Abstract
The higher plants of today array a large number of small chloroplasts in their photosynthetic cells. This array of small chloroplasts results from organelle division via prokaryotic binary fission in a eukaryotic plant cell environment. Functional abnormalities of the tightly coordinated biochemical event of chloroplast division lead to abnormal chloroplast development in plants. Here, we described an abnormal chloroplast phenotype in an ethylene insensitive ethylene response1-1 (etr1-1) of Arabidopsis thaliana. Extensive transgenic and genetic analyses revealed that this organelle abnormality was not linked to etr1-1 or ethylene signaling, but linked to a second mutation in ACCUMULATION AND REPLICATION3 (ARC3), which was further verified by genetic complementation analysis. Despite the normal expression of other plastid division-related genes, the loss of ARC3 caused the enlargement of chloroplasts as well as the diminution of a photosynthetic protein Rubisco in etr1-1. Our study has suggested that the increased size of the abnormal chloroplasts may not be able to fully compensate for the loss of a greater array of small chloroplasts in higher plants.
Keywords: arc3-3, etr1-1, giant chloroplast
INTRODUCTION
Chloroplasts in the higher plants of today provide strong evidence for endosymbiosis between prokaryotic and eukaryotic cells (Kutschera and Niklas, 2005; Margulis, 1970; McFadden, 2001; Sagan, 1967). Whole-genome sequencing of Arabidopsis and other plant species has further revealed that a large portion of the organelle genome was transferred to the nuclear genome or lost after the first symbiosis event (The Arabidopsis Genome Initiative, 2000).
Genetic factors that originated in prokaryotic genomes have many roles in eukaryotic cellular activities. For example, a bacterial two-component system (Hoch, 2000; Stock et al., 1989) contributes to intracellular signaling pathways for plant hormones such as cytokinin (Ferreira and Kieber, 2005; Hwang et al., 2002) and ethylene (Mason and Schaller, 2005; Mount and Chang, 2002). Many of these genes also encode machinery proteins for organelle division and coordinate the biological activities of the two merged organisms (Leon et al., 1998). In mesophyll cells of mature leaves of higher plants, chloroplasts are propagated from 10–15 plastid progenitors into 50–120 organelles through prokaryotic binary fission (Sakamoto et al., 2008). Any functional alteration of the proteins involved in plastid division leads to the generation of extremely large chloroplasts that often occupy a considerable portion of the cytoplasm. This abnormal plastid phenotype has been termed Accumulation and Replication of Chloroplast (ARC) and is used to screen for regulatory molecules involved in organelle division (Marrison et al., 1999; Pyke and Leech, 1994).
ARC proteins are involved in positioning of plastid division sites and pressing against chloroplast membranes to execute division process (Supplementary Fig. S1; Aldridge et al., 2005; Maple and Møller, 2010; Osteryoung and McAndrew, 2001). A Ca2+-dependent ATPase, ARC11 is the Arabidopsis equivalent of bacterial MinD (Fujiwara et al., 2004). ARC3 and ARC5, respectively, locate at the stroma side and the cytosolic side of chloroplasts (Gao et al., 2003; Maple et al., 2007; Shimada et al., 2004). Both ARC6 and PARALOG of ARC6 (PARC6) are found at the inner membrane of plastids (Glynn et al., 2008; 2009). PLASTID DIVISION (PDV) 1 and PDV2 are constituents of plastid division–associated protein complexes at the outer membrane of chloroplasts (Miyagishima et al., 2006; Okazaki et al., 2009).
In spite of abnormal chloroplast shapes and arrangements, arc mutants sustain a relatively normal life span (Sakamoto et al., 2008). Thus, it has been proposed that the enlarged size of the chloroplasts of arc mutants may compensate for the loss of organelle numbers and maintain a constant chloroplast volume for normal plant growth and development (Pyke and Leech, 1994; Pyke et al., 1994; Stokes et al., 2000). Recently, this view was challenged, because arc6 and other arc mutations affect light-harvesting capacity and cause the loss of adaptability toward light alteration (Ii and Webber, 2005). The decreased photosynthetic efficiency in arc6 was partly explained by changes in thylakoid architecture.
In this study we described a giant chloroplast phenotype in etr1-1, a dominant ethylene-insensitive mutant of ETHYLENE RESPONSE1 (ETR1; AT1G66340). The abnormal chloroplast phenotype was also observed in etr1-7 that was an intragenic-suppressor of etr1-1, but not in other ethylene response mutants. Here, we elucidated that neither ETR1 nor ethylene signaling was relevant to the abnormal chloroplast phenotype; rather, a second mutation on ARC3 was responsible for the abnormal chloroplast phenotype in etr1-1 and etr1-7. Although other plastid-division related genes were expressed normally, the loss of ARC3 caused impaired plastid division and generated giant chloroplasts in etr1-1.
MATERIALS AND METHODS
Plant materials and isolation of mesophyll protoplasts
Plants were grown in soil at 23°C for 20 to 22 days under 60 μmol/m2/s with a 13 h photoperiod. Protoplast isolation was carried out as previously described (Yoo et al., 2007). Arabidopsis thaliana Columbia served as the wild-type and etr1-1, etr1-2, etr1-7, ctr1-1, ein2-1, ein3-1, and arc3-2 (SALK_057144) mutants were used for experiments.
Fine mapping of the second mutation in etr1-1
To map the second mutation in etr1-1 that was responsible for enlarged chloroplasts, F2 population was generated from a cross between Ler and etr1-1. Among 183 F2 plants, 44 mutants were selected based on the chloroplast phenotype under microscopy and subjected for further molecular marker analysis.
Gene expression analysis
For gene expression analysis, seedlings were grown in B5 plates (Duchefa) containing 1% sucrose under a 16 h photoperiod. Total RNA was isolated by the Trizol method (Invitrogen) and 1 μg of total RNA was used for cDNA synthesis. Semiquantitative reverse transcriptase PCR analysis was carried out with gene-specific primers for ARC3, PDV1, and PDV2. Detailed primer sequence information can be found in Supplementary Table S1. Experiments were repeated three times with consistent results.
RESULTS AND DISCUSSION
Accumulation and replication of chloroplast phenotype in etr1-1
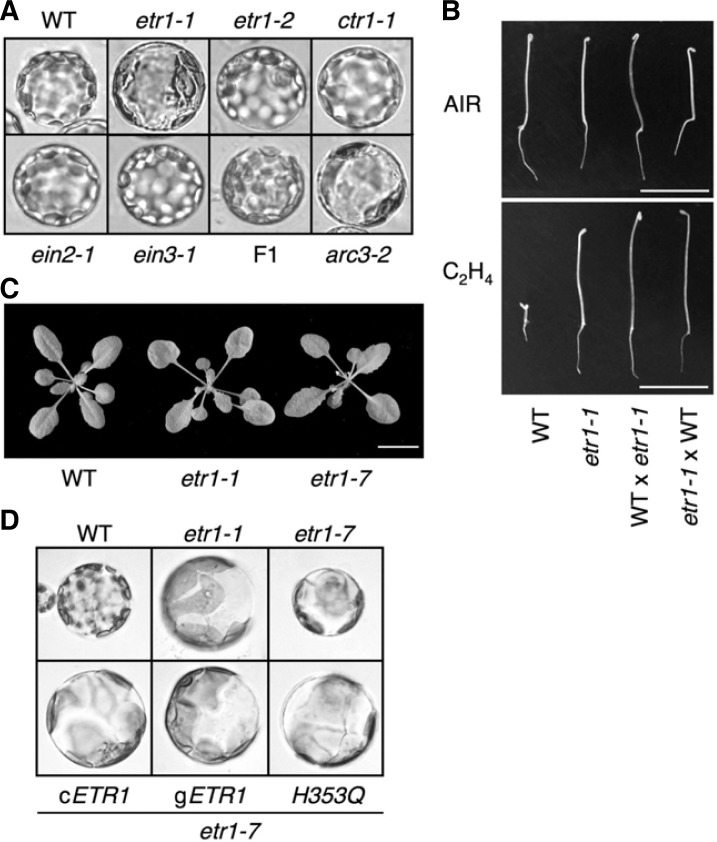
Leaf mesophyll cells of higher plants typically contain about 50 to 120 small chloroplasts (Sakamoto et al., 2008), as shown in Arabidopsis wild-type (Col) protoplasts freshly isolated from mature leaf tissue (Fig. 1A). In contrast, we observed an aberrant chloroplast phenotype in the dominant ethylene-insensitive mutant etr1-1 (Fig. 1A). This mutant had enlarged chloroplasts that were much reduced in number compared to WT.
Fig. 1.
Accumulation and replication of chloroplast phenotype in etr1-1. (A) Chloroplast phenotypes in leaf mesophyll protoplasts isolated from different genetic backgrounds. F1 indicates the F1 lines of etr1-1 × WT (Col). Image was taken under a microscope (200×). (B) Analysis of triple response manifested by apical hook formation, hypotocyl and root growth inhibition and hypocotyl thickening for 3.5-day-old etiolated seedlings in the absence and presence of 1-aminocylopropane-1-carboxylic acid (ACC, 10 μM). Scale bar, 10 mm. (C) Arabidopsis WT, etr1-1, and etr1-7 plants grew normally in soil at 23°C under a 13 h photoperiod (60 μmol/m2/s). Scale bar, 10 mm. (D) Chloroplasts in leaf mesophyll protoplasts of etr1-1, etr1-7, and transgenic etr1-7 expressing wild-type ETR1 (cETR1 or gETR1) or His-kinase inactive ETR1 (H353Q).
To investigate whether ETR1 function in ethylene signaling is involved in chloroplast phenotype, we observed the chloroplast morphology of another dominant allele of etr1, etr1-2, together with other ethylene-response mutants, including the ethylene constitutively responsive mutant, ctr1-1 and ethylene-insensitive mutants, ein2-1 and ein3-1. The abnormal phenotype was not observed in any of these mutants (Fig. 1A), indicating that ethylene signaling is irrelevant to the chloroplast phenotype of etr1-1. We next generated F1 plants by crossing etr1-1 with WT in a reciprocal manner; F1 plants were ethylene insensitive due to the dominant nature of the etr1-1 allele in ethylene signaling (Fig. 1B). However, the leaf cells showed a normal chloroplast shape (Fig. 1A), providing another evidence that the ethylene signaling function of ETR1 is irrelevant to the abnormal chloroplast phenotype in etr1-1.
We noted that the chloroplast abnormality in etr1-1 resembled the arc phenotype resulting from defects in organelle division as in arc3-2 (Fig. 1A; Marrison et al., 1999; Pyke and Leech, 1994). To further examine whether the ethylene signaling-independent arc phenotype was specific to the etr1-1 allele, we observed the chloroplast phenotype of etr1-7, an intragenic suppressor of etr1-1 that has no obvious developmental defect compared to WT and etr1-1 (Fig. 1C) (Hua and Meyerowitz, 1998). Unexpectedly, the arc phenotype was also observed in etr1-7 (Fig. 1D), even though ETR1 expression was greatly diminished in the mutant (Cho and Yoo, 2007). A peculiarity of chloroplast division is that both loss- and gain-of-function alleles involved in chloroplast division often result in the same arc phenotype. This indicates that the molecular ratio among the machinery proteins is important for the precise control of plastid division (Maple et al., 2007). To test if this was the case in chloroplast aberrations in gain-of-function etr1-1 and loss-of-function etr1-7 mutants, we isolated and observed leaf mesophyll protoplasts from transgenic etr1-7 lines that expressed ETR1 cDNA, genomic DNA, or cDNA with a histidine kinase mutation (H353Q) in ETR1 under the control of the ETR1 promoter (Cho and Yoo, 2007). The arc phenotype in etr1-7 was again present in all of the transgenic etr1-7 lines (Fig. 1D). These results indicated that ETR1 itself was not the genetic factor underlying the chloroplast phonotype; instead, there had to be another mutation that was cosegregating with ETR1 and causing the arc phenotype in etr1-1.
A second mutation in ARC3 of etr1 mutants
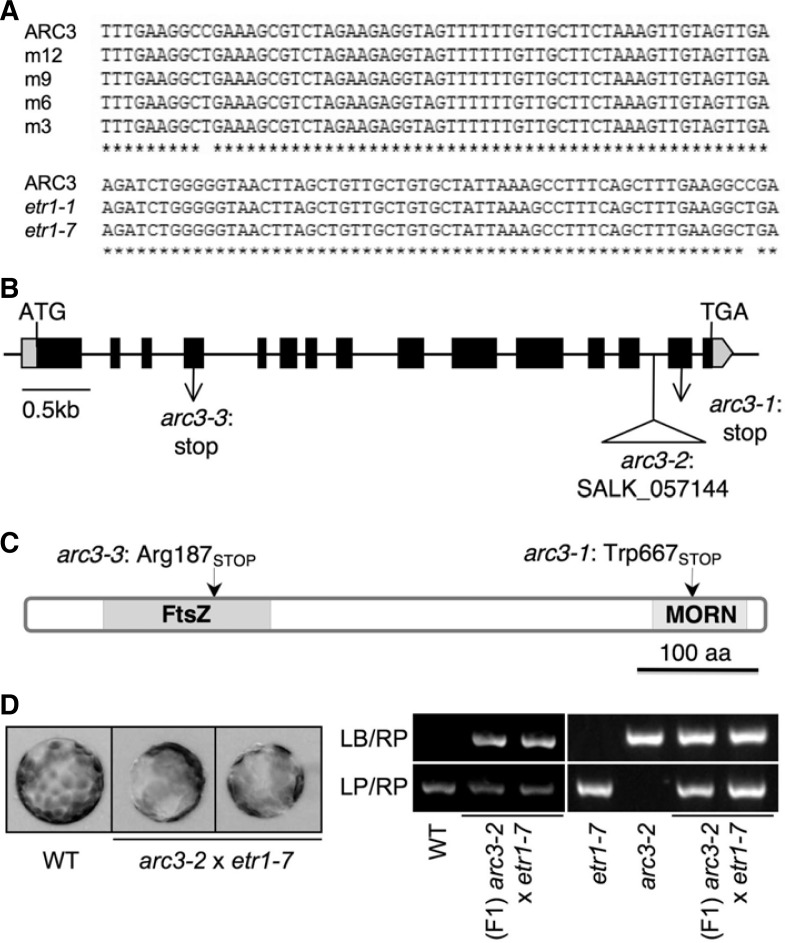
To isolate and characterize the genetic factor causing the arc phenotype in etr1-1 (Fig. 1A), we crossed etr1-1 with Lands-berg erectra (Ler) and mapped a second mutation by marker-assisted allele positioning. The mutation was placed near ETR1 after the first round of marker analysis. In all of the mapping lines with arc phenotype (m12, m9, m6, and m3), the second mutation was identified as a single nucleotide change in ARC3 (AT1G75010) that caused a premature translation stop at Arg187 (Fig. 2A). We named this allele arc3-3, complying with previously described alleles (Fig. 2B). As expected, the same mutation was found on ARC3 in etr1-7, because etr1-1 and etr1-7 share genetic backgrounds (Fig. 2A) and showed the same arc phenotype (Fig. 1D).
Fig. 2.
ARC3 mutation cosegregation with etr1-1. (A) Nucleotide sequences of arc3 isolated from four independent mapping lines (m12, m9, m6, and m3), as well as etr1-1 and etr1-7, were aligned with WT sequence. (B) arc3-3 was identified from the mapping population of etr1-1 × Ler. The genome structure of ARC3 was shown with previously (arc3-1 and arc3-2) and newly (arc3-3) identified alleles. (C) Schematic diagram of ARC3 protein was shown with arc3-1 and arc3-3 mutations. (D) Chloroplast phenotypes in leaf mesophyll protoplasts isolated from WT and two F1 lines of arc3-2 × etr1-7. Molecular marker analysis for arc3-2 was shown.
ARC3 is located on the stromal side of chloroplasts and is involved in FtsZ-ring positioning in the middle of the organelle (Supplementary Fig. S1; Maple et al., 2007; Shimada et al., 2004). arc3-1 has a point mutation that leads to a premature translation stop at Trp667, and arc3-2 has T-DNA insertion at the 13th intron (Fig. 2B). Both alleles cause defects in the MORN domain at the C-terminal end of ARC3 (Fig. 2C), and mutants carrying either allele have average 13 chloroplasts (Fig. 1A; Maple et al., 2007; Shimada et al., 2004). The new allele arc3-3 had a defect in the FtsZ domain at the N-terminal of the protein, which caused much shorter ARC3 peptide synthesis, if any, compared to those in arc3-1 and arc3-2 (Fig. 2C).
To further verify our discovery, etr1-7 was crossed with arc3-2 and F1 plants were used for genetic complementation test. As shown in Fig. 2D, F1 plants were heterozygous for arc3-2, but kept arc phenotype due to arc3-3 in etr1-7 (Fig. 2D). This result conclusively demonstrated that the loss of ARC3 in etr1-7 caused the giant chloroplast phenotype.
At the plastid division site of chloroplasts, PDV1 and PARC6 interact in the inner membrane space and PARC6 may associate with ARC3 in the stroma (Supplementary Fig. S1; Okazaki et al., 2009; Osteryoung and McAndrew, 2001). Likewise, PDV2 and ARC6 interact each other and ARC6 also associates with FtsZ1 and FtsZ2 (Glynn et al., 2008; Osteryoung and McAndrew, 2001). All of these protein-protein interactions collectively support the precise localization of the plastid division ring to the outer membrane and the Z-ring to the inner membrane at the chloroplast division site.
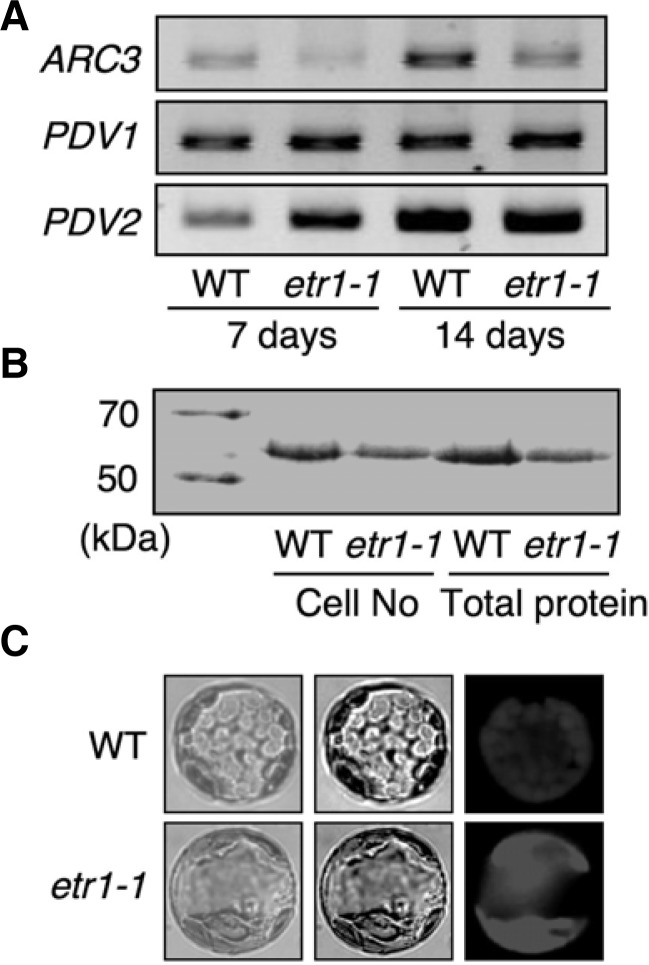
To examine if arc3-3 affects other plastid division machineries in etr1-1, we measured the transcript levels of PDV1 and PDV2 together with ARC3. When we quantified the transcript levels with cDNA generated from seedlings grown on full-strength B5 agar media containing 1% sucrose, relatively lower levels of ARC3 were detected in etr1-1 compared to WT (Fig. 3A). Perhaps because of the premature stop codon, ARC3 transcript was relatively unstable in etr1-1 (Fig. 2C). In b oth young and mature seedlings, PDV1 transcript levels were more or less similar in WT and etr1-1. PDV2 accumulation was only slightly higher in young etr1-1 seedlings compared to WT. The higher accumulation of PDV2 may reflect its early compensatory response with respect to the defect in the PDV1 complex caused by the arc3-3 allele in etr1-1 mutants during the early stage of plastid division (see the topological model of the chloroplast division site in Supplementary Fig. S1).
Fig. 3.
arc3-3 related molecular and chloroplast phenotypes in etr1-1. (A) ARC3, but not PDV1 and PDV2, transcript level was reduced in etr1-1. Transcript levels were monitored using semi-quantitative PCR with cDNA templates generated from whole seedlings. (B) Rubisco protein was visualized by Coommassie Blue staining. (C) Chloroplast phenotypes in leaf mesophyll protoplasts. Image was taken under a fluorescence microscope (200×).
We then measured the protein levels of rubisco (RBC) in WT and etr1-1 and monitored the effects of chloroplast structure on photosynthetic machinery. In the same number (4 × 104 cells) of leaf mesophyll cells, RBC proteins in etr1-1 were relatively less abundant than in WT (Fig. 3B). In the same amount of total protein (5 μg), RBC protein accumulation was also reduced in etr1-1 compared to WT (Fig. 3B). These results indicated that the smaller number of chloroplasts was not fully compensated for by their enlarged size in etr1-1 arc3-3 mutants.
Chlorophyll fluorescence from leaf protoplasts delineated chloroplasts as many small structures in WT and a few larger structures in etr1-1 (Fig. 3C). The quantitative analysis of chlorophyll fluorescence has been utilized to acquire information about the photosynthetic yields of photosystem II, the relative level of the reduced quinone pool, and the extent of non-photochemical quenching during photosynthetic light reactions (Akimoto and Mimuro, 2007; Karukstis and Sauer, 1983). It will be interesting to further examine if the altered structure of plastids can cause changes in chlorophyll fluorescence and photosynthesis activity.
In summary, we have unravelled that an unexpected chloroplast phenotype in etr1-1 is caused not by etr1-1, but by a second mutation, arc3-3, that cosegregates with etr1-1. It appears that the increased size of the abnormal chloroplasts may not be able to fully compensate for the loss of a greater array of small chloroplasts.
Supplementary Material
Acknowledgments
We thank Yeun-il Park (Chungnam National University) for sharing information about etr1-1 phenotype and discussion. This work was supported by Korean National Research Foundation (NRF) grants to SDY (2011-0002896, 0003855), and YHC (2011-0004969).
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Akimoto S., Mimuro M. Application of time-resolved polarization fluorescence spectroscopy in the femtosecond range to photosynthetic systems. Photochem. Photobiol. 2007;83:163–170. doi: 10.1562/2006-02-28-IR-825. [DOI] [PubMed] [Google Scholar]
- Aldridge C., Maple J., Møller S.G. The molecular biology of plastid division in higher plants. J. Exp. Bot. 2005;56:1061–1077. doi: 10.1093/jxb/eri118. [DOI] [PubMed] [Google Scholar]
- Cho Y.H., Yoo S.D. ETHYLENE RESPONSE 1 histidine kinase activity of Arabidopsis promotes plant growth. Plant Physiol. 2007;143:612–616. doi: 10.1104/pp.106.091504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira F.J., Kieber J.J. Cytokinin signaling. Curr. Opin. Plant Biol. 2005;8:518–525. doi: 10.1016/j.pbi.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Fujiwara M.T., Nakamura A., Itoh R., Shimada Y., Yoshida S., Møller S.G. Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J. Cell Sci. 2004;117:2399–2410. doi: 10.1242/jcs.01092. [DOI] [PubMed] [Google Scholar]
- Gao H., Kadirjan-Kalbach D., Froehlich J.E., Osteryoung K.W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA. 2003;100:4328–4333. doi: 10.1073/pnas.0530206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Froehlich J.E., Osteryoung K.W. Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell. 2008;20:2460–2470. doi: 10.1105/tpc.108.061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Yang Y., Vitha S., Schmitz A.J., Hemmes M., Miyagishima S.Y., Osteryoung K.W. PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 2009;59:700–711. doi: 10.1111/j.1365-313X.2009.03905.x. [DOI] [PubMed] [Google Scholar]
- Hoch J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- Hua J., Meyerowitz E.M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hwang I., Chen H-C., Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–515. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii J.A., Webber A.N. Photosynthesis in Arabidopsis thaliana mutants with reduced chloroplast number. Photosynth. Res. 2005;85:373–384. doi: 10.1007/s11120-005-7708-x. [DOI] [PubMed] [Google Scholar]
- Karukstis K.K., Sauer K. Fluorescence decay kinetics of chlorophyll in photosynthetic membranes. J. Cell Biochem. 1983;23:131–158. doi: 10.1002/jcb.240230112. [DOI] [PubMed] [Google Scholar]
- Kutschera U., Niklas K.J. Endosymbiosis, cell evolution and speciation. Theory Biosic. 2005;124:1–24. doi: 10.1016/j.thbio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Leon P., Arroyo A., Mackenzie S. Nuclear control of plastid and mitochondrial development in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:453–480. doi: 10.1146/annurev.arplant.49.1.453. [DOI] [PubMed] [Google Scholar]
- Maple J., Møller S.G. The complexity and evolution of the plastid-division machinery. Biochem. Soc. Trans. 2010;38:783–788. doi: 10.1042/BST0380783. [DOI] [PubMed] [Google Scholar]
- Maple J., Volta L., Soll J., Møller S.G. ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 2007;8:293–299. doi: 10.1038/sj.embor.7400902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Origin of eukaryotic cells. New Haven: Yale University Press; 1970. [Google Scholar]
- Marrison J.L., Rutherford S.M., Robertson E.J., Lister C., Dean C., Leech R.M. The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J. 1999;18:651–662. doi: 10.1046/j.1365-313x.1999.00500.x. [DOI] [PubMed] [Google Scholar]
- Mason M.G., Schaller G.E. Histidine kinase activity and the regulation of ethylene signal transduction. Can. J. Bot. 2005;83:563–570. [Google Scholar]
- McFadden G.I. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 2001;37:951–959. [Google Scholar]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell. 2006;18:2517–2530. doi: 10.1105/tpc.106.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S.M., Chang C. Evidence for a plastid origin of plant ethylene receptor genes. Plant Physiol. 2002;130:10–14. doi: 10.1104/pp.005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K., Kabeya Y., Suzuki K., Mori T., Ichikawa T., Matsui M., Nakanishi H., Miyagishima S.Y. The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell. 2009;21:1769–1780. doi: 10.1105/tpc.109.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., McAndrew R.S. The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:315–333. doi: 10.1146/annurev.arplant.52.1.315. [DOI] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol. 1994;104:201–207. doi: 10.1104/pp.104.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Rutherford S.M., Robertson E.J., Leech R.M. arc6, a fertile Arabidopsis mutant with only two mesophyll cell chloroplasts. Plant Physiol. 1994;106:1169–1177. doi: 10.1104/pp.106.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L. On the origin of mitosing cells. J. Theoretical Biol. 1967;14:225–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Miyagishima S.-Y., Jarvis P. Chloroplast Biogenesis, Control of plastid development, protein import, division and inheritance. The Arabidopsis book. 0110. 2008. [DOI] [PMC free article] [PubMed]
- Shimada H., Koizumi M., Kuroki K., Mochizuki M., Fujimoto H., Ohta H., Masuda T., Takamiya K. ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 2004;45:960–967. doi: 10.1093/pcp/pch130. [DOI] [PubMed] [Google Scholar]
- Stock J.B., Ninfa A.J., Stock A.M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Mol. Biol. Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K.D., McAndrew R.S., Figueroa R., Vitha S., Osteryoung K.W. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 2000;124:1668–1677. doi: 10.1104/pp.124.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. Arabidopsis mesophyll protoplasts, A versatile cell system for transient gene expression analysis. Nat. Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.