Abstract
Human endogenous retroviruses (HERVs) mediate structural variation and genomic instability based on their multiple copy number, inherent ability to mobilize via reverse transcriptase, and high sequence similarity. Moreover, they undergo multiple amplification and retrotransposition events, resulting in the widespread distribution of complete or partial retroviral sequences throughout the primate genome. As such, HERV elements have played important biological roles in genome evolution, and their long terminal repeat (LTR) elements contain numerous regulatory sequences, including effective promoters, enhancers, polyadenylation signals, and transcription factor-binding sites. Lastly, HERV elements are capable of influencing the expression of neighboring genes, a process that also contributed to primate evolution.
Keywords: chromosomal distribution, copy number, HERV, LTR elements, promoter
INTRODUCTION
Human endogenous retroviruses (HERVs) have been subjected to numerous amplification and transposition events, resulting in the widespread distribution of complete or partial retroviral sequences throughout the human genome. Approximately 8% of the human genome is composed of HERVs, which were transmitted in a Mendelian fashion after reverse transcriptase-mediated germ-line infection by ancient active exogenous retroviruses (International Human Genome Sequencing Consortium, 2001; Lower et al., 1996). Most HERV families inserted into the primate genome and underwent amplification on several occasions at the time of the divergence of hominoids from Old World monkeys approximately 30–45 million years ago (Sverdlov, 2000). HERVs are present within full-length or incomplete sequences and contain multiple stop codons, insertions, deletions, and frame shift mutations (de Parseval et al., 2001; Kim, 2001). However, the envelope proteins of some HERV families are expressed preferentially in the human placenta (Kamat et al., 2002; Mi et al., 2000; Venables et al., 1995) as well as several cancer cells (Armbruester et al., 2002; Yi et al., 2004; 2006).
Retroviruses are capable of entering host genomes at many sites, implying a capacity to inactivate genes by physical disruption (Varmus, 1982), and retroviral LTR sequences are able to exert regulatory effects as promoters and enhancers of cellular genes (Domansky et al., 2000; Kovalskaya et al., 2006). Most LTRs have no influence on gene function and are not relevant to disease pathology (Lower et al., 1996). However, a small minority of these sequences does participate in the regulation of gene expression by serving as alternative promoters (Landry et al., 2002; Medstrand et al., 2001; Sin et al., 2006). They also provide polyadenylation signals that contribute to the formation of alternative transcripts (Mager et al., 1999; Sin et al., 2007). Therefore, LTRs could change the regulatory patterns of neighboring genes in evolutionary processes.
HERVs have been proposed to act as etiological cofactors in several chronic diseases, including cancer, autoimmunity, and neurological diseases (Karlsson et al., 2001; Lower, 1999; Perron et al., 2005). HERV elements have also been implicated in other disorders such as male infertility (Kamp et al., 2000). It has also been suggested that they are involved in carcinogenesis based on the expression of HERV mRNA and functional proteins (Sauter et al., 1995). Further, HERVs may be associated with the generation of new promoters or the activation of proto-oncogenes (Huh et al., 2008; 2009; Schulte et al., 1996). Therefore, it is very likely that some HERVs influence gene regulation by expressing their retroviral genes at the transcriptional level, inducing rearrangement events, and providing regulatory sequences. Accordingly, the present review mainly focuses on the chromosomal distribution and transcriptional regulation of HERV elements in primates.
Chromosomal distribution and copy numbers of the HERV family in humans and great apes
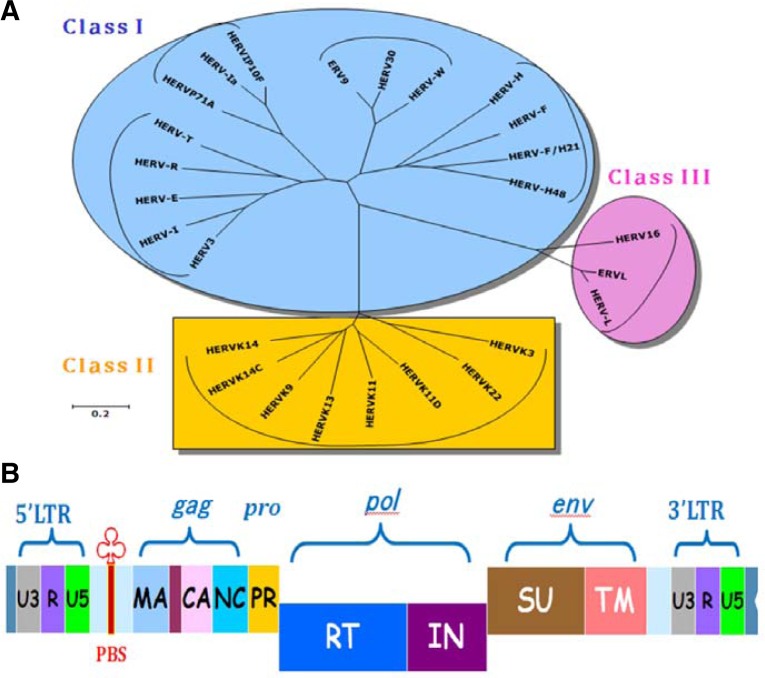
HERVs represent footprints of previous retroviral infection and can be separated into three classes according to the level of pol sequence homology. Such a classification is based on complementary binding between tRNA and the primer-binding site (PBS) (Larsson et al., 1989). A one-letter amino acid code is used to denote the corresponding HERV families: HERV-K, HERV-R, HERV-W, and HERV-H. The basic genes of HERVs are gag, pro, pol, and env. The gag gene codes for the structural matrix (MA), capsid (CA), and nucleocapsid (NC) proteins, pro codes for protease (PR) protein, pol codes for the enzymes of reverse transcriptase (RT) and integrase (IN), and env codes for the envelope surface (SU) and transmembrane (TM) proteins (Fig. 1). The human genome has been estimated to contain about 30–50 HERV-K proviruses (Ono, 1986), and some [HERV-K101, K102, K103, K104, K106, K107 (K10), K108 (HLM-2), K109] were previously found to be unique to humans, showing full-length open reading frames for retroviral protein precursors (Barbulescu et al., 1999). As an estimate, there are about 1,000 copies of HERV-K, including truncated forms, in total (Ha et al., 2011). The genomes of chimpanzee and orangutan contain 862 and 796 copies of HERV-K, respectively. In addition, in comparison with other HERV families (HERV-H, HERV-R, and HERV-W), the HERV-K family including solitary LTR elements was found to be abundantly present in both humans and great apes using the genome-wide browser for retroelements (GEBRET) database (Table 1). In a previous study, BLAST search of the DDBJ/EMBL/GenBank databases identified 140 sequences of HERV-W family viruses representing 39 HERV-W proviruses, 40 full-length HERV-W viruses, and 61 truncated HERV-W elements (Costas, 2000). Using the UCSC genome browser and Repbase, the copy number of HERV-W family viruses has been calculated, and 153 HERV-W elements and 209 solitary sequences were reported. According to this study, these are distributed in all chromosomes except for chromosomes 16 and 22, which is consistent with the Southern blot and PCR data (Kim 2001; Voisset et al., 2000). Similar copy numbers have also been detected in the chimpanzee genome by database and fluorescence in situ hybridization (FISH) analyses (Table 1) (Kim et al., 2008a). However, the copy numbers of Alu elements significantly differ between humans and great apes (Locke et al., 2011). This difference could be explained by a retrotransposition event during primate evolution. The decreased copy numbers of Alu elements could indicate a reduction in the reverse transcriptase activity of L1 elements, since both Alu and L1 elements are dependent on the same enzymatic machinery for propagation.
Fig. 1.
Phylogenetic tree obtained by the neighboring-joining method for pol sequence homology in humans. Branch lengths are proportional to the distances between the taxa (A). Genomic structure of HERV possessing the genes gag, pro, pol, and env (B). The gag gene codes for the structural matrix (MA), capsid (CA), and nucleocapsid (NC) proteins, pro codes for protease (PR) protein, pol codes for the enzymes of reverse transcriptase (RT) and integrase (IN), and env codes for the envelope surface (SU) and transmembrane (TM) proteins. LTR consists of U3, R, and U5 elements. PBS represents primer-binding sites.
Table 1.
Chromosomal distribution and copy numbers of HERV family in humans and great apes
| Chr | HERV-H / solitary LTR | HERV-R / solitary LTR | HERV-W / solitary LTR | HERV-K / solitary LTR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Hu | Ch | Or | Hu | Ch | Or | Hu | Ch | Or | Hu | Ch | Or | |
| 1 | 25 / 54 | 22 / 33 | 21 / 40 | 6 / 8 | 9 / 4 | 6 / 15 | 13 / 11 | 10 / 10 | 12 / 7 | 88 / 79 | 72 / 78 | 60 / 63 |
| 2 | 28 / 46 | 16 / 27 | 13 / 26 | 3 / 10 | 7 / 11 | 0 / 8 | 13 / 24 | 20 / 12 | 18 / 12 | 51 / 67 | 26 / 52 | 28 / 44 |
| 3 | 24 / 64 | 23 / 39 | 25 / 41 | 1 / 9 | 6 / 5 | 1 / 8 | 19 / 15 | 14 / 24 | 14 / 19 | 57 / 47 | 71 / 72 | 70 / 69 |
| 4 | 20 / 58 | 8 / 25 | 12 / 18 | 0 / 7 | 2 / 2 | 3 / 2 | 15 / 21 | 5 / 14 | 4 / 6 | 81 / 70 | 29 / 56 | 35 / 48 |
| 5 | 13 / 29 | 13 / 33 | 15 / 26 | 0 / 2 | 2 / 4 | 0 / 4 | 6 / 15 | 18 / 9 | 16 / 13 | 41 / 54 | 49 / 56 | 57 / 55 |
| 6 | 24 / 49 | 10 / 18 | 11 / 19 | 0 / 4 | 7 / 6 | 1 / 9 | 15 / 9 | 5 / 22 | 5 / 20 | 69 / 63 | 25 / 48 | 41 / 46 |
| 7 | 14 / 27 | 8 / 43 | 8 / 30 | 2 / 7 | 0 / 3 | 0 / 1 | 7 / 15 | 6 / 7 | 7 / 3 | 37 / 46 | 61 / 40 | 51 / 43 |
| 8 | 8 / 44 | 12 / 11 | 11 / 9 | 0 / 1 | 0 / 2 | 0 / 1 | 6 / 5 | 3 / 8 | 7 / 5 | 79 / 44 | 17 / 41 | 28 / 39 |
| 9 | 8 / 14 | 10 / 13 | 8 / 9 | 0 / 1 | 3 / 2 | 0 / 1 | 3 / 7 | 4 / 12 | 4 / 10 | 19 / 40 | 21 / 32 | 16 / 25 |
| 10 | 16 / 22 | 9 / 17 | 12 / 16 | 1 / 2 | 3 / 6 | 1 / 10 | 5 / 12 | 6 / 9 | 6 / 8 | 33 / 35 | 39 / 28 | 46 / 21 |
| 11 | 16 / 23 | 13 / 19 | 10 / 18 | 5 / 7 | 11 / 3 | 3 / 2 | 6 / 6 | 12 / 10 | 9 / 16 | 56 / 32 | 38 / 35 | 36 / 31 |
| 12 | 12 / 25 | 11 / 17 | 8 / 14 | 3 / 4 | 1 / 4 | 0 / 3 | 12 / 13 | 7 / 8 | 5 / 9 | 46 / 36 | 19 / 16 | 25 / 18 |
| 13 | 9 / 25 | 14 / 9 | 9 / 16 | 0 / 7 | 4 / 4 | 0 / 4 | 4 / 12 | 5 / 6 | 6 / 4 | 16 / 23 | 23 / 44 | 32 / 46 |
| 14 | 7 / 21 | 9 / 19 | 6 / 13 | 0 / 6 | 0 / 4 | 0 / 5 | 3 / 9 | 5 / 6 | 4 / 11 | 20 / 34 | 12 / 23 | 20 / 26 |
| 15 | 6 / 7 | 5 / 14 | 6 / 13 | 0 / 1 | 0 / 5 | 1 / 6 | 4 / 6 | 5 / 8 | 4 / 7 | 20 / 14 | 21 / 34 | 19 / 32 |
| 16 | 9 / 19 | 3 / 8 | 3 / 6 | 1 / 3 | 0 / 0 | 0 / 0 | 0 / 0 | 4 / 5 | 2 / 4 | 19 / 27 | 17 / 13 | 23 / 17 |
| 17 | 2 / 7 | 6 / 9 | 3 / 3 | 0 / 4 | 1 / 5 | 1 / 6 | 2 / 3 | 0 / 0 | 0 / 0 | 17 / 20 | 15 / 26 | 11 / 16 |
| 18 | 2 / 16 | 2 / 4 | 4 / 7 | 0 / 3 | 0 / 5 | 0 / 4 | 2 / 5 | 2 / 4 | 1 / 0 | 5 / 12 | 13 / 21 | 13 / 15 |
| 19 | 10 / 12 | 5 / 13 | 4 / 11 | 5 / 6 | 1 / 3 | 0 / 3 | 2 / 1 | 2 / 8 | 4 / 3 | 84 / 55 | 6 / 6 | 10 / 8 |
| 20 | 1 / 14 | 7 / 6 | 7 / 5 | 0 / 0 | 1 / 4 | 1 / 3 | 2 / 1 | 1 / 1 | 0 / 1 | 12 / 6 | 66 / 53 | 87 / 42 |
| 21 | 0 / 8 | 1 / 9 | 2 / 6 | 0 / 4 | 0 / 0 | 0 / 0 | 3 / 3 | 1 / 2 | 2 / 1 | 1 / 7 | 8 / 7 | 14 / 9 |
| 22 | 3 / 2 | 10 / 3 | 1 / 6 | 0 / 4 | 1 / 5 | 0 / 4 | 0 / 1 | 3 / 3 | 2 / 2 | 9 / 14 | 0 / 8 | 0 / 7 |
| 23 | 2 / 2 | 1 / 2 | 1 / 2 | 0 / 2 | 3 / 1 | 0 / 1 | 5 / 14 | 4 / 8 | ||||
| X | 18 / 59 | 9 / 30 | 16 / 30 | 2 / 16 | 4 / 15 | 2 / 15 | 10 / 15 | 9 / 13 | 0 / 15 | 60 / 60 | 72 / 50 | 70 / 51 |
| Y | 7 / 23 | 7 / 15 | 0 / 19 | 2 / 16 | 12 / 15 | 0 / 15 | 1 / 0 | 1 / 0 | 8 / 0 | 81 / 60 | 137 / 50 | 0 / 51 |
|
| ||||||||||||
| Sum | 282 / 668 | 235 / 436 | 216 / 403 | 31 / 132 | 76 / 119 | 20 / 131 | 153 / 209 | 151 / 202 | 140 / 177 | 1001 / 945 | 862 / 903 | 796 / 830 |
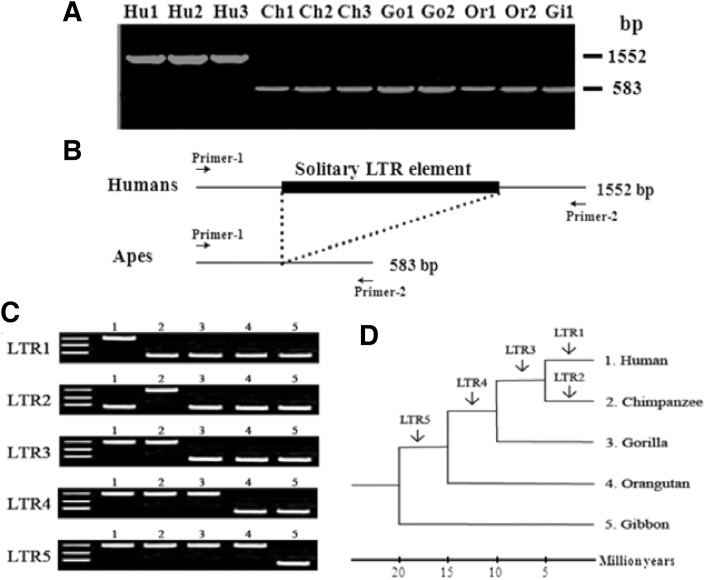
Sequence divergence between 5′ LTR and 3′ LTR sequences constitutes a viable molecular clock in terms of evolution (Fig. 2). According to the mechanism of reverse transcription, the two LTRs must be identical. However, with time, they could undergo divergence due to various mutations (substitutions, insertions, deletions, rearrangement, and inversion) acquired during cellular DNA replication, thus indicating that divergence between the two LTRs could be used as a molecular clock to understand the evolutionary radiation of the HERV family (Mager and Freeman, 1995). The 5′LTR and 3′LTR sequences of HERV-K in long C4 genes of human, orangutan, and African green monkey display similar sequence divergence values of 9.1–10.5% (Dangel et al., 1995). These data indicate that the HERV family is stabilized in the primate genome during evolutionary radiation, and could serve as a reliable reference point for studies on gene duplication and evolution.
Fig. 2.
Schematic representation of the molecular marker for resolving phylogeny and evolution among closely related species. Homologous recombination between the 5′- and 3′-LTRs of a provirus HERV family results in excision of the structural genes and leaves behind a solitary LTR element. The solitary LTR element could be detected by PCR amplification and agarose gel electrophoresis (A, B). Differences in band patterns of the PCR products allow to calculate integration time of solitary LTR element in primate genomes (C, D). Species names are abbreviated as follows: Hu, human; Ch, chimpanzee; Go, gorilla; Or, orangutan. The branching times of the dendrogram should be considered as approximate. Integration event of the solitary LTR element of HERV-K family was modified from Medstrand and Mager (1998) and Buzdin et al. (2003).
Intraelement homologous recombination between the 5′LTR and 3′LTR of HERVs results in the excision of structural genes (gag-pol-env), leaving a solitary LTR element (Mager and Goodchild, 1989). Multiple copy numbers of solitary LTR elements of the HERV-K family (GenBank accession no. AC002350, AC002400, AC002508, L47334, U47924, Z80898, AL034407) have been identified as being unique to humans (Buzdin et al., 2003; 2005; Medstrand and Mager, 1998). Short interspersed element, SINE-R.C2 retroposon derived from the solitary HERV-K LTR is also a human-specific element. Even more, two similar retroposons, HS307 and HS408, were previously identified in the Xq21.3 region, which is linked to schizophrenia and schizoaffective disorder (Kim et al., 1999). These specific insertion sites for HERV elements with relevance to human disease could be useful in resolving phylogeny and evolution among closely related species. In fact, they exhibit insertional polymorphisms in the human population (Hughes and Coffin, 2004; Macfarlane and Simmonds, 2004), indicating that they inserted into the human genome approximately 1.2 million years ago. Therefore, solitary LTR elements could provide valuable information regarding the evolutionary history of retroelements within the primate genome. Further investigation into HERV families will cast light on their biological function in relation to the gene regulatory network system and transcriptional activity.
Transcript variants and alternative promoters of functional genes of the HERV family
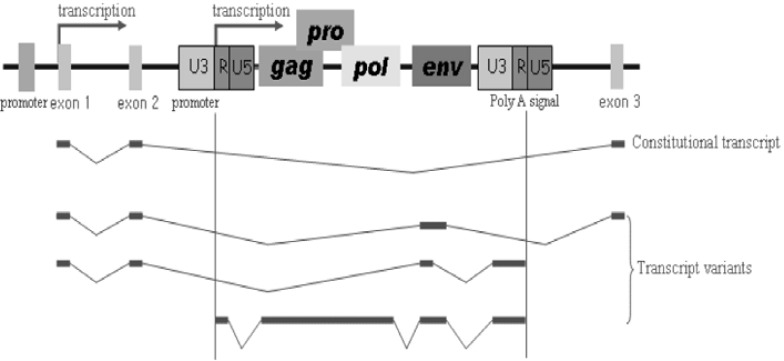
Transcript variants of functional genes are induced by HERV elements, which are capable of altering expression (Fig. 3). Further, HERV LTR elements have been suggested to be alternative gene promoters, as alternatively spliced variants of genes show different expression patterns depending on tissue or cell type. Segmental duplication of LTR fusion genes (LTR element and functional gene) can result in the generation of an alternative promoter and transcriptional variants. Recently, placenta-specific expression of IL-2 receptor by the HERV LTR was detected (Cohen et al., 2011). Other examples include the placenta-specific, LTR-driven transcription of endothelin B receptor (EDNRB) gene by tissue-specific transcription factors (Landry and Mager, 2003) as well as placenta-specific transcription driven by the alternative LTR promoter of CYP19 gene located 100 kb upstream of the coding region (Kamat et al., 2002). The HERV-E LTR element has also been used as an alternative promoter for the endothelin B receptor and apolipoprotein C-I genes in humans (Medstrand et al., 2001), whereas the alternative promoter of the HERV-H LTR for the gasdermin-like (GSDML) gene was shown to be present in human tissues and cancer cells (Sin et al., 2006). The HERV-K LTR element acts as a switch of alternative splicing and encodes the 67 carboxy-terminal amino acid residues in human leptin receptor protein (Kapitonov and Jurka, 1999). The human NDUFV1 gene containing a solitary HERV-K LTR element was demonstrated to have strong tissue-specific promoter activity in germ-derived cells (Tera-1 and EP2102), whereas it has no apparent activity in HEK293 cells (Kuzmin et al., 2010). On the other hand, insertion of HERV-H LTR element provides the polyadenylation signals HHLA2 and HHLA3 (Mager et al., 1999). In the case of NAD synthetase 1(NADSYN1) gene, the HERV-H LTR integrates into the 3′ flanking region. Thereafter, the polyadenylation signal within the LTR element encourages the formation of alternative transcripts (Sin et al., 2007).
Fig. 3.
Schematic representation of genomic structures of HERV elements and functional genes. Alternative promoter of HERV LTR element (U3-R-U5) creates various transcript variants. The structural genes of HERVs are gag, pro, pol, and env. The gag codes for the structural matrix (MA), capsid (CA), and nucleocapsid (NC) proteins, pro codes for protease (PR) protein, pol codes for the enzymes of reverse transcriptase (RT) and integrase (IN), and env codes for the envelope surface (SU) and transmembrane (TM) proteins.
Functional LTR transcription start sites are located between the R and U5 region (3′ termini of the R region) (Kovalskaya et al., 2006). LTR elements have strong bidirectional promoter activity and contain a negative regulatory element in their U5 region (Akopov et al., 1998; Domansky et al., 2000; Schon et al., 2001). The ERV1 solitary LTR element acts as a bidirectional promoter for the human Down syndrome critical region 4 (DSCR4) and DSCR8 genes. Their shared LTR promoter is more active in the sense than antisense orientation (Dunn et al., 2006). In the case of the Opitz syndrome gene Mid1, it has been shown to be transcribed from the HERV-E LTR promoter, indicating that the HERV-E LTR element plays functional roles as both an alternative tissue-specific promoter and enhancer in placenta and embryonic kidney cell lines for the Mid1 gene (Landry et al., 2002). A human-specific solitary LTR element (L47334) was previously shown to have enhancer activity in Tera-1 human testicular carcinoma cells (Ruda et al., 2004). Promoter activities of human-specific LTR elements may depend on the epigenetic status of their CpG dinucleotides (Ego et al., 2005; Park et al., 2010; Swindle et al., 2004). The varying genetic structure of alternative promoters results in different functional effects, which include stimulating different transcription patterns depending on the tissue type or developmental stage (Gerlo et al., 2006). Use of alternative promoters also could regulate alternative transcript variants, resulting in the production of different protein isoforms (Xin et al., 2008). These changes may result in loss or gain of alternative functional domains. Further, the use of multiple promoters could be an important evolutionary mechanism that provides flexibility in the regulation of gene expression. Specifically, multiple promoters compete with the host promoter for the use of its transcriptional machinery. Thereafter, the integrated proviral elements evolve new biological functions during evolution, thereby regulating transcriptional potential. Therefore, expression of these elements varies significantly among cell lines, in some cases showing strict cell type specificity. Accumulated changes in the LTR elements involved in gene regulation are likely to have consequences with regard to diversification, speciation, and evolution.
CONCLUSIONS
HERV elements have undergone insertion into the primate genome and constitute a substantial part of the human genome. These LTRs change the transcriptional regulation of neighboring genes by supplying new promoters, and definitely contribute to genomic plasticity during primate evolution, which indicates that they could be used as genetic markers for understanding evolutionary history. Multiple copy numbers of HERV elements can create new functional exons, alternative splicing products, and miRNAs via integration and adaption events, which are implicated in several human diseases such as male infertility, schizophrenia, and cancer. It was also determined that human-specific HERV or solitary LTR elements are involved in the specific activation and antisense regulation of neighboring functional genes. Alternative promoters and enhancers provided by LTR elements contribute to species diversity and transcriptional gene regulation, and their expression tends to be specific depending on cell type, tissue, and developmental stage. Further, tissue-specific promoters may be utilized to develop cell culture models or gene therapies. In some cases, LTR elements do not act as a promoter or enhancer and instead may be implicated in epigenetic regulation. LTR consensus sequences contain multiple binding sites for transcription factors such as p53, CTCF, Pou5F1-Sox2, and ESR1 (Bourque et al., 2008; Cohen et al., 2009). As such, epigenetic regulation of these binding sites is directly correlated with gene silencing and genomic instability. Tissue-specific epigenetic modifications in the LTR promoter region are associated with the formation of transcript variants and gene expression. Therefore, epigenome analyses of the LTR promoter region will revolutionize our understanding of the biological function of HERVs and neighboring genes. Taken together, elucidation of the various roles of HERVs and solitary LTR elements will provide insights into hominid genetic traits in relation to segmental duplication, copy number variation, alternative promoters, epigenetic events, and transcriptional regulation.
Acknowledgments
This work was supported by the Financial Supporting Project of Long-term Overseas Dispatch of PNU’s Tenure-track Faculty, 2009.
REFERENCES
- Akopov S.B., Nikolaev L.G., Khil P.P., Lebedev Y.B., Sverdlov E.D. Long terminal repeats of human endogenous retrovirus K family (HERV-K) specifically bind host cell nuclear proteins. FEBS Lett. 1998;421:229–223. doi: 10.1016/s0014-5793(97)01569-x. [DOI] [PubMed] [Google Scholar]
- Barbulescu M., Turner G., Seaman M.I., Deinard A.S., Kidd K.K., Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 1999;9:861–868. doi: 10.1016/s0960-9822(99)80390-x. [DOI] [PubMed] [Google Scholar]
- Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G., Chew J.L., Ruan Y., Wei C.L., Ng H.H., et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin A., Ustyugova S., Khodosevich K., Mamedov I., Lebedev Y., Hunsmann G., Sverdlov E. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages. Genomics. 2003;81:149–156. doi: 10.1016/s0888-7543(02)00027-7. [DOI] [PubMed] [Google Scholar]
- Buzdin A., Vinogradova T., Lebedev Y., Sverdlov E. Genome-wide experimental identification and functional analysis of human specific retroelements. Cytogenet. Genome Res. 2005;110:468–474. doi: 10.1159/000084980. [DOI] [PubMed] [Google Scholar]
- Cohen C.J., Lock W.M., Mager D.L. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Cohen C.J., Rebollo R., Babovic S., Dai E.L., Robinson W.P., Mager D.L. Placenta-specific expression of the interleukin-2 (IL-2) receptor β subunit from an endogenous retroviral promoter. J. Biol. Chem. 2011;286:35543–35552. doi: 10.1074/jbc.M111.227637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas J. Characterization of the intragenomic spread of the human endogenous retrovirus family HERV-W. Mol. Biol. Evol. 2000;19:526–533. doi: 10.1093/oxfordjournals.molbev.a004108. [DOI] [PubMed] [Google Scholar]
- Dangel A.W., Baker B.J., Mendoza A.R., Yu C.Y. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K(C4) are a molecular clock of evolution. Immunogenetics. 1995;42:41–52. doi: 10.1007/BF00164986. [DOI] [PubMed] [Google Scholar]
- de Parseval N., Casella J., Gressin L., Heidmann T. Characterization of the three HERV-H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology. 2001;279:558–569. doi: 10.1006/viro.2000.0737. [DOI] [PubMed] [Google Scholar]
- Domansky A.N., Kopantzev E.P., Snezhkov E.V., Lebedev Y.B., Leib-Mosch C., Sverdlov E.D. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 2000;472:191–195. doi: 10.1016/s0014-5793(00)01460-5. [DOI] [PubMed] [Google Scholar]
- Dunn C.A., Romanish M.T., Gutierrez L.E., van de Lagemaat L.N., Mager D.L. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene. 2006;366:335–342. doi: 10.1016/j.gene.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Ego T., Tanaka T., Shimotohno K. Interaction of HTLV-1 Tax and methyl-CpG-binding domain 2 positively regulates the gene expression from the hypermethylated LTR. Oncogene. 2005;24:1914–1923. doi: 10.1038/sj.onc.1208394. [DOI] [PubMed] [Google Scholar]
- Gerlo S., Davis J.R., Mager D.L., Kooijman R. Prolactin in man: a tale of two promoters. Bioessays. 2006;28:1051–1055. doi: 10.1002/bies.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H.S., Chung W.K., Ahn K., Bae J.H., Park S.J., Moon J.W., Nam K.H., Han K., Cho H., Kim H.S. Development of GEBRET: a web-based analysis tool for retroelements in primate genomes. Genes Genomics. 2011;33:679–684. [Google Scholar]
- Hughes J.F., Coffin J.M. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA. 2004;101:1668–1672. doi: 10.1073/pnas.0307885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh J.W., Ha H.S., Kim D.S., Kim H.S. Placenta-restricted expression of LTR-derived NOS3. Placenta. 2008;29:602–608. doi: 10.1016/j.placenta.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Huh J.W., Kim Y.H., Lee S.R., Kim H., Kim D.S., Kim H.S., Kang H.S., Chang K.T. Gain of new exons and promoters by lineage-specific transposable elements-integration and conservation event on CHRM3 gene. Mol. Cells. 2009;28:111–117. doi: 10.1007/s10059-009-0106-z. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kamat A., Hinshelwood M.M., Murry B.A., Mendelson C.R. Mechanisms in tissue-specific regulation of estrogen biosynthesis in humans. Trends Endocrinol. Metab. 2002;13:122–128. doi: 10.1016/s1043-2760(02)00567-2. [DOI] [PubMed] [Google Scholar]
- Kamp C., Hirschmann P., Voss H., Huellen K., Vogt P.H. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum. Mol. Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encode an additional carboxy-terminal sequence in the human leptin receptor. J. Mol. Evol. 1999;48:248–251. doi: 10.1007/pl00013153. [DOI] [PubMed] [Google Scholar]
- Kim H.S. Sequence and phylogeny of HERV-W pol fragments. AIDS Res. Hum. Retroviruses. 2001;17:1665–1671. doi: 10.1089/088922201753342086. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Wadekar R.V., Takenaka O., Winstanley C., Mitsunaga F., Kageyama T., Hyun B.H., Crow T.J. SINER. C2 (a Homo sapiens specific retroposon) is homologous to CDNA from postmortem brain in schizophrenia and to two loci in the Xq21.3/Yp block linked to handedness and psychosis. Am. J. Med. Genet. 1999;88:560–566. [PubMed] [Google Scholar]
- Kovalskaya E., Buzdin A., Gogvadze E., Vinogradova T., Sverdlov E. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology. 2006;346:373–378. doi: 10.1016/j.virol.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Kuzmin D., Gogvadze E., Kholodenko R., Grzela D.P., Mityaev M., Vinogradova T., Kopantzev E., Malakhova G., Suntsova M., Sokov D., et al. Novel strong tissue specific promoter for gene expression in human germ cells. BMC Biotechnol. 2010;17:58–68. doi: 10.1186/1472-6750-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J.R., Mager D.L. Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J. Virol. 2003;77:7459–7466. doi: 10.1128/JVI.77.13.7459-7466.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J.R., Rouhi A., Medstrand P., Mager D.L. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 2002;19:1934–1942. doi: 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- Larsson E., Kato N., Cohen M. Human endogenous proviruses. Curr. Top. Microbiol. Immunol. 1989;148:115–132. doi: 10.1007/978-3-642-74700-7_4. [DOI] [PubMed] [Google Scholar]
- Locke D.P., Hillier L.W., Warren W.C., Worley K.C., Nazareth L.V., Muzny D.M., Yang S.P., Wang Z., Chinwalla A.T., Minx P., et al. Comparative and demographic analysis of orangutan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower R., Lower J., Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane C., Simmonds P. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J. Mol. Evol. 2004;59:642–656. doi: 10.1007/s00239-004-2656-1. [DOI] [PubMed] [Google Scholar]
- Mager D.L., Freeman J.D. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology. 1995;213:395–404. doi: 10.1006/viro.1995.0012. [DOI] [PubMed] [Google Scholar]
- Mager D.L., Hunter D.G., Schertzer M., Freeman J.D. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3) Genomics. 1999;59:255–263. doi: 10.1006/geno.1999.5877. [DOI] [PubMed] [Google Scholar]
- Medstrand P., Mager D.L. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P., Landry J.R., Mager D.L. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 2001;276:1896–1903. doi: 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J. Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Huh J.W., Kim D.S., Ha H.S., Jung Y.D., Ahn K., Oh K.B., Park E.W., Chang K.T., Kim H.S. Analysis of the molecular and regulatory properties of active porcine endogenous retrovirus gamma-1 long terminal repeats in kidney tissues of the NIH-miniature pig. Mol. Cells. 2010;30:319–325. doi: 10.1007/s10059-010-0121-0. [DOI] [PubMed] [Google Scholar]
- Sauter M., Schommer S., Kremmer E., Remberger K., Dölken G., Lemm I., Buck M., Best B., Neumann-Haefelin D., Mueller-Lantzsch N. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon U., Seifarth W., Baust C., Hohenadl C., Erfle V., Leib-Mosch C. Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats. Virology. 2001;279:280–291. doi: 10.1006/viro.2000.0712. [DOI] [PubMed] [Google Scholar]
- Schulte A.M., Lai S., Kurtz A., Czubayko F., Riegel A.T., Wellstein A. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germline insertion of an endogenous retrovirus. Proc. Natl. Acad. Sci. USA. 1996;93:14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin H.S., Huh J.W., Kim D.S., Kang D.W., Min D.S., Kim T.H., Ha H.S., Kim H.H., Lee S.Y., Kim H.S. Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells. Arch. Virol. 2006;151:1985–1994. doi: 10.1007/s00705-006-0764-5. [DOI] [PubMed] [Google Scholar]
- Sin H.S., Huh J.W., Kim W.Y., Kim D.S., Ahn K., Ha H.S., Kim H.S. Long terminal repeats provide alternative polyadenylation signals to NADSYN1 gene. Korean J. Genet. 2007;29:395–401. [Google Scholar]
- Sverdlov E.D. Retroviruses and primate evolution. Bioessays. 2000;22:161–171. doi: 10.1002/(SICI)1521-1878(200002)22:2<161::AID-BIES7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Swindle C.S., Kim H.G., Klug C.A. Mutation of CpGs in the murine stem cell virus retroviral vector long terminal repeat represses silencing in embryonic stem cells. J. Biol. Chem. 2004;279:34–41. doi: 10.1074/jbc.M309128200. [DOI] [PubMed] [Google Scholar]
- Varmus H.E. Form and function of retroviral proviruses. Science. 1982;216:812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Venables P.J., Brookes S.M., Griffiths D., Weiss R.A., Boyd M.T. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology. 1995;211:589–592. doi: 10.1006/viro.1995.1442. [DOI] [PubMed] [Google Scholar]
- Voisset C., Bouton O., Bedin F., Duret L., Mandrand B., Mallet F., Paranhos-Baccala G. Chromosomal distribution and coding capacity of the human endogenous retrovirus HERV-W family. AIDS Res. Hum. Retroviruses. 2000;16:731–740. doi: 10.1089/088922200308738. [DOI] [PubMed] [Google Scholar]
- Xin D., Hu L., Kong X. Alternative promoters influence alternative splicing at the genomic level. PLoS One. 2008;3:e2377. doi: 10.1371/journal.pone.0002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.M., Kim H.M., Kim H.S. Expression of the human endogenous retrovirus HERV-W family in various human tissues and cancer cells. J. Gen. Virol. 2004;85:1203–1210. doi: 10.1099/vir.0.79791-0. [DOI] [PubMed] [Google Scholar]
- Yi J.M., Kim H.M., Kim H.S. Human endogenous retrovirus HERV-H family in human tissues and cancer cells: expression, identification, and phylogeny. Cancer Lett. 2006;231:228–239. doi: 10.1016/j.canlet.2005.02.001. [DOI] [PubMed] [Google Scholar]