Abstract
The BNIPs (BCL2 and adenovirus E1B 19 kDa interacting proteins) are a subfamily of BCL2 family proteins typically containing a single BCL2 homology 3 (BH3) domain. BNIPs exert important roles in two major degradation processes in cells - apoptosis and autophagy. Although it is known that the function of BNIPs is transcriptionally regulated under hypoxic conditions in tumors, their regulation in the developing brain and neurons following the induction of apoptosis/autophagy is largely unknown. In this study, we demonstrate that three members of the BNIP family, BNIP1, BNIP3 and BNIP3L, are expressed in the developing brain with distinct brain region specificity. BNIP3 mRNA was especially enriched in the entorhinal cortex, raising a possibility that it may have additional biological functions in addition to its apoptotic and autophagic functions. Following starvation-induced autophagy induction, BNIP1 mRNA was selectively increased in cultured neurons. However, the apoptogenic chemical staurosporine failed to modulate the expression of BNIPs, which is in contrast to the marked induction of all BNIPs by glucose-oxygen deprivation. Finally, neonatal nerve axotomy, which triggers apoptosis in motoneurons, selectively enhanced BNIP3 mRNA expression. Collectively, these results suggest that the expression of BNIPs is differentially regulated depending on the stimuli, and BNIPs may exert unique biological functions.
Keywords: apoptosis, autophagy, axotomy, BNIP, development
INTRODUCTION
Degradation of cells and organelles is an essential processes of normal life in eukaryotic organisms. For instance, cell death is a major requirement for the systems-matching of the developing nervous system (Buss et al., 2006). During the innervation of motoneurons (MNs) into the target muscle, approximately 50% of the initially generated MNs undergo programmed cell death. The survival of MNs is largely dependent on target-derived neurotrophic signals, and the deprivation of target-derived signals by nerve axotomy promotes massive MN death within 1 week in early post-natal animals (Sun and Oppenheim, 2003). Another degradation process in organisms is autophagy, which is the removal of organelles. Autophagy is also a catabolic program for maintaining metabolic homeostasis in response to starvation or changing nutrient conditions. Additional functions of autophagy in differentiation, development and cell death have been increasingly identified, indicating that autophagy is an essential biological process (Mizushima and Komatsu, 2011).
Both apoptosis and autophagy are regulated by BCL2 family molecules. BCL2 can inhibit apoptosis and modulate autophagy (Levine et al., 2008). BCL2 interacts with BH3-only proteins, and the functions of BCL2 are associated with the diverse functions of BH3-only family proteins. For instance, many BH3-only proteins, such as tBID, BIM, PUMA, NOXA and HRK, promote apoptosis (Lomonosova and Chinnadurai, 2008). In contrast, BECLIN is also a member of the BH3-only protein family and is involved in the induction of autophagy but not in the regulation of apoptosis (Liang et al., 1998).
Intriguingly, BNIPs (BCL2 and adenovirus E1B 19 kDa interacting proteins) are a third class of BH3-only proteins that can induce both apoptosis and autophagy (Zhang and Ney, 2009). At least 4 members have been identified; these proteins are BNIP1-3 and BNIP3L (also known as NIX). BNIP1-3 were first identified by a yeast two-hybrid screen using adenovirus protein E1B 19 kDa as bait (Boyd et al., 1994). The E1B 19 kDa protein protects cells from apoptosis after adenoviral infection, and anti-apoptotic BCL2 protein can functionally replace E1B 19 kDa protein. In this context, BNIP1-3 are pro-apoptotic proteins that suppress the anti-apoptotic function of the E1B 19 kDa or BCL2 protein. BNIP3L was isolated from human placenta cDNA based on sequence homology with BNIP3 (Matsushima et al., 1998). BNIP3L also has a BH3 domain and is a proapoptotic protein.
In addition to the pro-apoptotic function of BNIPs, at least two members of the BNIP family, BNIP3 and BNIP3L, also play roles in autophagy (Zhang and Ney, 2009). Overexpression of BNIP3 or BNIP3L promotes autophagy in several cellular contexts (Daido et al., 2004; Hamacher-Brady et al., 2007; Sandoval et al., 2008). Furthermore, the expression of BNIP3 and BNIP3L are increased in glioma cells during chemical- or hypoxia-induced autophagy, suggesting that their functional activation is controlled at the transcriptional level (Daido et al., 2004; Kanzawa et al., 2005). However, BNIP regulation and function in the nervous system during neuronal apoptosis and autophagy are less well understood. Here, we examined the expression patterns of BNIP1, BNIP3, and BNIP3L during brain development, following the induction of autophagy and/or apoptosis in vitro, and after nerve axotomy in vivo. Our current observations suggest that the expression levels of BNIPs are differentially regulated depending on the cell type and apoptogenic/autophagic stimuli, suggesting that they are differentially involved in cellular degradation processe.
MATERIALS AND METHODS
Cell culture and treatments
Rat embryonic cerebral cortex neurons were obtained from embryonic day 17 (E17) Sprague-Dawley rats. Cerebral cortex cells were dissociated with 0.25% trypsin-EDTA (Gibco BRL, USA), and the dissociated cells were plated on poly-D-lysine (50 μg/ml)-coated coverslips at a density of 2 × 105 cells/cm2 in growth medium composed of Neurobasal medium (Gibco BRL, USA), B27 supplement (Gibco BRL, USA), 200 mM L-glutamine (Gibco BRL, USA), 5 mM glutamic acid (Sigma-Aldrich, USA) and antibiotics (Gibco BRL, USA). The culture medium was changed to a medium having no glutamate at 3 days in vitro (DIV3). On DIV4, media were replaced with Hank’s Balanced Salt Solution (Gibco BRL, USA) for starvation, or 1 μM staurosporine (Calbiochem, Germany) was applied for the induction of apoptosis.
Oxygen-glucose deprivation (OGD) was performed as described previously (Noh et al., 2006). On DIV10, the culture medium was replaced with deoxygenated glucose-free DMEM (Gibco BRL, USA) in an anaerobic chamber maintaining 5% CO2, 10% H2, and 85% N2, and the cells were incubated under oxygen-glucose deprived conditions (OGD) for 60 min. OGD was terminated by removing the cultures from the chamber and returning them to normal growth medium. Control cultures were incubated for the same period in normoxic conditions. The start of reoxygenation (reperfusion) was assigned as the 0 h.
Animals and treatments
For nerve axotomy, post-natal day 2 (P2) rat pups were anesthetized on ice, and the facial nerve was cut at the exit position (Sun and Oppenheim, 2003). Following surgery, the skin incision was sutured and warmed, and the animals were returned to their mother. All experiments were carried out in accordance with the regulations and approval of the Animal Care and Use Committee of the Korea University.
Reverse transcription-polymerase chain reaction (RT-PCR)
RNAs were extracted using the Qiagen RNeasy kit following the manufacturer’s protocol. RNA (2 μg) was reverse-transcribed by M-MLV reverse transcriptase (Promega, USA) with random primers (Promega, USA). For the RT-PCR of BNIP1, BNIP3 and BNIP3L, BNIP1-F (5′-CCA CAA AAA GCA GAT GCT CA-3′) and BNIP1-R (5′-AAG AGG CGC TTT TTC ACA AT-3′); BNIP3-F (5′-GCT CCC AGA CAC CAC AAG AT-3′) and BNIP3-R (5′-GCT ACA ATA GGC ATC AGT CTG ACA-3′); and BNIP3L-F (5′-GAG CTA CCC ATG AAC AGC AG-3′) and BNIP3L-R (5′-GGT GTG CTC AGT CGT TTT CC-3′) primer sets were used.
RNA in situ hybridization
Rat brains were obtained at the E16, P12 and adult stages and rapidly frozen in pre-chilled isopentane (−80°C). The frozen brains were cut (12 μm thick), thaw-mounted onto (3-aminopropyl) triethoxysilane (Sigma-Aldrich, USA) coated slides, and fixed in 4% paraformaldehyde. Sections were treated with 0.25% acetic anhydride in 0.1 M triethanolamine/0.9% NaCl (pH 8.0), dehydrated and defatted in ethanol and chloroform, and then air-dried. Riboprobes used in this study were directed against bases 260–685 of rat BNIP1 (NM_080897), 309–909 of rat BNIP3 (NM_053420), and 115–644 of rat BNIP3L (NM_080888). The antisense probes were labeled by in vitro transcription in the presence of [35S]-UTP (Perkin Elmer, USA), and the sections were hybridized overnight at 63°C with labeled probes. On the following day, the sections were washed in 4× SSC, treated with 0.02 mg/ml RNase A (USB, USA) in SSC for 30 minutes at 37°C, washed in graded serial dilutions of SSC (2× SSC, 1× SSC, 0.5× SSC), incubated in 0.1× SSC at 62°C for 30 min, and dehydrated in ethanol. The hybridized sections were then exposed to X-ray film.
RESULTS
Expression patterns of BNIPs during development and in the adult brain
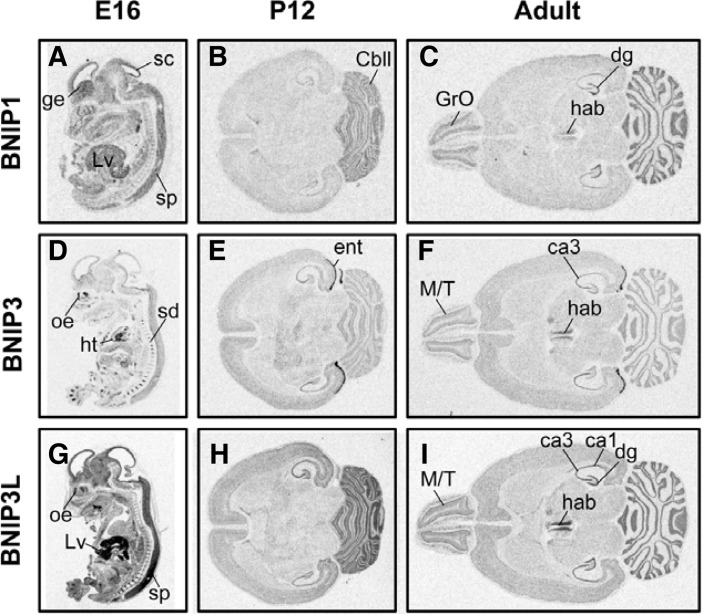
The expression patterns of BNIP1, BNIP3, and BNIP3L were examined in E16 embryos, and P12, and adult brains (Fig. 1). BNIP1 expression was ubiquitously observed in embryos, with moderate-to-strong signals in the nervous system. BNIP1 mRNA signals were especially strong in the ganglion eminence, superior colliculus and spinal cord in the central nervous system (Fig. 1A). Expression of BNIP1 was also strong in the embryonic liver. At stage P12, BNIP1 mRNA expression was strong in the cerebellum granule cell layer (Fig. 1B). In the adult brain, habenular nuclei, granule cell layers in the olfactory bulb, dentate gyrus, and cerebellum exhibited significantly stronger BNIP1 mRNA signals than other brain regions (Fig. 1C).
Fig. 1.
The expression patterns of BNIP1 (A–C), BNIP3 (D–F) and BNIP3L (G–I) in developing embryos (E16, A, D, G), post-natal day 12 (P12, B, E, and H) and adult (C, F, and I) brains. Embryos were parasagitally cut, and post-natal/adult brains were horizontally cut. At least two independent sections from different individuals were examined, and typical images are shown. The abbreviations used are as follows: ge, ganglion eminence; Lv, liver; sc, superior colliculus; sp, spinal cord; cbll, cerebellum; Gro, granule cell layer of olfactory bulb; hab, habenular nuclei; dg, dentate gyrus of hippocampal formation; oe, olfactory epithelia; ht, heart; ent, entorhinal cortex; M/T, mitral/tufted cell layer of olfactory bulb; ca1, CA1 region of hippocampus; ca3, CA3 region of hippocampus.
BNIP3 mRNA expression was more confined to restricted regions including the developing cartilage in the spinal disc, cardiac muscle, and olfactory epithelia in the embryo, whereas BNIP3 mRNA expression in the central nervous system was only weak-to-marginal (Fig. 1D). At stage P12, weak BNIP3 mRNA signal was observed in most brain regions. Interestingly, a strong signal was observed in the entorhinal cortex (Fig. 1E). A similar expression pattern was maintained in the adult brain, with moderate signals in the CA3 region of the hippocampus, habenular nuclei, and mitral/tufted cell layers in the olfactory bulb (Fig. 1F).
BNIP3L mRNA expression was strong and ubiquitous throughout the whole embryo, with the strongest expression in the liver, olfactory epithelium, and spinal cord (Fig. 1G). Strong signals were found in the hippocampus and cerebellum at stage P12 (Fig. 1H). In the adult, the mitral/tufted cell layer of olfactory bulb, habenular nuclei, CA1-3 and dentate gyrus of the hippocampus, and cerebellum exhibited strong BNIP3L mRNA signals (Fig. 1I).
Induction of BNIPs mRNA in cultured neurons during autophagy and apoptosis
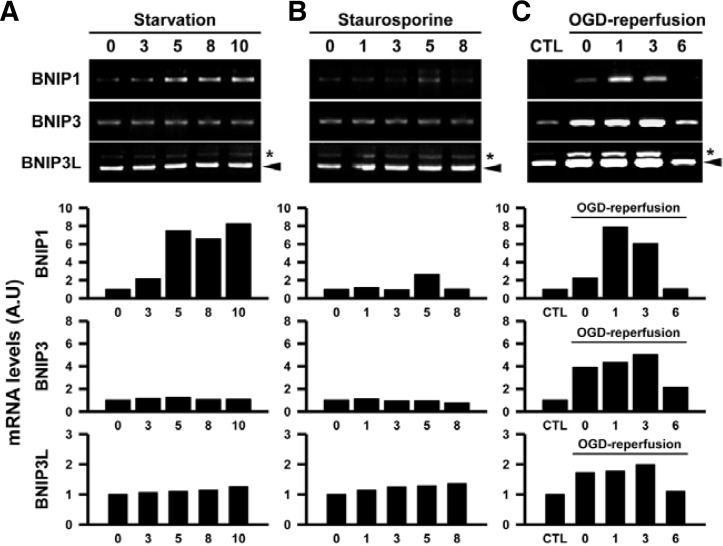
The action of a large subset of BH3-only proteins is regulated by transcriptional induction upon stimulation (Youle and Strasser, 2008). Because BNIPs are related to autophagy and apoptosis, we next tested whether the mRNA expression levels of BNIPs are regulated during starvation-induced autophagy and the staurosporine-induced apoptosis (Figs. 2A and 2B). Following starvation (switching growth media to minimal conditions), we found that level of LC3-II, which is associated with autophagosomes as the cleaved form of LC3-I, was increased, and the intracellular localization of LC3 showed enhanced puncta formation (Data not shown). Interestingly, BNIP1 mRNA expression was progressively increased, whereas BNIP3 and BNIP3L expression levels were unchanged by starvation (Fig. 2A). On the other hand, staurosporine treatment did not affect the expression of BNIPs in cultured cerebral cortex neurons (Fig. 2B), although Caspase-3 was activated (Data not shown).
Fig. 2.
Induction of BNIPs mRNAs after starvation (A), staurosporine treatment (B), and oxygen-glucose deprivation (OGD)-reperfusion (C) in cultured rat cerebral cortex neurons. Expression levels of mRNAs were detected by RT-PCR, and typical agarose gel (1.5%) images are shown in the upper panels. The arrowheads indicate the BNIP3L cDNA PCR product (530 bps), and the asterisks indicate a non-specific PCR product. Experiments were duplicated, and similar results were obtained. Graphs in the lower panels show the quantification of DNA band intensities.
It is known that the expression of BNIP family mRNAs is induced by hypoxia in many tumor cells (Chinnadurai et al., 2008). Similarly, the expression of BNIPs was also affected by oxygen-glucose deprivation (OGD) in cultured cerebral cortex neurons (Fig. 2C). BNIP1 mRNA expression was slightly induced by the 0 hour of reperfusion time when cells were exposed to OGD for 1 h. It increased by 1 h after reperfusion, and returned to basal levels by 6 h. On the other hand, BNIP3 and BNIP3L mRNA expressions rapidly reached peak levels by OGD and sustained at a high level until 3 h after OGD. The induced expression returned to the basal level by 6 h after OGD-reperfusion.
Induction of BNIP3 mRNA in facial motoneurons after nerve axotomy
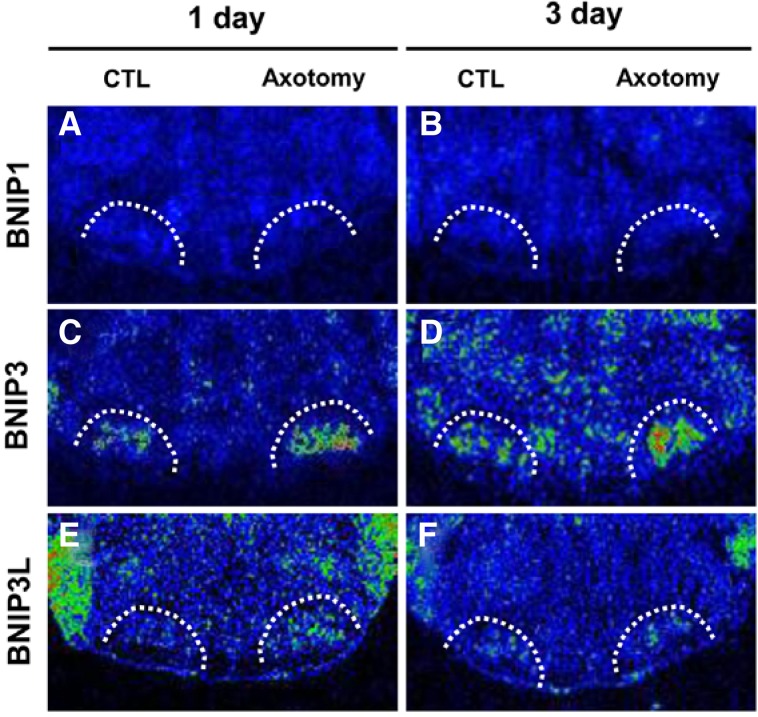
Transection of the facial nerve in the neonatal rat triggers the apoptosis of MNs (Oppenheim et al., 1995; Sendtner et al., 1992; Sun and Oppenheim, 2003; Yan et al., 1995). To test whether BNIPs are involved in apoptosis in vivo, we examined the induction of BNIP mRNAs following facial nerve axotomy (Fig. 3). Among the three BNIP family members, the mRNAs of BNIP3 and BNIP3L were found in the control facial nuclei, whereas BNIP1 mRNA was not detected in the facial nuclei (Figs. 3A and 3B). Following axotomy, BNIP3 mRNA was rapidly induced in the axotomized facial nuclei within 1–3 days (Figs. 3C and 3D). However, BNIP3L mRNA was only marginally induced 1 day after axotomy, but returned to the basal level by day 3 (Figs. 3E and 3F).
Fig. 3.
Changes in the expression of BNIP1 (A, B), BNIP3 (C, D) and BNIP3L (E, F) mRNAs on 1 day (A, C, and E) or 3 days (B, D, and F) after unilateral facial nerve axotomy in neonatal (P2) rat pups. Facial nuclei are marked by white dotted lines. The in situ hybridized mRNA signals are pseudocolored, and the levels of BNIPs mRNAs are visualized as green (low level) to red (high level).
DISCUSSION
In this study, we found that the expression levels of BNIP1, 3 and 3L were differentially regulated during development and upon apoptosis- and/or autophagy-inducible signals. In the rat brain, moderate and significant expression of BNIP1 and BNIP3L mRNAs was observed. Considering that all neurons expressing BNIPs do not undergo apoptosis, the basal expression of BNIPs in the developing brain is not directly related to apoptosis. However, autophagy is an essential cellular event in healthy neurons for the maintenance of homeostasis (Mizushima and Komatsu, 2011), and basally expressed BNIPs may play significant roles in autophagy. Interestingly, we found that BNIP1 and BNIP3L expression was strong in the adult cerebellum, and BNIP3 expression appeared to be induced in the post-natal brain, especially in the entorhinal cortex. However, these brain regions exhibit neither spontaneous apoptosis nor enhanced autophagy in the adult brain. Therefore, these unexpected expression patterns may suggest that BNIPs have other roles in addition to the control of apoptosis/autophagy. It is known that BCL2 family proteins are also involved in many biological processes, such as mitochondrial dynamics (Cleland et al., 2011; Karbowski et al., 2006) and intracellular calcium regulation (Oakes et al., 2005; Scorrano et al., 2003). Considering that BNIPs interact with BCL-2/BAX proteins, these BNIPs may also have roles in these additional processes. Consistent with this hypothesis, recent reports have demonstrated that BNIPs are involved in the regulation of mitochondrial dynamics. BNIP1 induces the expression of DRP1, a mitochondrial fission molecule, resulting in mitochondrial fragmentation (Ryu et al., 2012). BNIP3 also promotes mitochondrial fragmentation through mitochondrial recruitment of DRP1 and inhibitory interaction with OPA1, a mitochondrial fusion molecule (Landes et al., 2010; Lee et al., 2011).
We tested whether these BNIP mRNA expression levels are regulated by several stimuli that induce autophagy or apoptosis. Interestingly, BNIPs differentially responded to these stimuli. Starvation that promotes autophagy in neurons evoked mild and progressive transcriptional activation of BNIP1. It has been reported that BNIP3 is induced during ceramide-induced autophagy in glioma cells (Daido et al., 2004), and forced expression of BNIP3 evoked autophagic cell death (Kanzawa et al., 2005). In this respect, it is surprising that only BNIP1 mRNA was induced in the starvation-induced autophagic condition, suggesting that BNIPs may differentially respond according to the stimuli or cell types. Although BNIPs are a class of proapoptotic BH3-only proteins (Galvez et al., 2006; Imazu et al., 1999), staurosporine, an apoptogenic chemical, failed to induce BNIP mRNA expression. It is well known that the transcriptional induction of BH3-only proteins is highly stimuli-dependent. For instance, deprivation of NGF triggers BIM transcription via FOXO transcription factors (Gilley et al., 2003), whereas DNA damage triggers PUMA and NOXA transcription via p53 induction (Villunger et al., 2003). In this respect, staurosporine-induced apoptosis does not appear to be mediated by BNIP activation. Axotomy of the neonatal facial nerve promotes apoptosis of MNs in vivo. Apoptosis after nerve axotomy is mediated by the pro-apoptotic gene BAX (Park et al., 2007; Sun and Oppenheim, 2003), and BAX activation is mediated by several BH3-only proteins, such as HRK, NOXA and BIM (Kiryu-Seo et al., 2005; Wakabayashi et al., 2002; 2005). BNIP3 expression was also prominently induced after nerve axotomy, and BNIP3 may be involved in the apoptosis of axotomized MNs in concert with other activating BH3-only proteins.
However, OGD, that triggers autophagy and apoptosis, markedly enhanced the expression of all three BNIPs. BNIP3 and BNIP3L expression levels are reportedly increased by hypoxia in tumor cells (Birse-Archbold et al., 2005; Hamacher-Brady et al., 2007; Tracy et al., 2007), and the transcription factor HIF is responsible for the transcriptional induction of BNIP3 (Lee and Paik, 2006; Leist and Jaattela, 2001; Sowter et al., 2001). HIF expression is also induced in cortical neurons by hypoxia (Halterman et al., 1999). BNIP1 mRNA induction during hypoxic conditions has not yet been reported, but our current observation demonstrated that BNIP1 expression is also regulated by similar stimuli.
Collectively, these results indicate that 1) the basal expression of BNIPs in the developing brain per se does not promote cell death, and 2) the induction of BNIPs is selective depending on the type of stimuli.
Acknowledgments
This research was supported by the Mid-career Researcher Program (2008-0057782) and the Original Technology Research Program for Brain Science (2011-0019212) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology. This study was also supported by a grant of the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A090311).
REFERENCES
- Birse-Archbold J.L., Kerr L.E., Jones P.A., McCulloch J., Sharkey J. Differential profile of Nix upregulation and translocation during hypoxia/ischaemia in vivo versus in vitro. J. Cereb. Blood Flow Metab. 2005;25:1356–1365. doi: 10.1038/sj.jcbfm.9600133. [DOI] [PubMed] [Google Scholar]
- Boyd J.M., Malstrom S., Subramanian T., Venkatesh L.K., Schaeper U., Elangovan B., D’Sa-Eipper C., Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]
- Buss R.R., Sun W., Oppenheim R.W. Adaptive roles of programmed cell death during nervous system development. Annu. Rev. Neurosci. 2006;29:1–35. doi: 10.1146/annurev.neuro.29.051605.112800. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G., Vijayalingam S., Gibson S.B. BNIP3 subfamily BH3-only proteins: mitochondrial stress sensors in normal and pathological functions. Oncogene. 2008;27(Suppl 1):S114–127. doi: 10.1038/onc.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland M.M., Norris K.L., Karbowski M., Wang C., Suen D.F., Jiao S., George N.M., Luo X., Li Z., Youle R.J. Bcl-2 family interaction with the mitochondrial morphogenesis machinery. Cell Death Differ. 2011;18:235–247. doi: 10.1038/cdd.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daido S., Kanzawa T., Yamamoto A., Takeuchi H., Kondo Y., Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- Galvez A.S., Brunskill E.W., Marreez Y., Benner B.J., Regula K.M., Kirschenbaum L.A., Dorn G.W., 2nd Distinct pathways regulate proapoptotic Nix and BNip3 in cardiac stress. J. Biol. Chem. 2006;281:1442–1448. doi: 10.1074/jbc.M509056200. [DOI] [PubMed] [Google Scholar]
- Gilley J., Coffer P.J., Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman M.W., Miller C.C., Federoff H.J. Hypoxia-inducible factor-1alpha mediates hypoxia-induced delayed neuronal death that involves p53. J. Neurosci. 1999;19:6818–6824. doi: 10.1523/JNEUROSCI.19-16-06818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A., Brady N.R., Logue S.E., Sayen M.R., Jinno M., Kirshenbaum L.A., Gottlieb R.A., Gustafsson A.B. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- Imazu T., Shimizu S., Tagami S., Matsushima M., Nakamura Y., Miki T., Okuyama A., Tsujimoto Y. Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene. 1999;18:4523–4529. doi: 10.1038/sj.onc.1202722. [DOI] [PubMed] [Google Scholar]
- Kanzawa T., Zhang L., Xiao L., Germano I.M., Kondo Y., Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Norris K.L., Cleland M.M., Jeong S.Y., Youle R.J. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- Kiryu-Seo S., Hirayama T., Kato R., Kiyama H. Noxa is a critical mediator of p53-dependent motor neuron death after nerve injury in adult mouse. J. Neurosci. 2005;25:1442–1447. doi: 10.1523/JNEUROSCI.4041-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landes T., Emorine L.J., Courilleau D., Rojo M., Belenguer P., Arnaune-Pelloquin L. The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep. 2010;11:459–465. doi: 10.1038/embor.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Paik S.G. Regulation of BNIP3 in normal and cancer cells. Mol. Cells. 2006;21:1–6. [PubMed] [Google Scholar]
- Lee Y., Lee H.Y., Hanna R.A., Gustafsson A.B. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1924–1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M., Jaattela M. Triggering of apoptosis by cathepsins. Cell Death Differ. 2001;8:324–326. doi: 10.1038/sj.cdd.4400859. [DOI] [PubMed] [Google Scholar]
- Levine B., Sinha S., Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Kleeman L.K., Jiang H.H., Gordon G., Goldman J.E., Berry G., Herman B., Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosova E., Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27(Suppl 1):S2–19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima M., Fujiwara T., Takahashi E., Minaguchi T., Eguchi Y., Tsujimoto Y., Suzumori K., Nakamura Y. Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosomes Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Noh M.R., Kim S.K., Sun W., Park S.K., Choi H.C., Lim J.H., Kim I.H., Kim H.J., Kim H., Eun B.L. Neuroprotective effect of topiramate on hypoxic ischemic brain injury in neonatal rats. Exp. Neurol. 2006;201:470–478. doi: 10.1016/j.expneurol.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Oakes S.A., Scorrano L., Opferman J.T., Bassik M.C., Nishino M., Pozzan T., Korsmeyer S.J. Proapoptotic BAX and BAK regulate the type 1 inositol trisphosphate receptor and calcium leak from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2005;102:105–110. doi: 10.1073/pnas.0408352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim R.W., Houenou L.J., Johnson J.E., Lin L.F., Li L., Lo A.C., Newsome A.L., Prevette D.M., Wang S. Developing motor neurons rescued from programmed and axotomy-induced cell death by GDNF. Nature. 1995;373:344–346. doi: 10.1038/373344a0. [DOI] [PubMed] [Google Scholar]
- Park O.H., Lee K.J., Rhyu I.J., Geum D., Kim H., Buss R., Oppenheim R.W., Sun W. Bax-dependent and - independent death of motoneurons after facial nerve injury in adult mice. Eur. J. Neurosci. 2007;26:1421–1432. doi: 10.1111/j.1460-9568.2007.05787.x. [DOI] [PubMed] [Google Scholar]
- Ryu S.W., Choi K., Kim S., Choi C. Endoplasmic reticulum-specific BH3-only protein BNIP1 induces mitochondrial fragmentation in a Bcl-2- and Drp1-dependent manner. J. Cell Physiol. 2012;227:3027–3035. doi: 10.1002/jcp.23044. [DOI] [PubMed] [Google Scholar]
- Sandoval H., Thiagarajan P., Dasgupta S.K., Schumacher A., Prchal J.T., Chen M., Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L., Oakes S.A., Opferman J.T., Cheng E.H., Sorcinelli M.D., Pozzan T., Korsmeyer S.J. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- Sendtner M., Holtmann B., Kolbeck R., Thoenen H., Barde Y.A. Brain-derived neurotrophic factor prevents the death of motoneurons in newborn rats after nerve section. Nature. 1992;360:757–759. doi: 10.1038/360757a0. [DOI] [PubMed] [Google Scholar]
- Sowter H.M., Ratcliffe P.J., Watson P., Greenberg A.H., Harris A.L. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- Sun W., Oppenheim R.W. Response of motoneurons to neonatal sciatic nerve axotomy in Bax-knockout mice. Mol. Cell. Neurosci. 2003;24:875–886. doi: 10.1016/s1044-7431(03)00219-7. [DOI] [PubMed] [Google Scholar]
- Tracy K., Dibling B.C., Spike B.T., Knabb J.R., Schumacker P., Macleod K.F. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A., Michalak E.M., Coultas L., Mullauer F., Bock G., Ausserlechner M.J., Adams J.M., Strasser A. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Kosaka J., Hommura S. Up-regulation of Hrk, a regulator of cell death, in retinal ganglion cells of axotomized rat retina. Neurosci. Lett. 2002;318:77–80. doi: 10.1016/s0304-3940(01)02487-9. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Kosaka J., Oshika T. JNK inhibitory kinase is up-regulated in retinal ganglion cells after axotomy and enhances BimEL expression level in neuronal cells. J. Neurochem. 2005;95:526–536. doi: 10.1111/j.1471-4159.2005.03389.x. [DOI] [PubMed] [Google Scholar]
- Yan Q., Matheson C., Lopez O.T. In vivo neurotrophic effects of GDNF on neonatal and adult facial motor neurons. Nature. 1995;373:341–344. doi: 10.1038/373341a0. [DOI] [PubMed] [Google Scholar]
- Youle R.J., Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell. Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]