory proteins |
| Unknown function | NA | orf19.4596 | 3.4 | UD | Unknown |
| NA | orf19.894 | 2.3 | 1.5 | Unknown | |
| NA | orf19.7043 | 2.3 | UD | Unknown | |
NA, not assigned; UD, Undetermined; since the P-value is over 0.05.
These contrasting numbers of up- or down-regulated genes between the two conditions suggested that C. albicans responded differently to different H2O2 concentrations. The fold-change of sub-toxic up-regulated genes (maximum value of 3.6) was generally lower than that of toxic up-regulated genes (maximum > 60) (Table 2 and Supplementary Table 2). These data indicate that C. albicans cells exposed to the toxic condition responded by markedly changing their transcriptome profile within 30 min to survive or adapt to the oxidative stress, compared with cells exposed to the sub-toxic condition under which normal cellular proliferation occurs. The microarray data was validated by the accordance with the expression levels of eight genes (PRX1, SOD5, DDR48, MET3, MET14, MET15, and PSA2), which represent a cell wall protein gene, HSG, and genes involved in oxidative stress response and sulfate assimilation among 31 up-regulated genes with P value of < 0.1 at 0, 1, and 10 mM H2O2, and 10% FBS as well (Supplementary Fig. 1B).
Functional classification of regulated genes
Thirty-one up-regulated sub-toxic genes were categorized based on function assigned by the Candida Genome Database (Supplementary Table 1 and Supplementary Fig. 1C). Eight genes were functionally unknown. The remainder could be classified into 11 functional groups, including metabolism, cell wall integrity, methionine biosynthesis and sulfate assimilation pathway, oxidative stress response, oxidoreductase, DNA replication, hyphal induction, transport, iron metabolism, stress response, and pathogenesis. Many of the genes were involved in cell proliferation and metabolism, as demonstrated in other species (Biswas et al., 2007). Up-regulation of some cell wall genes (ALS2, ALS4, PSA2, and PGA7) was expected, since the composition of cell wall in C. albicans is apparently altered during morphogenesis to modulate cellular proliferation or morphogenesis in response to external signals (Castillo et al., 2008).
Excessive extracellular oxidative stress induces the cellular defense and repair systems in other organisms (Davies, 2005). C. albicans displayed a similar behavior (Supplementary Fig. 1D). Of the 270 genes that were up-regulated in the presence of the toxic condition of H2O2, 46 genes (17%) were determined to belong to a group of protein modification and degradation. The others included genes involved in diverse functions, including stress response, oxidative stress, signaling cascade, DNA repair, transport, amino acid biosynthesis, cell cycle checkpoint, and autophagy. When cells respond to internal or external stimuli, suppression (or down-regulation) of some genes is important as much as activation (or up-regulation) of other genes at the transcriptional level. For example, suppression of negative regulators such as TUP1, NRG1, RFG1, and MIG1 is required for hyphal differentiation induced by elevated temperature and serum in C. albicans (Murad et al., 2001a; 2001b). The functional categories of the genes include translation, DNA replication, cell cycle, cell wall biosynthesis, transport, and aerobic respiration (Supplementary Fig. 1E). Under the toxic condition, cells seemed to selectively restrict the expression of genes required for growth and proliferation.
Implication of CPH1 in the pseudohyphal induction by H2O2
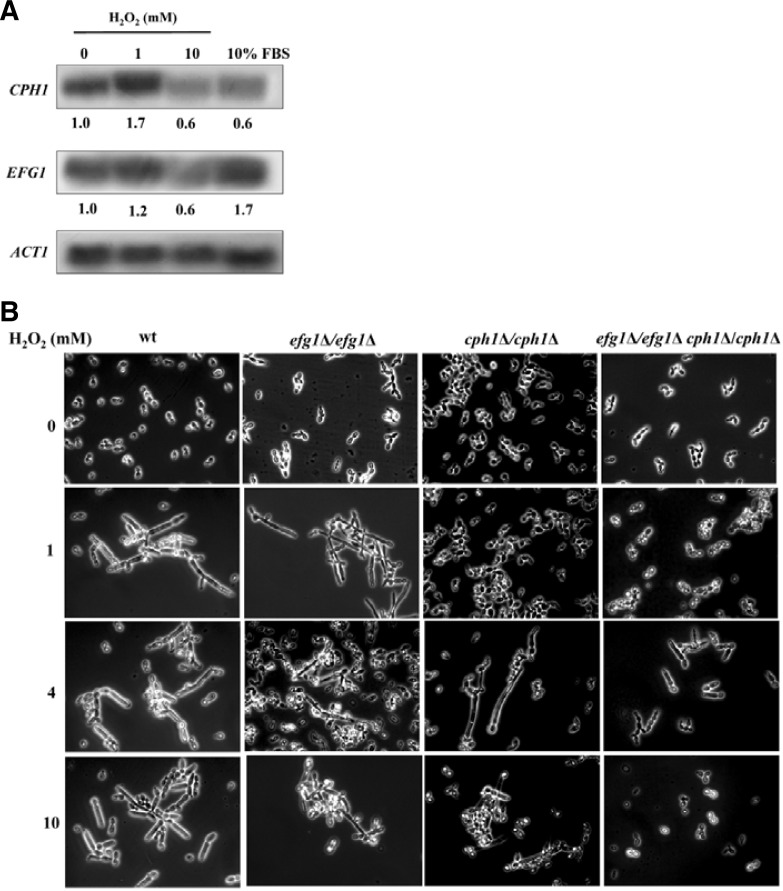
Although no difference was evident in HSGs gene expression and in morphology between pseudohyphal cells induced by 1 and 10 mM H2O2, transcription profiles distinguished these cells from each other. However, no particular transcription factors were found to be regulated under those two conditions. So, we directly examined the expression levels of two major transcription factors responsible for hyphal differentiation signaling pathways in C. albicans: Efg1 for the cAMP pathway and Cph1 for the MAPK pathway. Northern analysis was performed with total RNAs extracted from yeast cells and cells treated with 1 and 10 mM H2O2 and 10% FBS. Following normalization based on actin, the expression levels were determined relative to yeast cells (Fig. 3A). CPH1 was increased 1.7-fold in the presence of 1 mM H2O2 but was decreased by about half at 10 mM H2O2 and FBS. On the other hand, EFG1 was increased 1.7-fold-in 10% FBS, as expected, while being barely increased at 1 mM H2O2 or decreased by about half at 10 mM H2O2. These data suggest that CPH1 might be important for H2O2-induced pseudohyphae.
Fig. 3.
Effect of CPH1 and EFG1 on pseudohyphal induction by H2O2. (A) Expression of CPH1 and EFG1. Total RNAs were prepared from cells cultured in YPD for yeast cells or YPD containing 1 mM H2O2 for 180 min, 10 mM H2O2 for 30 min, and 10% serum for 180 min at 37°C. Fifteen micrograms of RNA were loaded per lane for Northern analysis. Transcripts for CPH1 and EFG1 were detected using probes amplified with gene-specific primers. (B) Effect of CPH1 and EFG1 on hyphal induction by H2O2. Wild type and cph1-null (cph1Δ/cph1Δ), efg1-null (efg1Δ/efg1Δ), and double null (cph1Δ/cph1Δ efg1Δ/efg1Δ) were cultured in YPD containing 0, 1, 4, and 10 mM H2O2. Culture time was 180 min except for 10 mM (30 min).
To address this issue, the pseudohyphal differentiation in the wild type and three mutant (efg1Δ/efg1Δ, cph1Δ/cph1Δ, efg1Δ/efg1Δcph1Δ/cph1Δ) cells treated with 0, 1, 4, and 10 mM H2O2 was assessed. As shown in Fig. 3B, efg1Δ/efg1Δ cells, which are defective in hyphal differentiation by FBS, behaved very similarly to wild type cells, forming pseudohyphae at all H2O2 concentrations. Meanwhile, neither cph1Δ/cph1Δ nor efg1Δ/efg1Δcph1Δ/cph1Δ cells displayed pseudohyphae at 1 mM H2O2. At 4 mM, however, some cph1Δ/cph1Δ cells developed into pseudohyphae and efg1Δ/efg1Δcph1Δ/cph1Δ cells were some-what elongated. At 10 mM H2O2, efg1Δ/efg1Δcph1Δ/cph1Δ seemed to be too severely damaged to define the cell type, but filamentous forms of cph1Δ/cph1Δ were observed. These data again suggests that Cph1 might be implicated in the pseudohyphal induction by H2O2.
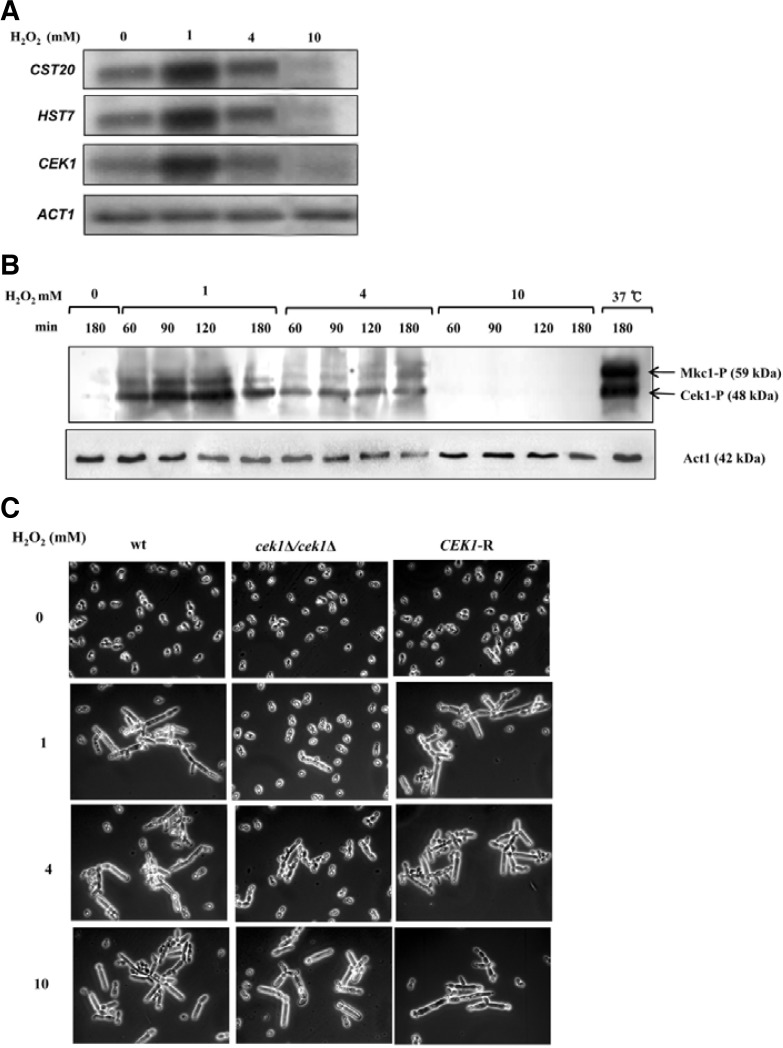
To assess this at the molecular level, the expression of CEK1 a MAPK immediately upstream of Cph1 in the MAPK pathway and its two immediately upstream genes (HST7 and CST20), and phosphorylation of Cek1 were investigated in cells treated in the same way as in Fig. 4B. As shown in Fig. 4A, CEK1, HST7, and CST20 were up-regulated by a H2O2 condition of 1 mM but not by 4 mM, and where down-regulated by 10 mM H2O2. The phosphorylation of Cek1 was consistent with this expression pattern: compared to untreated cells, the level of phosphorylated Cek1 increased in cells treated with 1 mM H2O2, whereas the level was decreased in the cells treated with 4 or 10 mM H2O2 (Fig. 4B). Cells grown at high temperature (37°C), which is known to induce pseudohyphae, was included as control. Both phosphorylated Cek1 and Mkc1 were prominently expressed at 37°C. Suggesting complexity of pathway might involve in the induction of pseudohyphae. The pseudohyphal differentiation was assessed in the wild type, a mutant cek1Δ/cek1Δ and a revertant of cek1Δ/CEK1 (CEK1-R) under the same conditions as used in Fig. 3B. As expected, cek1Δ/cek1Δ cells behaved exactly like cph1Δ/cph1Δ cells at all H2O2 concentrations and, in particular, exhibited a yeast form at 1 mM H2O2. However, CEK1-R cells developed into pseudohy-phae at this concentration (Fig. 4C). These data, together with the observations summarized in Fig. 3B, indicate that Cek1 MAPK through the transcription factor Cph1 was shown to be important for sub-toxic induced pseudohyphae. However, the absence of expression of several hyphae-specific genes known to be downstream targets of Cph1-signaling pathway for true hyphae formation suggests that the Cek1-mediated signaling pathway is not solely responsible for pseudohyphal formation by H2O2 and, instead complexity of networking pathway exists by activation of different regulators.
Fig. 4.
CEK1 is responsible for the pseudohyphal induction by H2O2. The experimental conditions were same as in Fig. 4. (A) Expression of CST20, HST17, and CEK1. CST20 and HST17 are two genes immediately upstream of CEK1 in the MAPK pathway. (B) Phosphorylation of Cek1p. Western analysis was performed with total cell extracts. An anti-phospho-p44/p42 MAPK (Erk1/2) (Thr202/ Tyr204) antibody was used as a primary antibody that recognizes the phosphorylated form of Mkc1 and Cek1 kinases. Bands were visualized with an enhanced chemiluminescence system. (C) Effect of CEK1 on hyphal induction by H2O2. Wild type and cek1-null (cek1Δ/cek1Δ), and CEK1-revertant (CEK1-R) were used.
DISCUSSION
A clue for the pseudohyphal induction by H2O2 was obtained from the observation that the expression patterns of HSGs in cells treated with H2O2 was greatly different from those serum-induced true hyphae (Fig. 2A). In addition to a few environmental factors that induce pseudohyphae (Andaluz et al., 2006; Boisnard et al., 2008; Hwang et al., 2003; Kunze et al., 2007), overexpression of CPH1 (Lane et al., 2001) and RFG1 (Cleary et al., 2010; Kunze et al., 2007), or deletion of TUP1 (Braun et al., 1997), NRG1 (Murad et al., 2001b), RFG1 (Kadosh et al., 2001), FKH2 (Bensen et al., 2002), GRR1 (Butler et al., 2006), and several cell cycle-related genes (Bachewich et al., 2005; Berman et al., 2006; Wightman et al., 2004) and references therein] also results in the formation of pseudohyphae. Interestingly, HSGs (mostly HWP1 and ECE1) are expressed in strains tup1Δ, nrg1Δ, and rfg1Δ, in which one of three typical negative regulators of hyphae formation is deleted (Murad et al., 2001b), indicating that the pseudohyphal formation is independent of the expression of HSGs in some cases. Thus, the lack of HSGs expression is not a molecular maker for defining pseudohyphae, although it has helped to initiate the characterization of the filamentous form induced by H2O2.
Presently, a genome-wide transcriptional profiling analysis was done to investigate whether or not the transcriptional machinery of cells responded differently to the sub-toxic and toxic condition of H2O2. The number, expression level, and functional category of regulated genes contrasted greatly between the two conditions (Supplementary Figs. 1A, 1C–1E). Very recently, a similar study described the transcriptional response of CAF2 cells to 5 mM H2O2 for 10 min at 37°C (Alonso-Monge et al., 2010), reporting that 119 and 124 genes are up- and down-regulated, respectively. Many of the regulated genes (56 up-regulated and 26 down-regulated), including MRF1, overlapped with those induced by 10 mM H2O2, whereas only MRF1 overlapped with the genes up-regulated by 1 mM H2O2. Thus, MRF1 is up-regulated under the three conditions compared (2.7-fold at 1 mM, 23-fold at 5 mM, and 51-fold at 10 mM). Interestingly, PRX1, CSH1, DDR48 were downregulated when treated with 5 mM H2O2 for 10 min at 37°C, which were upregulated in subtoxic H2O2 treated cells. Measurement of the intracellular concentration of H2O2 (Rhee et al., 2010) would help to explain the difference of its biological activities under various experimental conditions such as temperature, exposed time, and extracellular concentration.
Despite such huge differences in the global transcriptional response, the morphological transition of both sub-toxic and toxic conditions was same; pseudohyphae formation. A few environmental conditions, including high phosphate (Hornby et al., 2004), induce pseudohyphae in C. albicans. However, the responsible regulators remain undefined. In S. cerevisiae, both cAMP and MAPK pathways are involved in pseudohyphal formation (Rupp et al., 1999). In the present study, it was demonstrated that the downstream regulator CEK1 through the transcription factor Cph1 might be important for H2O2-induced pseudohyphae. Since several hyphae-specific genes known to be downstream targets of CPH1 in true hyphae were not expressed (Fig. 2A), it is suspected that the CEK1-mediated signaling pathway was not solely responsible for the pseudohyphal formation induced by H2O2. Cek1/Cph1 might target additional regulators or new regulator might be involved in the pseudohyphal induction by H2O2, which needs to be defined.
An unbearable level of H2O2 is deleterious to cells. In fact, in a mutant strain in which the thiol-specific antioxidant gene TSA1 was deleted, the effect of 1 mM was the same as that of 10 mM in a wild type (Nasution et al., 2008). Accordingly, cells should be equipped with the machinery to convert H2O2 to H2O to maintain intracellular H2O2 at a proper concentration. H2O2 is decomposed mainly by catalases (CAT1), peroxidases such as glutathione peroxidase (GPX1), and peroxiredoxins (PRX1) (Aguirre et al., 2005). PRX1 requires additional redox proteins such as thioredoxin (TRX1) and sulfiredoxin (SRX1) for reduction of oxidized states generated from the reactions with H2O2. Table 3 lists various H2O2 decomposers or related genes that were up-regulated under either the sub-toxic or toxic condition. PRX1 was the only gene up-regulated at 1 mM (but below the cutoff value at 10 mM), whereas all other genes listed were up-regulated at 10 mM. Their induction levels were considerably high, for example, as much as 55-fold for orf19.3537 with sulfiredoxin activity. The variety in members and the degree of up-regulation may imply that one member communicates with other member(s) in functioning, at least in part, in H2O2 degradation, which may be necessary for fine-tuning the concentration of intracellular H2O2.
Table 3.
The expression level of H2O2 decomposers
| Gene | ORF ID | Fold change
|
Function of gene product | |
|---|---|---|---|---|
| Sub-toxic | Toxic | |||
| PRX1 | orf19.5180 | 2.2 | 1.3 | Putative cysteine peroxidase |
| TTR1 | orf19.6059 | 1.4 | 27.0 | Disulfide oxidoreductase |
| CAT1 | orf19.6229 | 1.7 | 21.0 | Catalase activity |
| CCP1 | orf19.238 | 1.3 | 19.0 | Cytochrome-c peroxidase |
| TSA1 | orf19.7417 | 1.6 | 17.1 | Thioredoxin peroxidase |
| TRR1 | orf19.4290 | 1.2 | 14.0 | Putative thioredoxin reductase |
| GLR1 | orf19.4147 | 1.0 | 9.0 | Glutathione reductase |
| TSA1B | orf19.7398.1 | 1.4 | 7.5 | Putative peroxidase |
| TRX1 | orf197611. | 1.1 | 7.1 | Putative thioredoxin |
| AHP1 | orf19.2762 | 1.5 | 2.9 | Alkyl hydroperoxide reductase |
| DOT5 | orf19.5417 | 1.2 | 2.6 | Thioredoxin peroxidase activity |
| NA | orf19.3537 | 1.2 | 55.0 | Sulfiredoxin activity |
| NA | orf19.86 | 1.6 | 17.0 | Glutathione peroxidase |
NA, not assigned
One feature of the transcriptional profile revealed in the toxic condition is that many genes were highly up- or down-regulated compared with the sub-toxic condition. The numbers of up- and down-regulated genes with over a 5-fold change were 74 out of 270 and 27 out of 151, respectively (Supplementary Tables 2 and 3). Most of those highly regulated genes were found to be related with either DNA replication or translation in the case of down-regulation or oxidative stress response in the case of up-regulation, as expected. At present, attention was given to extremely highly up-regulated genes (> 50-fold increase), hoping that they could be used as oxidative stress markers in C. albicans and other species including higher organisms. These include MRF1 (similar to mitochondrial respiratory protein, 51-fold), orf19.3537 (sulfiredoxin, 55-fold), and orf19.2165 (unknown function, 80-fold). MRF1 and orf19.3537 and are evolutionarily conserved in humans. In particular, orf19.3537, a homolog of S. cerevisiae sulfiredoxin SRX1 that reduces cysteine-sulfinic acid groups in peroxiredoxins Tsa1 and Ahp1 (Biteau et al., 2003), exhibited 34% identity and 60% positivity with human sulfiredoxin. MRF1, which is similar to mitochondrial respiratory protein, exhibited 33% identity and 55% positivity with human trans-2-enoyl-CoA reductase. In fact, a recent study reported that thioredoxin released from cells can be used as an oxidative stress marker in various human disorders (Nakamura, 2005). Therefore, it is worth investigating if those genes can be used as novel oxidative stress markers in humans.
Although apoptosis is induced when C. albicans is exposed to a variety of environmental stimuli such as acetic acid, H2O2, as well as amphotericin B (Phillips et al., 2003), little is known about the effector molecule required for the onset of apoptosis. A recent study suggested a Ras-cAMP-PKA signaling in the apoptotic response to weak acid exposure, but demonstrated that cells treated with 25 mM H2O2 for 30 min rarely undergo apoptosis (Phillips et al., 2006). This finding implies that it may be difficult to find candidate effector genes at an early stage of apoptosis among toxic up-regulated genes in our experimental condition. According to the C. albicans genome database, 14 genes appear to be associated with apoptosis, including ASF1 (apoptosis stimulating factor), RAS1, CYR1 (adenylyl cyclase), MCA1 (putative caspase), MCD1 (DNA damage response gene), NMA111 (serine-type peptidase), TDH3 (glutaraldehyde 3-phosphate dehydrogenase), orf19.2541 (3′-5′ exonuclease), orf19.3926 (endoribonuclease), orf19.967 (endonuclease), and several other apoptosis-related open reading frames (orf19.4423, orf19.643, orf19.713, orf19.2175). Among these, only orf19.713 increased 2.3-fold at 10 mM H2O2 (Supplementary Table 2). Orf19.713 is a homolog of human PDCD5 and S. cerevisiae YMR074C, and its overexpression promotes H2O2-induced apoptosis (Hong et al., 2009). The present finding that orf19.713 was up-regulated in the toxic condition (10 mM, 30 min), which is a much milder concentration than that where no apoptotic phenotype is observed (25 mM, 30 min), suggests that the protein product of orf19.713 might be a molecule sensing oxidative stress at the earliest stage of the H2O2-induced apoptotic pathway (Hong et al., 2009). This hypothesis can be easily verified by analyzing a mutant in which orf19.713 is deleted.
Acknowledgments
We are grateful to Drs. W. Fonzi, S. Kang, and M. Whiteway for kindly providing strains. This work was supported by Korea Research Foundation Grant (KRF-2003-005-E00007) to WK and Korea Science and Engineering Foundation grant funded by the Korea government (Ministry of Science and Technology) (No. 2006-0063-2) and the Pioneer Research Center Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (No. 2009-0081512) to WC. KS was a recipient of Ewha Global Partnership Program 2006.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Aguirre J., Rios-Momberg M., Hewitt D., Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Alonso-Monge R., Roman E., Arana D.M., Prieto D., Urrialde V., Nombela C., Pal J. The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet. Biol. 2010;47:587–601. doi: 10.1016/j.fgb.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Andaluz E., Ciudad T., Gomez-Raja J., Calderone R., Larriba G. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 2006;59:1452–1472. doi: 10.1111/j.1365-2958.2005.05038.x. [DOI] [PubMed] [Google Scholar]
- Bachewich C., Whiteway M. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell. 2005;4:95–102. doi: 10.1128/EC.4.1.95-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen E.S., Filler S.G., Berman J. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell. 2002;1:787–798. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Van Dijck P., Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Labarre J., Toledano M.B. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- Boisnard S., Ruprich-Robert G., Florent M., Da Silva B., Chapeland-Leclerc F., Papon N. Role of Sho1p adaptor in the pseudohyphal development, drugs sensitivity, osmotolerance and oxidant stress adaptation in the opportunistic yeast Candida lusitaniae. Yeast. 2008;25:849–859. doi: 10.1002/yea.1636. [DOI] [PubMed] [Google Scholar]
- Braun B.R., Johnson A.D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Buggisch M., Ateghang B., Ruhe C., Strobel C., Lange S., Wartenberg M., Sauer H. Stimulation of ES-cell-derived cardiomyogenesis and neonatal cardiac cell proliferation by reactive oxygen species and NADPH oxidase. J. Cell Sci. 2007;120:885–894. doi: 10.1242/jcs.03386. [DOI] [PubMed] [Google Scholar]
- Butler D.K., All O., Goffena J., Loveless T., Wilson T., Toenjes K.A. The GRR1 gene of Candida albicans is involved in the negative control of pseudohyphal morphogenesis. Fungal Genet. Biol. 2006;43:573–582. doi: 10.1016/j.fgb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Calderone R.A., Fonzi W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001;9:327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- Castillo L., Calvo E., Martinez A.I., Ruiz-Herrera J., Valentin E., Lopez J.A., Santandreu R. A study of the Candida albicans cell wall proteome. Proteomics. 2008;8:3871–3881. doi: 10.1002/pmic.200800110. [DOI] [PubMed] [Google Scholar]
- Cleary I.A., Mulabagal P., Reinhard S.M., Yadev N.P., Murdoch C., Thornhill M.H., Lazzell A.L., Monteaqudo C., Thomas D.P., Saville S.P. Pseudohyphal regulation by the transcription factor Rfg1p in Candida albicans. Eukaryot Cell. 2010;9:1363–1373. doi: 10.1128/EC.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottier F., Muhlschlegel F.A. Sensing the environment: response of Candida albicans to the X factor. FEMS Microbiol. Lett. 2009;295:1–9. doi: 10.1111/j.1574-6968.2009.01564.x. [DOI] [PubMed] [Google Scholar]
- Csank C., Schroppel K., Leberer E., Harcus D., Mohamed O., Meloche S., Thomas D.Y., Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K.J. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- Dhillon N.K., Sharma S., Khuller G.K. Signaling through protein kinases and transcriptional regulators in Candida albicans. Crit. Rev. Microbiol. 2003;29:259–275. doi: 10.1080/713610451. [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Harcus D., Nantel A., Marcil A., Rigby T., Whiteway M. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell. 2004;15:4490–4499. doi: 10.1091/mbc.E04-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J., Zhang J., Liu Z., Qin S., Wu J., Shi Y. Solution structure of S. cerevisiae PDCD5-like protein and its promoting role in H(2)O(2)-induced apoptosis in yeast. Biochemistry. 2009;48:6824–6834. doi: 10.1021/bi900488n. [DOI] [PubMed] [Google Scholar]
- Hornby J.M., Dumitru R., Nickerson K.W. High phosphate (up to 600 mM) induces pseudohyphal development in five wild type Candida albicans. J. Microbiol Methods. 2004;56:119–124. doi: 10.1016/j.mimet.2003.09.021. [DOI] [PubMed] [Google Scholar]
- Hwang C.S., Oh J.H., Huh W.K., Yim H.S., Kang S.O. Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol. Microbiol. 2003;47:1029–1043. doi: 10.1046/j.1365-2958.2003.03353.x. [DOI] [PubMed] [Google Scholar]
- Kadosh D., Johnson A.D. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze D., MacCallum D. Odds FC, Hube B. Multiple functions of DOA1 in Candida albicans. Microbiology. 2007;153:1026–1041. doi: 10.1099/mic.0.2006/002741-0. [DOI] [PubMed] [Google Scholar]
- Lane S., Birse C., Zhou S., Matson R., Liu H. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 2001;276:48988–48996. doi: 10.1074/jbc.M104484200. [DOI] [PubMed] [Google Scholar]
- Leberer E., Harcus D., Broadbent I.D., Clark K.L., Dignard D., Ziegelbauer K., Schmidt A., Gow N.A., Brown A.J., Thomas D.Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng P., Lee P.R., Wu H., Brown A.J. Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J. Bacteriol. 2001;183:4090–4093. doi: 10.1128/JB.183.13.4090-4093.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Stouffs M., Serrander L., Banfi B., Bettiol E., Charnay Y., Steger K., Krause K.H., Jaconi M.E. The NADPH oxidase NOX4 drives cardiac differentiation: Role in regulating cardiac transcription factors and MAP kinase activation. Mol. Biol. Cell. 2006;17:3978–3988. doi: 10.1091/mbc.E05-06-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kohler J., Fink G.R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo H.J., Kohler J.R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G.R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Bender J.A., Fink G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K.R., Barrett J.C. Reactive oxygen species as double-edged swords in cellular processes: low-dose cell signaling versus high-dose toxicity. Hum. Exp. Toxicol. 2002;21:71–75. doi: 10.1191/0960327102ht213oa. [DOI] [PubMed] [Google Scholar]
- Mesquita F.S., Dyer S.N., Heinrich D.A., Bulun S.E., Marsh E.E., Nowak R.A. Reactive oxygen species mediate mitogenic growth factor signaling pathways in human leiomyoma smooth muscle cells. Biol. Reprod. 20092009 doi: 10.1095/biolreprod.108.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A.M., d’Enfert C., Gaillardin C., Tournu H., Tekaia F., Talibi D., Marechal D., Marchais V., Cottin J., Brown A.J. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 2001a;42:981–993. doi: 10.1046/j.1365-2958.2001.02713.x. [DOI] [PubMed] [Google Scholar]
- Murad A.M., Leng P., Straffon M., Wishart J., Macaskill S., MacCallum D., Schnell N, Talibi D., Marechal D., Tekaia F., et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001b;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H. Thioredoxin and its related molecules update. Antioxid. Redox. Signal. 2005;7:823–828. doi: 10.1089/ars.2005.7.823. [DOI] [PubMed] [Google Scholar]
- Nantel A., Dignard D., Bachewich C., Harcus D., Marcil A., Bouin A.P., Sensen C.W., Hogues H., van het Hoog M., Gordon P., et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasution O., Srinivasa K., Kim M., Kim Y.J., Kim W., Jeong W., Choi W.J. Hydrogen peroxide induces hyphal differentiation in Candida albicans. Eukaryot Cell. 2008;7:2008–2011. doi: 10.1128/EC.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Garcia F., Sanchez M., Pla J., Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol. Cell. Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba M., Shibanuma M., Kuroki T., Nose K. Production of hydrogen peroxide by transforming growth factor-beta 1 and its involvement in induction of egr-1 in mouse osteoblastic cells. J. Cell Biol. 1994;126:1079–1088. doi: 10.1083/jcb.126.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.J., Sudbery I., Ramsdale M. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci USA. 2003;100:14327–14332. doi: 10.1073/pnas.2332326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.J., Crowe J.D., Ramsdale M. Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci USA. 2006;103:726–731. doi: 10.1073/pnas.0506405103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J., Findlay V.J., Dawson K., Millar J.B., Jones N., Morgan B.A., Toone W.M. Distinct regulatory proteins control the graded transcriptional response to increasing H(2)O(2) levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell. 2002;13:805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nat. Immunol. 2002;3:1129–1134. doi: 10.1038/ni1202-1129. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Bae Y.S., Lee S.R., Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci. STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Kang S.W., Jeong W., Chang T.S., Yang K.S., Woo H.A. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr. Opin. Cell. Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Chang T.S., Jeong W., Kang D. Methods for detection and measurement of hydrogen peroxide inside and outside of cells. Mol Cells. 2010;29:539–549. doi: 10.1007/s10059-010-0082-3. [DOI] [PubMed] [Google Scholar]
- Rocha C.R., Schroppel K., Harcus D., Marcil A., Dignard D., Taylor B.N., Thomas D.Y., Whiteway M., Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol Cell. 2001;12:631–643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman E., Arana D.M., Nombela C., Alonso-Monge R., Pla J. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 2007;15:181–190. doi: 10.1016/j.tim.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Rupp S., Summers E., Lo H.J., Madhani H., Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shin D.H., Jung S., Park S.J., Kim Y.J., Ahn J.M., Kim W., Choi W. Characterization of thiol-specific antioxidant 1 (TSA1) of Candida albicans. Yeast (Chichester, England) 2005;22:907–918. doi: 10.1002/yea.1283. [DOI] [PubMed] [Google Scholar]
- Stoldt V.R., Sonneborn A., Leuker C.E., Ernst J.F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.R., Yang S. Hydrogen peroxide: a signaling messenger. Antioxid. Redox. Signal. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- Sudbery P., Gow N., Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sundaresan M., Yu Z.X., Ferrans V.J., Irani K., Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- Thannickal V.J., Fanburg B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 2000;279:L1005–28. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- Veal E.A., Day A.M., Morgan B.A. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Vivancos A.P., Jara M., Zuin A., Sanso M., Hidalgo E. Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol. Genet Genomics. 2006;276:495–502. doi: 10.1007/s00438-006-0175-z. [DOI] [PubMed] [Google Scholar]
- Wightman R., Bates S., Amornrrattanapan P., Sudbery P. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell. Biol. 2004;164:581–591. doi: 10.1083/jcb.200307176. [DOI] [PMC free article] [PubMed] [Google Scholar]