Abstract
Dehydration-responsive element-binding proteins (DREBs) regulate plant responses to environmental stresses. In the current study, transcription of DREB2C, a class 2 Arabidopsis DREB, was induced by a superoxide anion propagator, methyl viologen (MV). The oxidative stress tolerance of DREB2C-overexpressing transgenic plants was significantly greater than that of wild-type plants, as measured by ion leakage and chlorophyll fluorescence under light conditions. The transcriptional activity of several ascorbate peroxidase (APX) genes as well as APX protein activity was induced in DREB2C overexpressors. Additionally, the level of H2O2 in the overexpressors was lower than in wt plants under similar oxidative stress conditions. An electrophoretic mobility shift assay and transient activator-reporter assay showed that APX2 expression was regulated by heat shock factor A3 (HsfA3) and that HsfA3 is regulated at the transcriptional level by DREB2C. These results suggest that DREB2C plays an important role in promoting oxidative stress tolerance in Arabidopsis.
Keywords: gene expression, signaling, transcription factor, transgenic plant
INTRODUCTION
DREBs belong to the AP2/ERF family of transcription factors (Yamaguchi-Shinozaki and Shinozaki, 1994), which confer stress endurance in plants and are the largest and most diverse family of proteins involved in the regulation of plant responses (Agarwal et al., 2007). In Arabidopsis, there are two classes of DREB genes, class 1 and class 2 (DREB1 and DREB2, respectively) (Sakuma et al., 2002). The deduced amino acid sequences of class 1 and class 2 genes show no significant level of sequence similarity except in their conserved DNA-binding domains (Jaglo-Ottosen et al., 1998). DREB1 genes are transcriptionally induced by low temperature, and the corresponding gene products induce the expression of multiple target genes that impart tolerance to freezing and drought in transgenic plants (Liu et al., 1998; Oh et al., 2007). DREB2 genes were initially identified as drought and high-salinity response genes (Liu et al., 1998), and were later shown to be induced in response to heat stress as well (Lim et al., 2007; Sakuma et al., 2006). Recent studies identified heat shock transcription factor A3 (HsfA3) as a highly up-regulated heat-inducible gene in transgenic plants over-expressing DREB2s (Chen et al., 2010; Yoshida et al., 2008). Moreover, transient promoter reporter assays using mesophyll protoplasts demonstrated that HsfA3 expression is directly regulated by DREB2s under conditions of heat stress (Chen et al., 2010; Schramm et al., 2008) and that DREB2-overexpressing transgenic plants have increased tolerance to heat stress (Almoguera et al., 2009; Lee et al., 2010; Lim et al., 2007; Matsukura et al., 2010). Recent work established that DREB2C physically interacts with ABF2, a bZIP protein regulating abscisic acid (ABA)-responsive gene expression, and its overexpression affected ABA sensitivity (Lee et al., 2010). Thus, the results suggesting that DREB2s may function as multi stimuli-response factor that interact with genes and/or proteins during different stress conditions (Lee et al., 2009). Despite the important roles played by DREB2s in abiotic stress responses, regulation with respect to oxidative stress has not been determined.
In the current study, oxidative damage triggered the expression of DREB2C at the mRNA level. The expression of DREB2C correlated with the up-regulation of HsfA3 and transcriptional activation of the antioxidant gene APX2 in Arabidopsis. The current results suggest a model in which DREB2C activates HsfA3 expression, and HsfA3 in turn regulates the expression of numerous oxidative stress-inducible genes, including APX2, to impart tolerance to oxidative stress.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) plants or DREB2C-overexpressing transgenic Arabidopsis Col-0 plants (Lim et al., 2007) were grown on 1× MS medium without any phytohormones (MSO, pH 5.8) with 2% sucrose under a 16 h light cycle (cool-white fluorescent light, photon flux of 70 μM m−2s−1) at 22°C. To induce synchronous germination, seeds were vernalized at 4°C for three days in the dark and then transferred to a growth chamber. All external stress treatments were performed on 2-week-old plants.
RNA extraction and reverse transcriptase (RT)-PCR
To analyze the expression of DREB2C, RNA was isolated from wt Arabidopsis Col-0 plants exposed to 10 μM MV. For RT-PCR analysis, cDNA was synthesized from 2 μg of total RNA. Each cDNA sample was diluted 1:10 and then 1 μl of the diluted cDNA was used as the template for PCR amplification, as described previously (Lim et al., 2007).
To examine the expression of Arabidopsis APX isoforms, total RNA was isolated from 2-week-old DREB2C-overexpressing transgenic lines grown under normal growth conditions. Complementary DNA synthesis and RT-PCR were performed as described above using APX gene primers (Supplementary Table S1). PCR products were sequenced directly to confirm that the amplified sequences were identical to the predicted sequences of the respective mRNAs based on Arabidopsis genomic data.
Ion leakage assay
Ion leakage was measured according to Scarpeci et al. (2008), with minor modifications. Two-week-old wt and DREB2C-overexpressing Arabidopsis plants grown on solid MSO were uprooted, thoroughly rinsed with de-ionized water, and then incubated in water (control) or in water supplemented with 0.5 μM MV for 1 to 4 days under dim light conditions at 22°C. After incubation, the conductivity of the suspension solution was measured with a conductance meter (Orion 3-Star Plus, Thermo Scientific, USA) before and after autoclaving at 121°C for 15 min to release all electrolytes. Relative ion leakage was expressed as a percentage of the total conductivity.
Chlorophyll fluorescence measurement
MV-treated Arabidopsis plants were dark-adapted for 15 min. In vivo chlorophyll fluorescence was measured at room temperature using a plant efficiency analyzer (Handy PEA, Hansatech Instruments, UK), according to the method described by Yu et al. (2002). Maximum quantum efficiency (Fv/Fm) was calculated as previously described (Baker and Rosenqvist, 2004).
Protein extraction and APX activity assay
To monitor the activity of APX, soluble protein fractions were extracted from 100 mg of 2-week-old wt or transgenic Arabidopsis plants as previously described (Panchuk et al., 2002). Protein concentrations were determined using a Bradford Protein Assay Kit (Bio-Rad, USA) and bovine serum albumin as a reference. The oxidation rate of AsA was measured by UV/Vis spectroscopy (Genesys 10 UV, Spectronic Unicam, USA) by monitoring the decrease in A290 1 min after the addition of protein extract.
3,3′-Diaminobenzidine (DAB) staining and H2O2 measurement
DAB staining was performed according to Thordal-Christensen et al. (1997), with slight modifications. Two-week-old wt and DREB2C-transgenic plants grown in 1× MSO medium were transferred to water or 0.5 μM MV and then incubated for 2 days under a 16 h light/8 h dark cycle at 22°C. Following this, plants were incubated for 9 h in 1 mg ml−1 DAB-HCl (pH 3.8) under dim light. Chlorophyll was removed by soaking the plants in a graded ethanol series (90, 80, 70, 60, 50, 30, 10%) for several hours. H2O2 causes polymerization of DAB, which results in a brown color.
Quantitative analysis of H2O2 was performed as described by Xing et al. (2007). Wt and transgenic plants were transferred to water (control) or 0.5 μM MV and then incubated for 2 days under a 16 h light/8 h dark cycle at 22°C. H2O2 was quantified using the Amplex Red Assay Kit (Invitrogen, USA) and a fluorescence microplate reader (SpectraMax Gemini XPS Molecular Devices, USA). Excitation wavelength was set at 530 nm and fluorescence was measured at 590 nm.
Promoter sequence analysis
Promoter sequences were analyzed to identify putative cis-elements in the APX2 (At3g09640) and HsfA3 (At5g03720) gene promoters. The upstream sequences of APX2 and HsfA3 (711- and 1094-bp, upstream of the transcription start site, respectively), were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org/tools/bulk/sequences/index.jsp). These sequences were analyzed for over-represented motifs using MEME (http://meme.sdsc.edu/meme/) (Bailey and Elkan, 1995).
Electrophoretic mobility shift assay (EMSA)
The EMSA was performed as described by Chen et al. (2010). The probes were as follows: an 85-bp [α-32P]dATP-labeled fragment (APX2-HSE-85) containing the heat shock element (HSE) within the APX2 promoter, and an 80-bp fragment (HsfA3-DRE-80) containing the dehydration responsive element /C-repeat (DRE/CRT) element within the HsfA3 promoter (Supplementary Fig. S1). Recombinant glutathione-S-transferase (GST)-DREB2C145–528 or GST-HsfA3 was expressed in E. coli BL21 (DE3) pLysE and purified using affinity chromatography as described by Lim et al. (2007). The specificity of cis element-binding was measured by competition assays using unlabeled APX2-HSE-85 and HsfA3-DRE-80 oligonucleotides (Chen et al., 2010).
Activator and reporter constructs used in the transient expression assays
To determine the levels of expression of DREB2C and HsfA3 in mesophyll protoplasts, PCR was carried out using cDNA isolated from Arabidopsis seedlings as the template and gene-specific primers (Supplementary Table S2). Amplified sequences were sub-cloned into a pUC19-derived plasmid between the CaMV 35S promoter and nopaline synthase terminator (Kim et al., 2007).
For promoter-reporter plasmid construction, the putative promoter sequences of APX2 (856-bp) and HsfA3 (1094-bp) (Supplementary Fig. S1) were amplified by PCR from genomic DNA using gene-specific primers (Supplementary Table S2). Amplified sequences were fused in-frame to the GUS gene sequence in a pUC19-derived plasmid (Kim et al., 2007). Protoplasts isolated from Arabidopsis mesophyll cells were transiently transformed using polyethylene glycol-calcium fusion, according to Yoo et al. (2007). GUS reporter assays and analysis of protein expression in Arabidopsis protoplasts were performed as described previously (Kim et al., 2007).
RESULTS AND DISCUSSION
DREB2C transcription is induced by oxidative stress
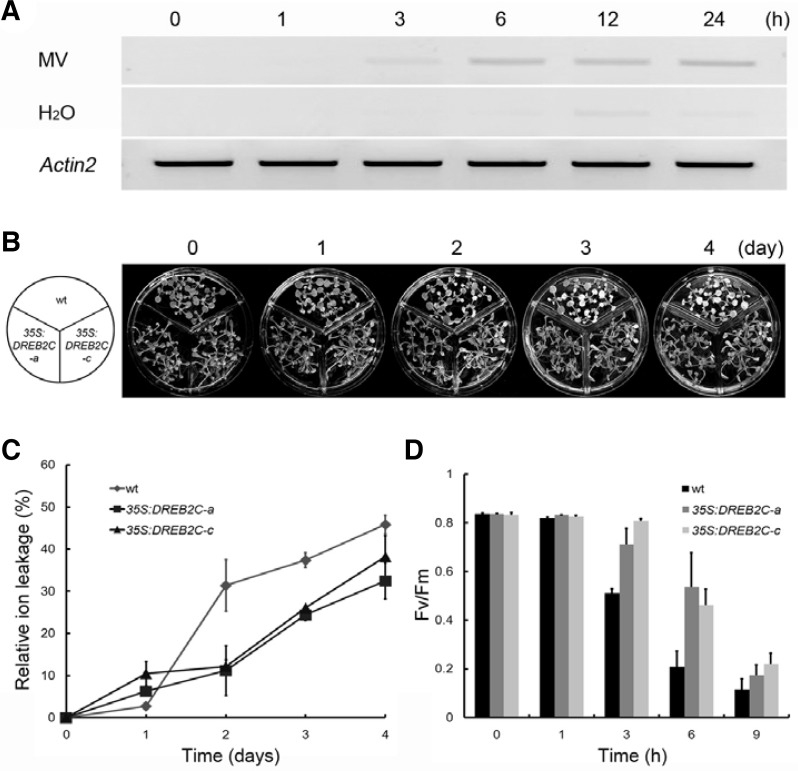
To determine the effect of oxidative stress on DREB2C expression, 2-week-old wild-type (wt) Arabidopsis plants were exposed to exogenous MV. Figure 1A shows that DREB2C transcripts were elevated in 3 h after treatment with MV, reached a peak at 6 h and remained elevated. These results clearly demonstrate that DREB2C is an oxidative stress-responsive gene, and suggest that DREB2C may be involved in the plant response to oxidative stress.
Fig. 1.
Enhanced oxidative stress tolerance of DREB2C overexpressors. (A) Changes in DREB2C transcript levels in response to oxidative stresses. RT-PCR analysis was used to determine mRNA transcript levels. Two-week-old plants were exposed to 10 μM MV for the indicated periods of time. Arabidopsis Actin2 (At3g18780) served as a control. (B) Two-week-old plants (wt and DREB2C overexpressors) grown at 22°C were exposed to 0.5 μM MV for the indicated time periods. Scale bar = 0.5 cm. (C) Ion leakage, an indicator of oxidative stress tolerance, was estimated as a percentage of total conductivity. Two-week-old plants grown at 22°C were exposed to 0.5 μM MV for the indicated periods of time. (D) PSII activity (Fv/Fm) was measured at 22°C after adaptation to the dark for 30 min. Two-week-old plants grown at 22°C were exposed to 0.5 μM MV for the indicated periods of time. Data represent the means ± SE of at least five independent experiments.
DREB2C-overexpressing plants exhibit increased tolerance to MV-induced oxidative stress
DREB2C transcription in wt plants was substantially elevated after treatment with MV (Fig. 1A). To investigate whether transgenic plants overexpressing DREB2C exhibited oxidative stress tolerance, DREB2C overexpressors (35S:DREB2C-a and -c) (Lim et al., 2007) and wt plants were placed in a solution of 0.5 μM MV under 16 h light/8 h dark conditions at 22°C. As shown Fig. 1B, wt plants exhibited more severe injury than 35S: DREB2C-a and -c plants two days after exposure to MV. Ion leakage and photosynthesis efficiency, which reflect the level of cellular damage due to oxidative stress (Noriega et al., 2007), were also measured. As shown Fig. 1C, MV caused severe ion leakage in wt plants, whereas DREB2C-transgenic plants were much less affected. PSII activity (Fv/Fm) gradually decreased in both the wt and transgenic plants (Fig. 1D). There was no difference in PSII activity between wt and DREB2C overexpressors exposed the plants for 1 h in MV solution, whereas after 3 h, PSII activity in the DREB2C overexpressors was significantly higher than wt plants. This conditional phenotype suggested that DREB2C plays a substantial role in oxidative stress tolerance, perhaps at the level of regulation of cellular antioxidant gene expression.
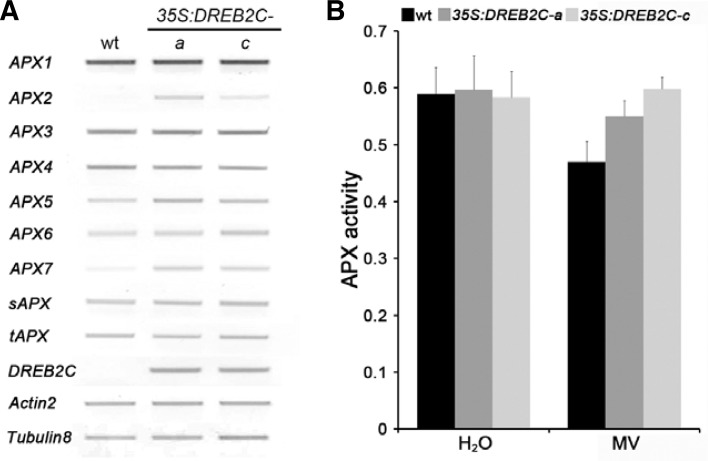
APX genes were induced in DREB2C overexpressors
Since DREB2C expression was regulated by oxidative stress (Fig. 1A) and DREB2C overexpressors exhibited an increased tolerance to oxidative stress (Figs. 1B–1D), we hypothesized that DREB2C may regulate oxidative stress-inducible antioxidant genes. Among antioxidants, APXs play a pivotal role in removing reactive oxygen species by catalyzing the conversion of highly damaging H2O2 into H2O (Giacomelli et al., 2007). Hence, we assessed the expression of nine Arabidopsis APX isoforms (Mittler et al., 2004) in DREB2C overexpressors by RT-PCR. As shown Fig. 2A, expression of the cytosolic (APX2) and mitochondrial (APX7) APX isoforms was induced in DREB2C overexpressors under non-stressed conditions, which suggested that DREB2C may activate the transcription of these antioxidant genes in Arabidopsis.
Fig. 2.
Expression of APX genes is elevated in DREB2C overexpressors. (A) Expression of nine Arabidopsis APX genes in DREB2C overexpressors was analyzed by RT-PCR using gene-specific primers (Supplementary Table 1). Arabidopsis Actin2 (At3g18780) and Tublin8 (At5g23860) were used as constitutively-expressed controls. Total RNA was isolated from 2-week-old plants (wt, 35S: DREB2C-a, and 35S:DREB2C-c). (B) APX activity in DREB2C overexpressors. Total soluble APX activity in 2-week-old wt and transgenic plants (35S:DREB2C-a, 35S: DREB2C-c) incubated in water (H2O) or 0.5 μM MV for two days. Data represent the means ± SE of at least five independent experiments.
Additionally, to determine whether DREB2C overexpression resulted in increased APX protein activity, APX activity was measured by soluble enzymatic activity assay (Panchuk et al., 2002). As shown in Fig. 2B, there was no significant difference in APX activity between wt and DREB2C overexpressors under control conditions. However, APX activity persisted in DREB2C overexpressors grown for two days in MV solution, but not in the wt. These data revealed that the transcriptional activity of APXs and intracellular levels of APX was sustained in DREB2C overexpressors.
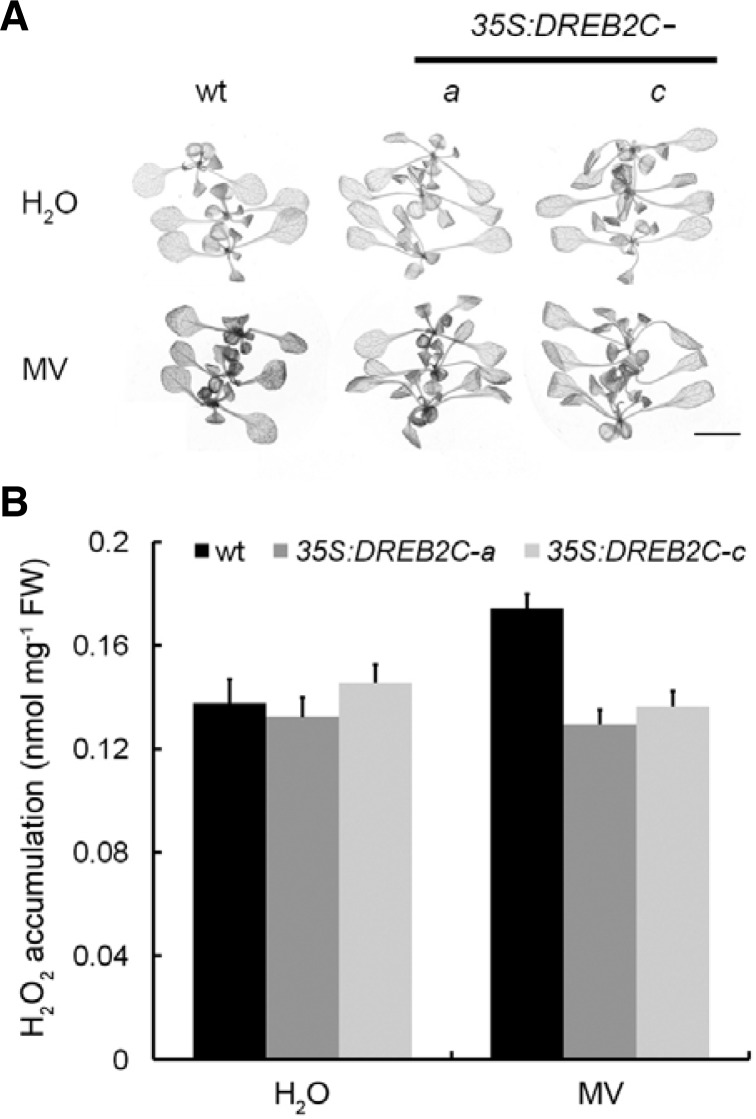
H2O2 accumulation is not increased in DREB2C overexpressors
H2O2 accumulation in DREB2C overexpressors was examined after exposure to 0.5 μM MV for 2 days. Under conditions of MV stress, more H2O2 accumulated in the leaves of wt plants than in DREB2C overexpressors (Fig. 3A). The pattern of H2O2 accumulation is similar to the appearance of bleaching in plants grown in MV solution (Fig. 1B), both in terms of timing and localization. H2O2 levels were also quantified (Fig. 3B). H2O2 production in wt leaves exposed to MV stress was higher than in DREB2C overexpressors; in fact, in the transgenic plants, similar levels of H2O2 were observed in water and MV. These data indicated that persisted levels of APX activity (Fig. 2) may lead to decreased accumulation of H2O2.
Fig. 3.
Accumulation of H2O2 in DREB2C overexpressors. (A) Histochemical detection of H2O2 production by DAB staining. Two-week-old plants (wt, 35S:DREB2C-a, and 35S:DREB2C-c) were incubated in water (H2O) or 0.5 μM MV for two days. Scale bar = 0.5 cm. (B) Hydrogen peroxide measurements were performed on plants grown under the conditions used in DAB staining. All experiments were repeated three times using samples from independent treatments.
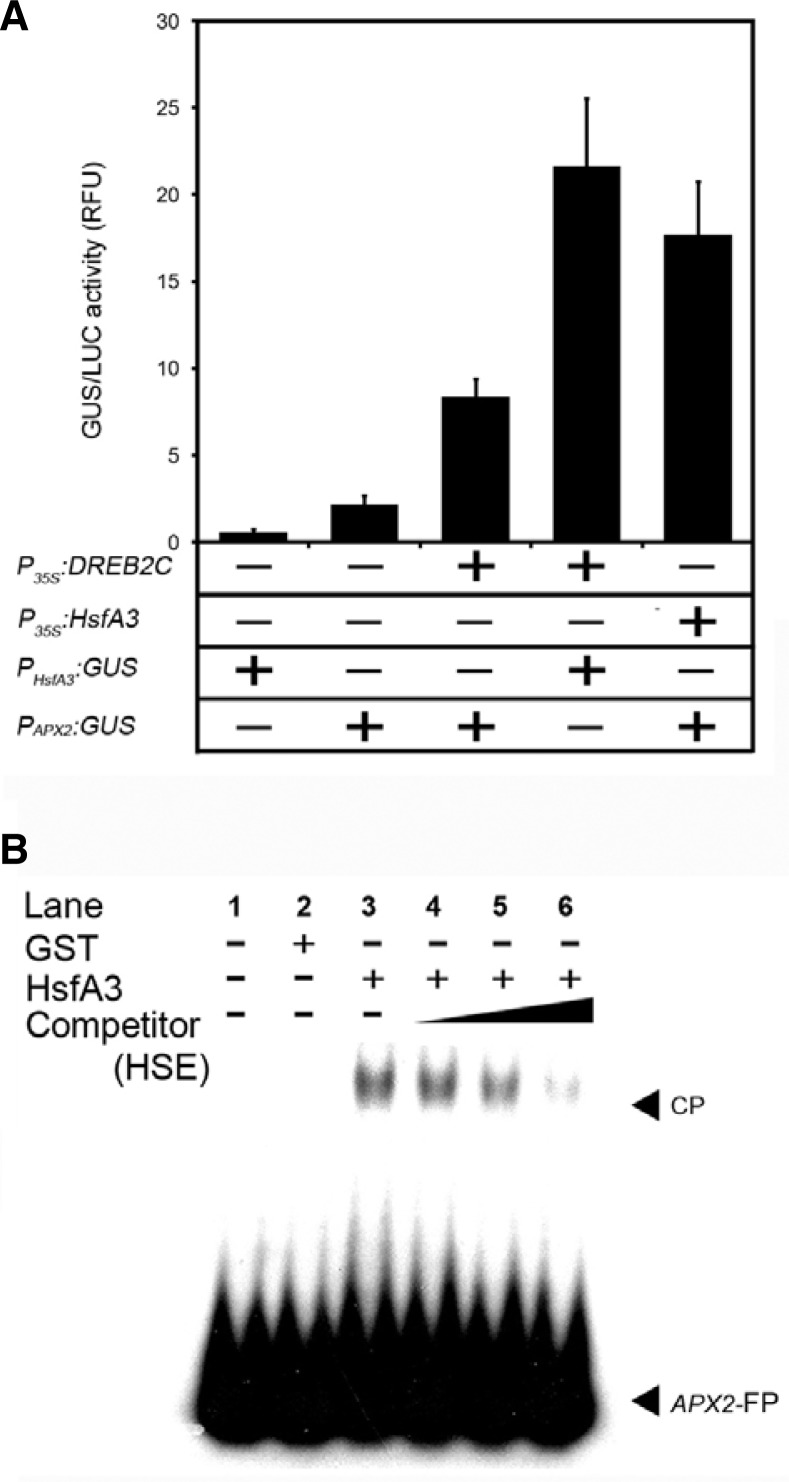
The APX2 promoter was activated by HsfA3
Based on the oxidative stress tolerance phenotype of DREB2C overexpressors (Fig. 1) and the fact that APX transcription and enzymatic activity were induced in these plants (Fig. 2), we were interested in the possibility that oxidative damage triggers the expression of DREB2C, and that DREB2C in turn induces the expression of APXs. To test this hypothesis, APX2 was selected for further analysis, since the APX2 promoter has been shown to be strongly induced by oxidative stresses (Karpinski et al., 1999). A 711-bp region of the APX2 promoter upstream of the putative transcriptional initiation site did not contain a DRE sequence (5′-ACCGAC-3′) but did contain a reversed DRE core sequence (5′-CAGCCA-3′). To determine whether DREB2C was able to bind to this reversed DRE core sequence in the APX2 promoter, in vitro EMSAs were carried out using a 72-bp 32P-dATP-labeled APX2 probe (APX2-rDRE-72, Supplementary Fig. S1A) and protein extracts from E. coli expressing recombinant DREB2C145–528 (Lim et al., 2007). There was no detectable binding of the APX2-rDRE-72 probe to DREB2C145–528 (Supplementary Fig. S2). To further explore this interaction between DREB2C and APX2 in vivo, transient activator-reporter assays were performed in Arabidopsis mesophyll protoplasts transiently expressing full-length DREB2C under the control of the CaMV35S promoter (P35S:DREB2C). The reporter construct consisted of an 856-bp fragment of the APX2 promoter (Supplementary Fig. S1A) fused to GUS (PAPX2:GUS). As shown in Fig. 4A, co-transformation with the P35S:DREB2C construct induced the expression of APX2, with an approximately 4-fold increase in GUS activity. These results supported the idea that there are other transcription factors downstream of DREB2C involved in the regulation of APX2 promoter activity.
Fig. 4.
HsfA3 regulates the expression of APX2 in a transcriptional cascade downstream of DREB2C. (A) Transient activator-reporter assay in Arabidopsis mesophyll protoplasts. The effector constructs consisted of the CaMV 35S promoter fused to full-length DREB2C (P35S:DREB2C) or HsfA3 (P35S:HsfA3) cDNA. The reporter constructs consisted of the HsfA3 (PHsfA3:GUS) or APX2 (PAPX2:GUS) promoter fused to GUS. GUS activity (relative fluorescence units, RFU) represents the means ± SD of three independent replicates. (B) EMSAs were performed using GST alone or GST-HsfA3 fusion protein and an 85-bp 32P-labeled fragment containing the HSE-like region from the APX2 promoter as a probe. Lane 1, APX2-free probe; lane 2, 10 μg GST; lane 3, 10 μg GST-HsfA3. For competition assays, 10 μg of GST-HsfA3 was pre-incubated with a 500-(lane 4), 1500-(lane 5) or 3000-(lane 6) fold molar excess of unlabeled APX2 probe prior to incubation with the 32P-labeled probe. Free (APX2-FP) and HsfA3 protein-complexed APX2 probes (CP) were separated on 5% polyacrylamide gels and visualized by autoradiography.
In previous efforts to identify transcription factors that interact with DREB2s in Arabidopsis, three research groups reported independently that HsfA3 functions downstream of the DREB2A stress-regulatory system (Schramm et al., 2008; Yoshida et al., 2008) and DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response (Chen et al., 2010). Hence, we hypothesized that DREB2C play a role in the activation of HsfA3, and that activated HsfA3 may then induce APX2 gene expression. To test this hypothesis, we explored this interaction between HsfA3 and APX2 in vivo, transient activator-reporter assays were performed in protoplasts expressing full-length HsfA3 (P35S:HsfA3) and a reporter construct containing an 856-bp region upstream of the APX2 promoter fused to GUS (PAPX2:GUS, Supplementary Fig. S1A). As shown in Fig. 4A, co-expression of HsfA3 resulted in an approximately 8-fold increase in the transactivation of the APX2 reporter gene (PAPX2:GUS) compared to protoplasts that did not express HsfA3. To further investigate whether the ability of recombinant HsfA3 to bind directly to the APX2 promoter was assessed by EMSA using an 85-bp APX2 probe (APX2-HSE-85, Supplementary Fig. S1A) containing an intact heat shock element (HSE; nGAAnnTTCn or nTTCnnGAAn). As shown in Fig. 4B, recombinant HsfA3 extracted from E. coli bound directly to the APX2 promoter sequence. These results, based on both GUS reporter assays and EMSAs, indicated that HsfA3 may regulate the expression of APX2 to promote oxidative stress tolerance in Arabidopsis.
In the current study, a novel interaction of HsfA3 with the APX2 promoter leading to the tolerance of oxidative stress was described. This cascade of regulatory interactions resulted in sustained APX activity and decreased accumulation of H2O2, thereby enhancing plant tolerance to oxidative damage. In light of these results, a reasonable model emerges for the role of DREB2C in the Arabidopsis oxidative stress response (Supplementary Fig. S3). DREB2C binds to the HsfA3 promoter and induces the expression of HsfA3. This in turn results in the activation of expression of other antioxidant genes, which leads to decreased levels of H2O2. This regulatory axis thereby functions to enhance the ability of the plant to adapt to oxidative stress.
The involvement of DREB2C and HsfA3 in the induction of APX2 expression indicates a crosstalk between heat and oxidative stress signaling (Panchuck et al., 2002). Although further studies are required to fully elucidate the DREB2C network in Arabidopsis, DREB2C appears to be a key regulator of heat inducible oxidative stress signaling.
Supplementary Material
Acknowledgments
This work was supported by the Next-Generation BioGreen 21 Program (SSAC, grant no. PJ008173, Rural Development Administration, Republic of Korea) and Basic Science Research Program (grant no. 20100009175, through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology, Republic of Korea). Je J. and Song C. were supported by Brain Korea 21 fellowship from the Ministry of Education, Science and Technology, Republic of Korea.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Agarwal P., Agarwal P.K., Nair S., Sopory S.K., Reddy M.K. Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol. Genet. Genomics. 2007;277:189–198. doi: 10.1007/s00438-006-0183-z. [DOI] [PubMed] [Google Scholar]
- Almoguera C., Prieto-Dapena P., Díaz-Martín J., Espinosa J.M., Carranco R., Jordano J. The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol. 2009;9:75. doi: 10.1186/1471-2229-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proceedings of the 3rd International Conference on Intelligent Systems for Molecular Biology; AAAI Press, Menlo Park. 1995. pp. 21–29. [PubMed] [Google Scholar]
- Baker N.R., Rosenqvist E. Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J. Exp. Bot. 2004;55:1607–1621. doi: 10.1093/jxb/erh196. [DOI] [PubMed] [Google Scholar]
- Chen H., Hwang J.E., Lim C.J., Kim D.Y., Lee S.Y., Lim C.O. Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Biophys. Res. Commun. 2010;401:238–244. doi: 10.1016/j.bbrc.2010.09.038. [DOI] [PubMed] [Google Scholar]
- Giacomelli L., Masi A., Ripoll D.R., Lee M.J., van Wijk K.J. Arabidopsis thaliana deficient in two chloroplast ascorbate peroxidases shows accelerated light-induced necrosis when levels of cellular ascorbate are low. Plant Mol. Biol. 2007;65:627–644. doi: 10.1007/s11103-007-9227-y. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Karpinski S., Reynolds H., Karpinska B., Wingsle G., Creissen G., Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Park B.O., Yoo J.H., Jung M.S., Lee S.M., Han H.J., Kim K.E., Kim S.H., Lim C.O., Yun D.J., et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 2007;282:36292–36302. doi: 10.1074/jbc.M705217200. [DOI] [PubMed] [Google Scholar]
- Lee K., Han K.S., Kwon Y.S., Lee J.H., Kim S.H., Chung W.S., Kim Y., Chun S.-S., Kim H.K., Bae D.-W. Identification of potential DREB2C targets in Arabidopsis thaliana plants overexpressing DREB2C using proteomic analysis. Mol. Cells. 2009;28:383–388. doi: 10.1007/s10059-009-0154-4. [DOI] [PubMed] [Google Scholar]
- Lee S.-J., Kang J.-Y., Park H.-J., Kim M.D., Bae M.S., Choi H.-I., Kim S.Y. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010;153:716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.J., Hwang J.E., Chen H., Hong J.K., Yang K.A., Choi M.S., Lee K.O., Chung W.S., Lee S.Y., Lim C.O. Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem. Biophys. Res. Commun. 2007;362:431–436. doi: 10.1016/j.bbrc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S., Mizoi J., Yoshida T., Todaka D., Ito Y., Maruyama K., Shinozaki K., Yamaguchi-Shinozaki K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics. 2010;283:185–196. doi: 10.1007/s00438-009-0506-y. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Breusegem F.V. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Noriega G.O., Yannarelli G.G., Balestrasse K.B., Batlle A., Tomaro M.L. The effect of nitric oxide on heme oxygenase gene expression in soybean leaves. Planta. 2007;226:1155–1163. doi: 10.1007/s00425-007-0561-8. [DOI] [PubMed] [Google Scholar]
- Oh S.J., Kwon C.W., Choi D.W., Song S.I., Kim J.K. Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol. J. 2007;5:646–656. doi: 10.1111/j.1467-7652.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- Panchuk I.I., Volkov R.A., Schöffl F. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002;129:838–853. doi: 10.1104/pp.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA. 2006;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpeci T.E., Zanor M.I., Carrillo N., Mueller-Roeber B., Valle E.M. Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol. Biol. 2008;66:361–378. doi: 10.1007/s11103-007-9274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F., Larkindale J., Kiehlmann E., Ganguli A., Englich G., Vierling E., von Koskull-Döring P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008;53:264–274. doi: 10.1111/j.1365-313X.2007.03334.x. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- Xing Y., Jia W., Zhang J. AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 2007;58:2969–2981. doi: 10.1093/jxb/erm144. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Sakuma Y., Todaka D., Maruyama K., Qin F., Mizoi J., Kidokoro S., Fujita Y., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 2008;368:515–521. doi: 10.1016/j.bbrc.2008.01.134. [DOI] [PubMed] [Google Scholar]
- Yu B., Xu C., Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA. 2002;99:5732–5737. doi: 10.1073/pnas.082696499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.